Abstract
Arabidopsis PISTILLATA (PI) encodes B-class MADS-box transcription factor (TF), and works together with APETALA3 (AP3) to specify petal and stamen identity. However, a small-scale gene duplication event of PI ortholog was observed in common buckwheat and resulted in FaesPI_1 and FaesPI_2. FaesPI_1 and FaesPI_2 were expressed only in the stamen of dimorphic flower (thrum and pin) of Fagopyrum esculentum. Moreover, intense beta-glucuronidase (GUS) staining was found in the entire stamen (filament and anther) in pFaesPI_1::GUS transgenic Arabidopsis, while GUS was expressed only in the filament of pFaesPI_2::GUS transgenic Arabidopsis. In addition, phenotype complementation analysis suggested that pFaesPI_1::FaesPI_1/pFaesPI_2::FaesPI_2 transgenic pi-1 Arabidopsis showed similar a flower structure with stamen-like organs or filament-like organs in the third whorl. This suggested that FaesPI_2 only specified filament development, but FaesPI_1 specified stamen development. Meanwhile, FaesPI_1 and FaesPI_2 were shown to function redundantly in regulating filament development, and both genes work together to require a proper stamen identity. The data also provide a clue to understanding the roles of PI-like genes involved in floral organ development during the early evolution of core eudicots and also suggested that FaesPI_1 and FaesPI_2 hold the potential application in bioengineering to develop a common buckwheat male sterile line.
1. Introduction
Common buckwheat (Fagopyrum esculentum) grains are gluten-free and with low-calories, but are rich in bioactive compounds (such as rutin, quercetin, polysaccharides, etc.) [1]. Hence, common buckwheat grains have increased demand for a great potential functional food with illness prevention and health benefits in recent years. However, common buckwheat is a heteromorphic self-incompatibility (SI) crop due to its distylous flowers (pin and thrum), with populations being equally composed of pin and thrum plants [2,3]. In pin plants, long styles are combined with short stamens and small pollen grains; in thrum plants, short styles are combined with long stamens and large pollen grains. Moreover, legitimate cross-pollinations occur strictly between pin and thrum flower, which results in low yield. Improving the yield stability and achene set rate requires a better understanding of the molecular basis of the heteromorphic SI and the development of distylous flowers in common buckwheat. Recent studies suggested that Primula GLO2, a PI-like MADS-box gene, is identified as an S-linked gene with its expression specific to S-morph flowers and is a strong candidate for the gene controlling anther height, but exactly how it regulates anther height is unclear [4,5].
The GLOBOSA (GLO)-/PISTILLATA (PI) like genes originated by duplication in the ancestral B gene of all extant angiosperms [6]. In almost all core eudicots, the PI-like genes work together with APETALA3 (AP3)-/DEFICIENS (DEF)-like genes to specify proper petal and stamen identity during flower development; both genes encode B-class MADS-box transcription factors which are functional as heterodimers with each other [7,8]. However, the flexibility of DEF/AP3- and GLO/PI-like protein interactions observed in early-diverging angiosperms may be one reason resulting in highly diverse flower morphologies in these species [7]. F. esculentum (Polygonaceae) belongs to the order Caryophyllales (an early-diverging core eudicots clade) and produces distylous flowers with single-whorl showy tepals, representing an obvious difference with most core eudicots flowers, which make it an ideal model for studying floral organ development and evolution [3,9].
Here, the genomic DNA of two PI-like MADS-box genes, FaesPI_1 and FaesPI_2, and their corresponding promoters were isolated from common buckwheat. Sequences alignment indicated that both genes show high identity in exon, but obvious differences in intron length and sequence. Furthermore, the promoters of FaesPI_1 and FaesPI_2 also found remarkable differences in length and distribution of cis-regulatory elements. A previous study indicated that FaesPI_1 is required only in stamen identity in F. esculentum [10]. In this study, the functional divergence of two common buckwheat PI-like genes was explored by analyzing their expression pattern, characterizing their promoters functions, and assessing the complementation phenotype of pFaesPI_1::FaesPI_1 and pFaesPI_2::FaesPI_2 transgenic pi-1 Arabidopsis. In addition, the possible roles of FaesPI_1 and FaesPI_2 genes regulating stamen development of distylous flowers were proposed in common buckwheat. The findings provide clues for understanding the structure and function evolution of GLO-/PI-like genes during early-diverging core eudicots.
2. Results
2.1. Isolation and Characterization of FaesPI_1 and FaesPI_2 from F. esculentum
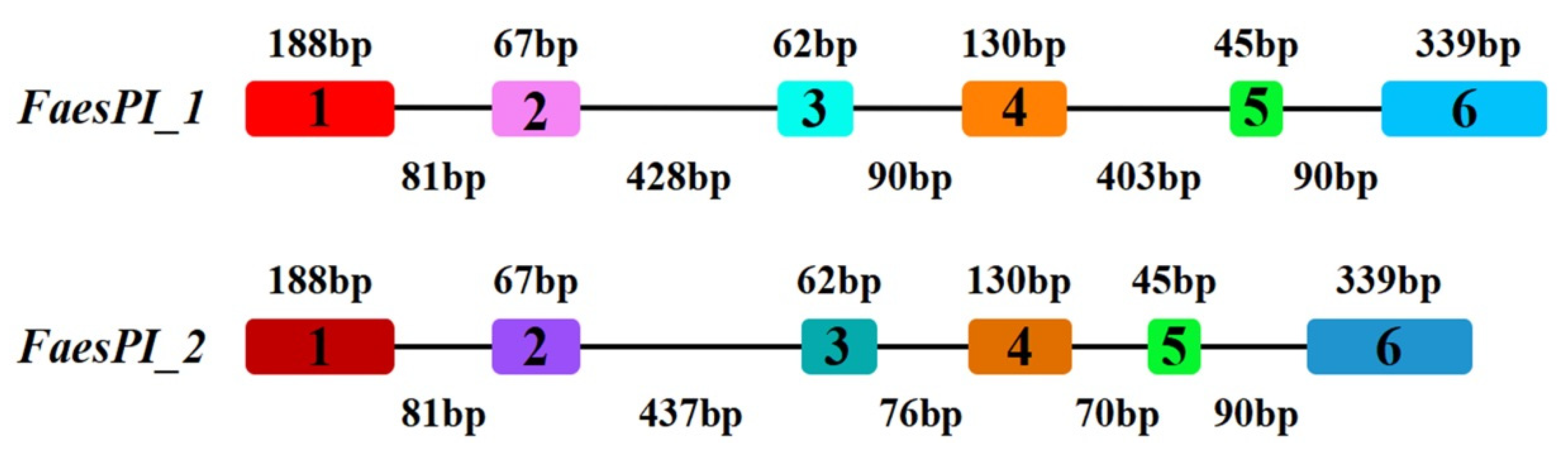
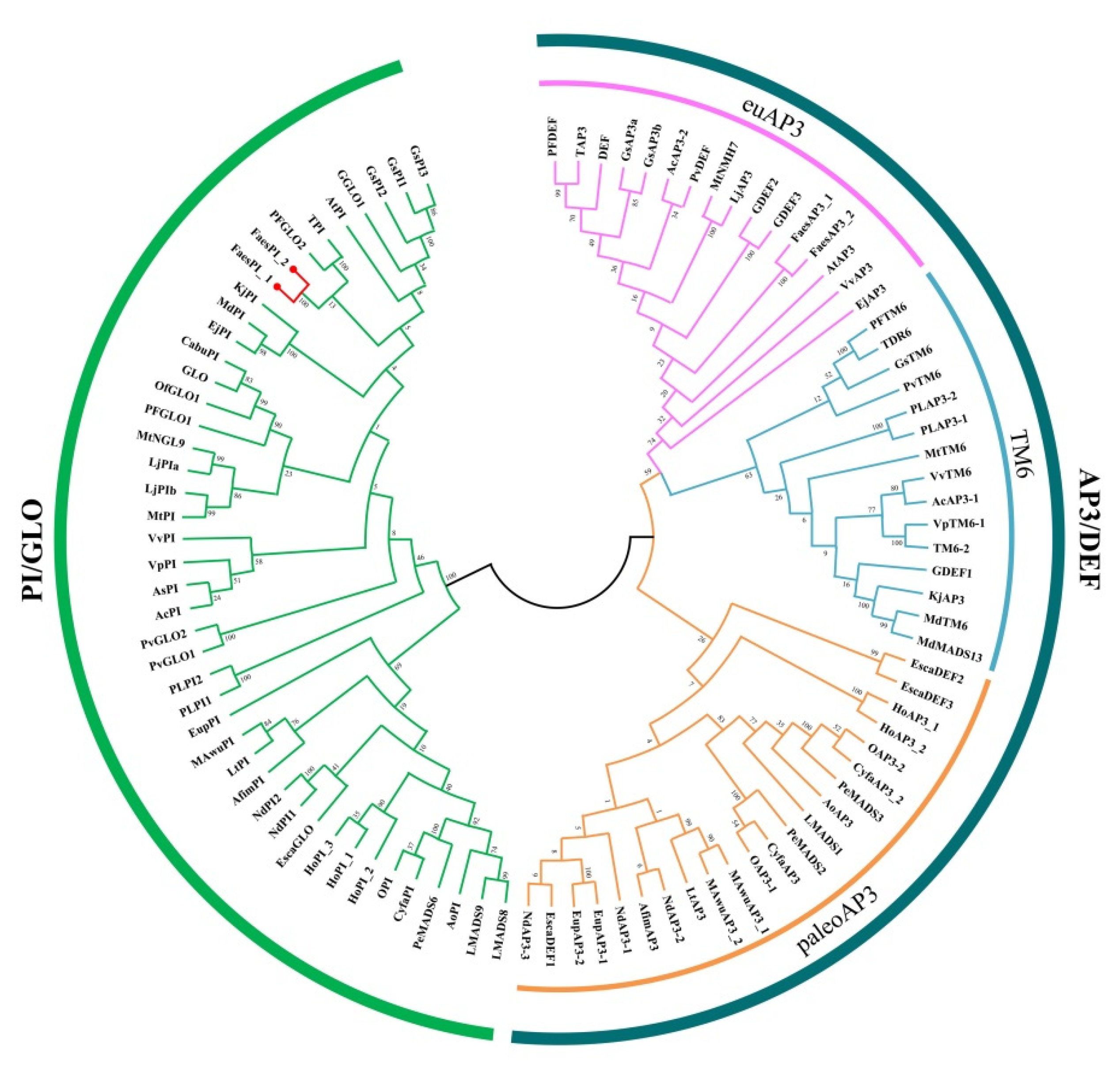
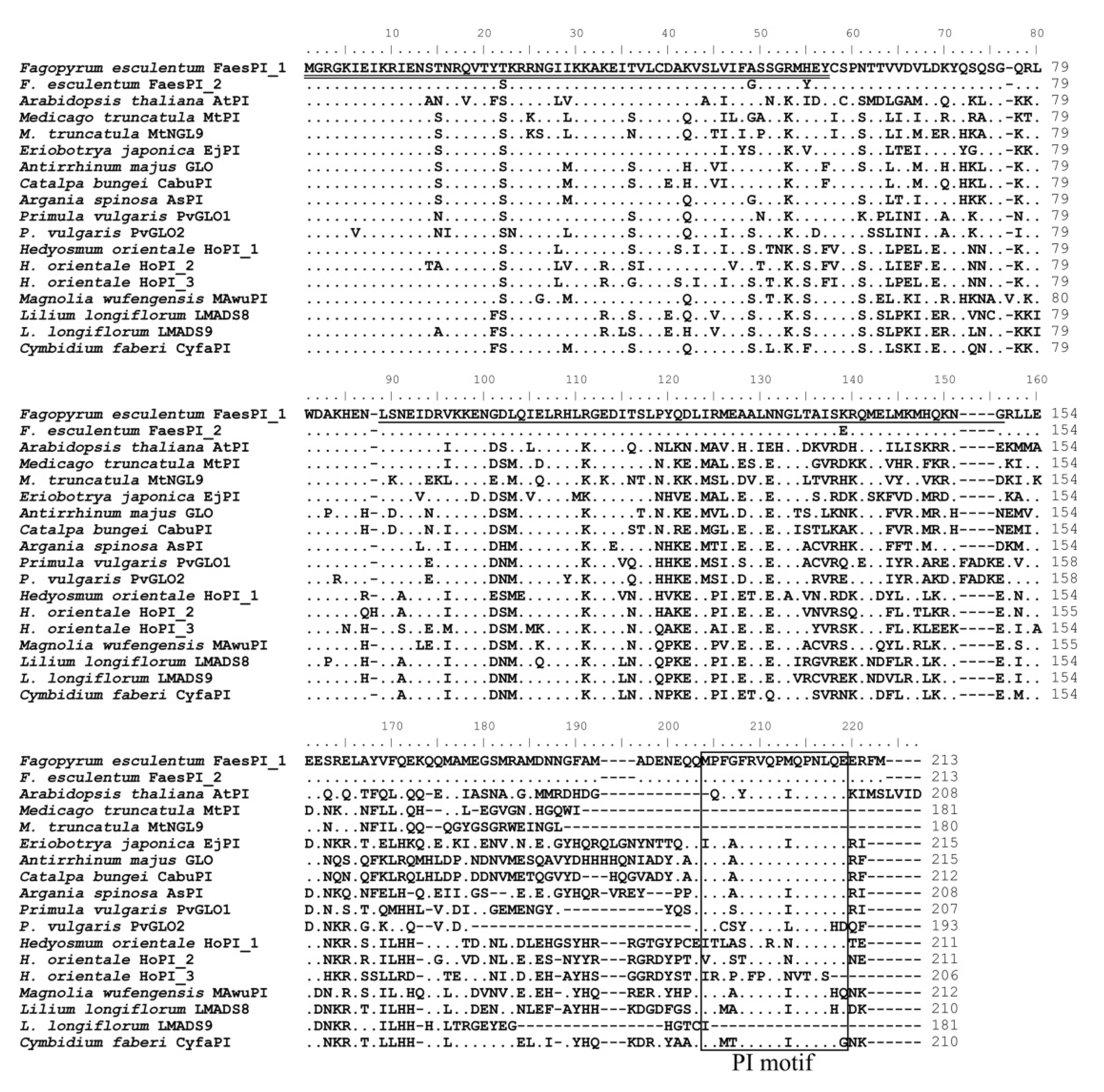
The genomic DNA sequence of FaesPI_1 (Genbank accession number: OM032616.1) is 1923 bp long and consists of six exons and five introns, while the genomic DNA sequence of FaesPI_2 (Genbank accession number: OM032617.1) is 1585 bp long and consists of six exons and five introns. Sequences alignment indicated that three introns (the second, the third, and the fourth) of FaesPI_1 and FaesPI_2 showed a remarkable difference in sequence and length. However, the sequence and length of the corresponding exon between FaesPI_1 and FaesPI_2 showed high conservation (Figure 1). For example, the CDS of FaesPI_1 and FaesPI_2 showed 98.18% identity and encoded 213 aa with 98.12% identity. Phylogenetic tree analysis grouped FaesPI_1 and FaesPI_2 into PI/GLO lineage of B-class MADS-box transcription factor (Figure 2), and both genes were separately designated as FaesPI_1 (Fagopyrum esculentum PISTILLATA_1) and FaesPI_2. In addition, proteins alignment shows that each buckwheat PI-like transcription factor comprises a 57 aa highly conserved MADS-box domain (1-57), a 64 aa weakly conserved K domain (87-50), and a highly conserved PI motif (194-209) lying at a variable C-terminal region (151-213) (Figure 3) [11].
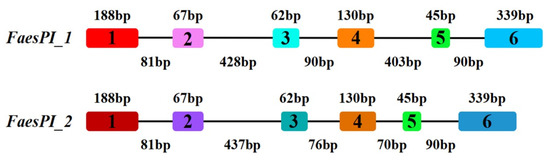
Figure 1.
Exon-intron structures of FaesPI_1 and FaesPI_2 genes. Color boxes present exons while black lines present introns.
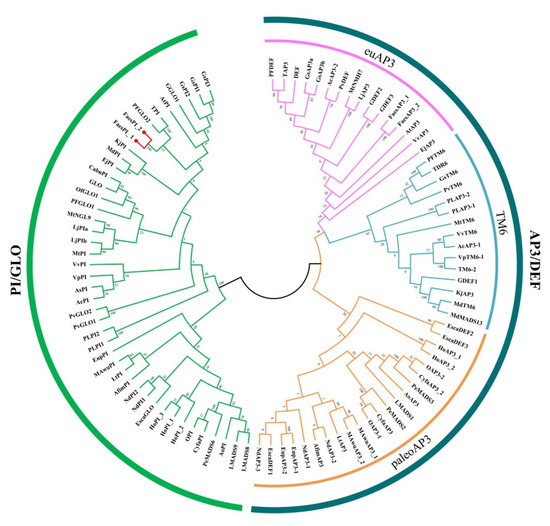
Figure 2.
Phylogenetic tree of FaesPI_1, FaesPI_2, and other B-class MADS-box proteins from different species.
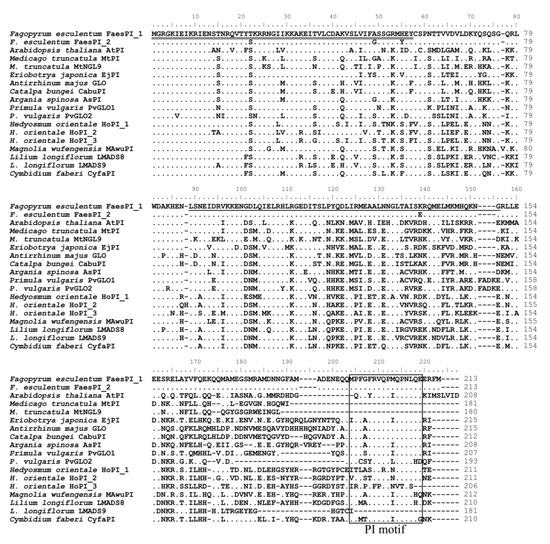
Figure 3.
Sequence alignments of FaesPI_1 and FaesPI_2 with other PI-like transcription factors (TF) of different species. The double underline refers to the MADS domain and the single underline the refers to K domain. The PI-derived motif lying at the variable C-terminal region is boxed. The dots refer to identical aa with FaesPI_1. Moreover, the dashes are introduced into sequences to improve the alignment.
2.2. Expression Analysis of FaesPI_1 and FaesPI_2
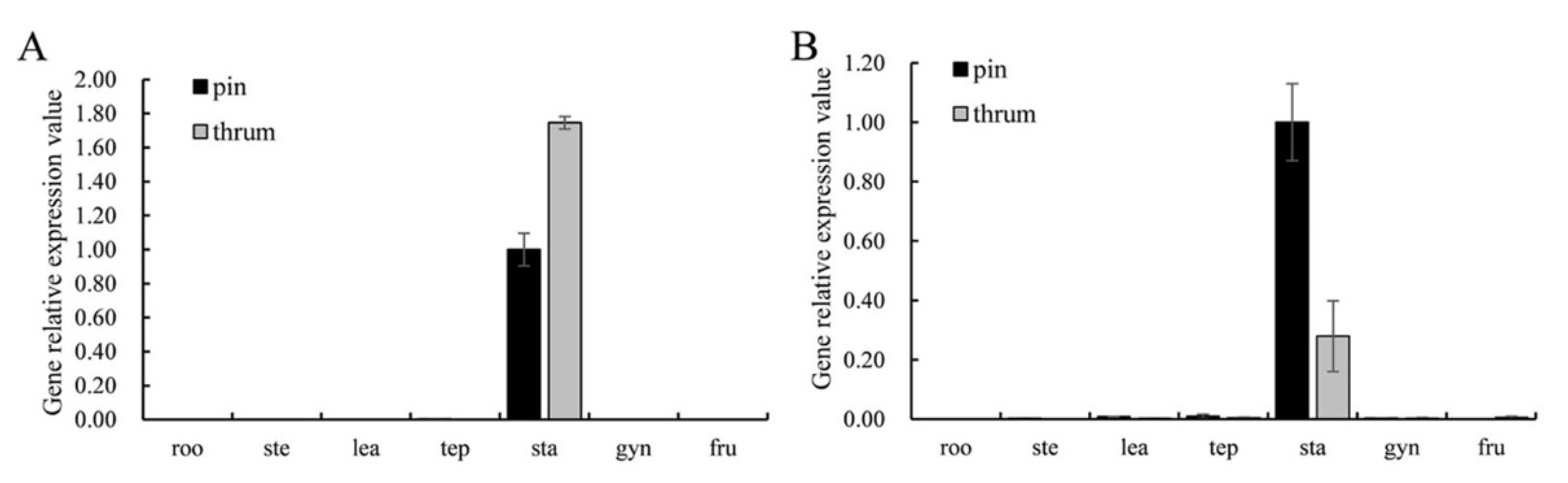
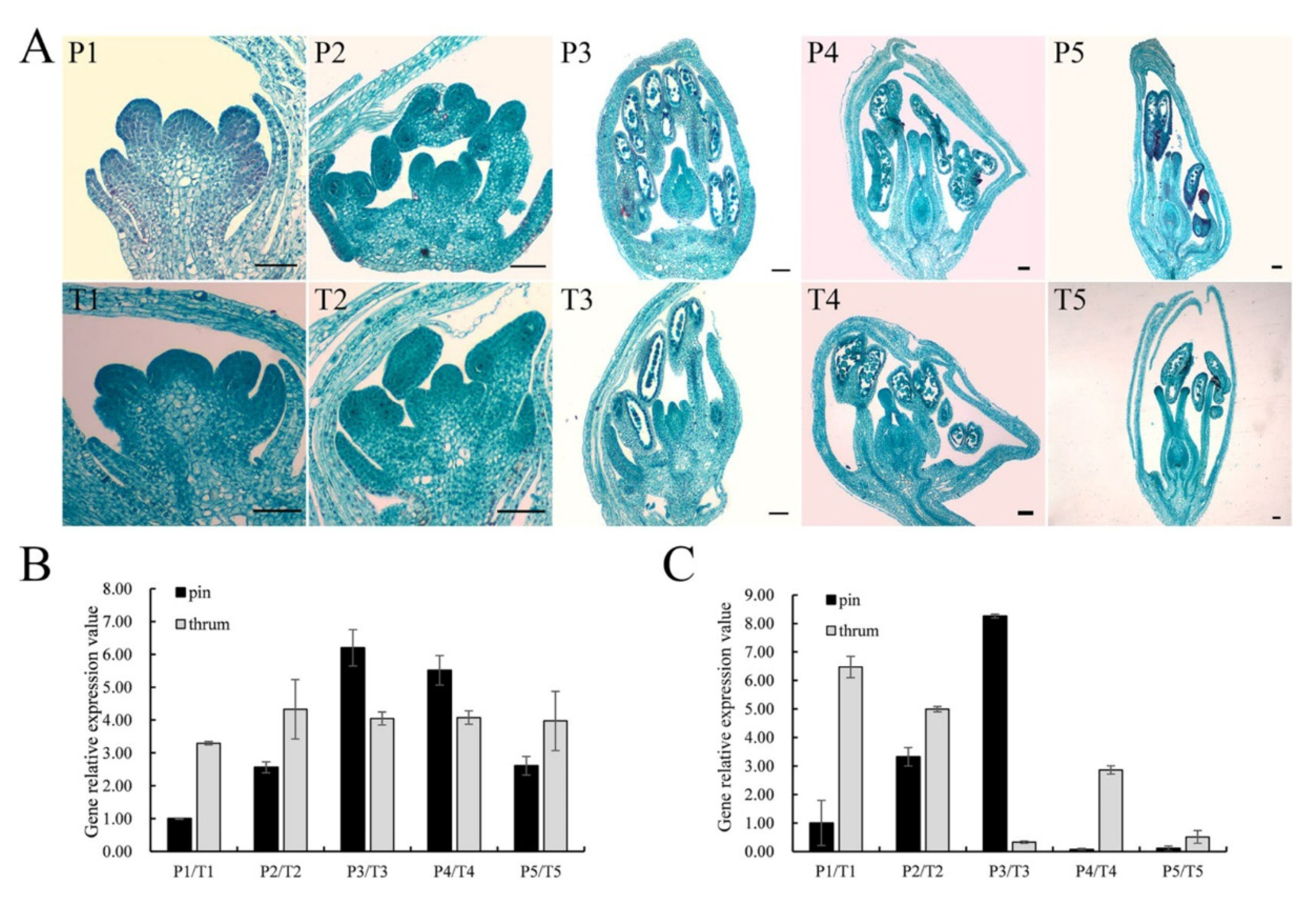
FaesPI_1 and FaesPI_2 were expressed only in the stamen of pin and thrum flower. However, the expression level of FaesPI_1 in thrum stamen was significantly higher than that of the pin stamen (p < 0.01). However, the expression level of FaesPI_2 in pin stamen was significantly higher than that of the thrum stamen (p < 0.05) (Figure 4A,B). FaesPI_1 and FaesPI_2 transcripts were detected after stamen primodium emergence in pin and thrum floral buds (Figure 5A–C). Moreover, FaesPI_1 expression increased constantly and achieved the peak until the microspore tetrad formation (Figure 5A,B(P3)) occurs in the pin flower, and then began to slowly decline until flower maturity. In addition, the FaesPI_1 expression level when the microspore tetrad formed (Figure 5A,B(P3)) or mononuclear microspore and tepal enclosed (Figure 5A,B(P4)) was significantly higher than that of its expression in other stage floral buds of pin flower (LSD, p < 0.05). However, FaesPI_1 expression reached a high level at the filament rapid elongation stage (Figure 5A,B(T2)) in the thrum flower and maintained at a high level until flower maturity. However, FaesPI_2 expression increased constantly and reached the peak until the microspore tetrad formation (LSD, p < 0.05) (Figure 5A,C(P3)) in the pin flower, and then there was a sharp drop, which was maintained at a very weak level until flower maturity (Figure 5A,C(P3)). Moreover, FaesPI_2 expression decreased constantly and reached the bottom until microspore mother cells begin to meiosis (Figure 5A,C(T3)) in the thrum flower, and then maintained at a very low level until flower maturity (Figure 5A,C(T3)).

Figure 4.
Expression of FaesPI_1 and FaesPI_2 in the root (roo), stem (ste), juvenile leaf (lea), tepal (tep), stamen (sta), gynoecium (gyn), and 6-day-old fruits (fru) were detected by qRT-PCR by using Faesactin as the internal control. (A) FaesPI_1 expression in seven organs of F. esculentum; (B) FaesPI_2 expression in seven organs of F. esculentum.

Figure 5.
Morphology, FaesPI_1, and FaesPI_2 expression in different development stage distylous flowers of F. esculentum. (A) Cytomorphological section of thrum and pin floral buds at different development stages; P1–P5: pin floral bud differentiation; P1: stamen primodium appearance; P2: filament rapid elongating; P3: microspore tetrad formation; P4: mononuclear microspore and tepal enclosing; P5: maturity floral bud with mature pollen and embryo sac before bloom; T1–T5: Cytomorphological section of the thrum floral buds; T1: stamen primodium appearance; T2: filament elongation; T3: meiosis of microspore mother cells; T4: periphery of mononuclear microspore, tepal enclosing; T5: maturity floral buds with mature pollen and embryo sac before bloom; (B) FaesPI_1 expression at pin and thrum floral buds were separately detected by qRT-PCR during floral bud differentiation; (C) FaesPI_2 expression at pin and thrum floral buds were separately detected by qRT-PCR during floral bud differentiation. Scale bar: 100 μm.
2.3. Characterization of FaesPI_1 and FaesPI_2 Promoters from F. esculentum
A 2.2 kb FaesPI_1 promoter (pFaesPI_1) (-2186/+68) (Genbank accession number: OM032614.1) and a 2.1kb FaesPI_2 promoter (pFaesPI_2) (-2057/+68) (Genbank accession number: OM032615.1) were separately cloned from common buckwheat. The putative cis-acting elements and transcription start site (TSS) of pFaesPI_1 and pFaesPI_2 were separately displayed in Figures S1 and S2. The pFaesPI_1 has a key CArG-box motif (-1231/-1222) for MADS-box TF recognizing and binding [12], and a CArG-box motif (-154/-145) is also found in the pFaesPI_2. Moreover, pFaesPI_1 has eight POLLEN1LELAT52-boxes and three GTGANTG10-boxes, which are cis-regulatory elements usually found in the promoter region of stamen-development regulated genes [13,14], while pFaesPI_2 contains eleven POLLEN1LELAT52-boxes and nine GTGANTG10-boxes. Moreover, pFaesPI_1 and pFaesPI_2 separately contains several AACAAA-/TTTGTT- motifs for floral homeotic protein APETALA2 recognizing and acting [15]. Furthermore, several MYCCONSENSUSAT-boxes are lying at pFaesPI_1 and pFaesPI_2, which indicates that the expression of both genes could be induced by freezing [16]. Some gibberellin-responsive elements are also lying at pFaesPI_1 (WRKY71OS-box, MYBGAHV-box, and PYRIMIDINEBOXOSRAMY1A-box) and pFaesPI_2 (WRKY71OS-box) [17,18,19]. Several esophyll-specific elements CACTFTPPCA1-boxes and a UP2ATMSD cis-element associated with gene expression during initiation of axillary bud outgrowth are separately found in pFaesPI_1 and pFaesPI_2, which indicated that the expression of both genes may extend to bud, leaf, and rachis [20,21]. However, two CONSTANS protein binding sites (CCAATBOX1) associated with flowering are only found in pFaesPI_1 [22], while a target binding site (LEAFYATAG-box) for regulators of floral identity LEAFY (LFY) associated with floral organ development is only lying at pFaesPI_2 [23]. In addition, four TCP-domain protein-binding elements (SITEIIATCYTC-box) are only found in pFaesPI_2 [24]. All of these suggested that pFaesPI_1 and pFaesPI_2 may drive the corresponding gene to regulate flowering and floral organ development in a different way.
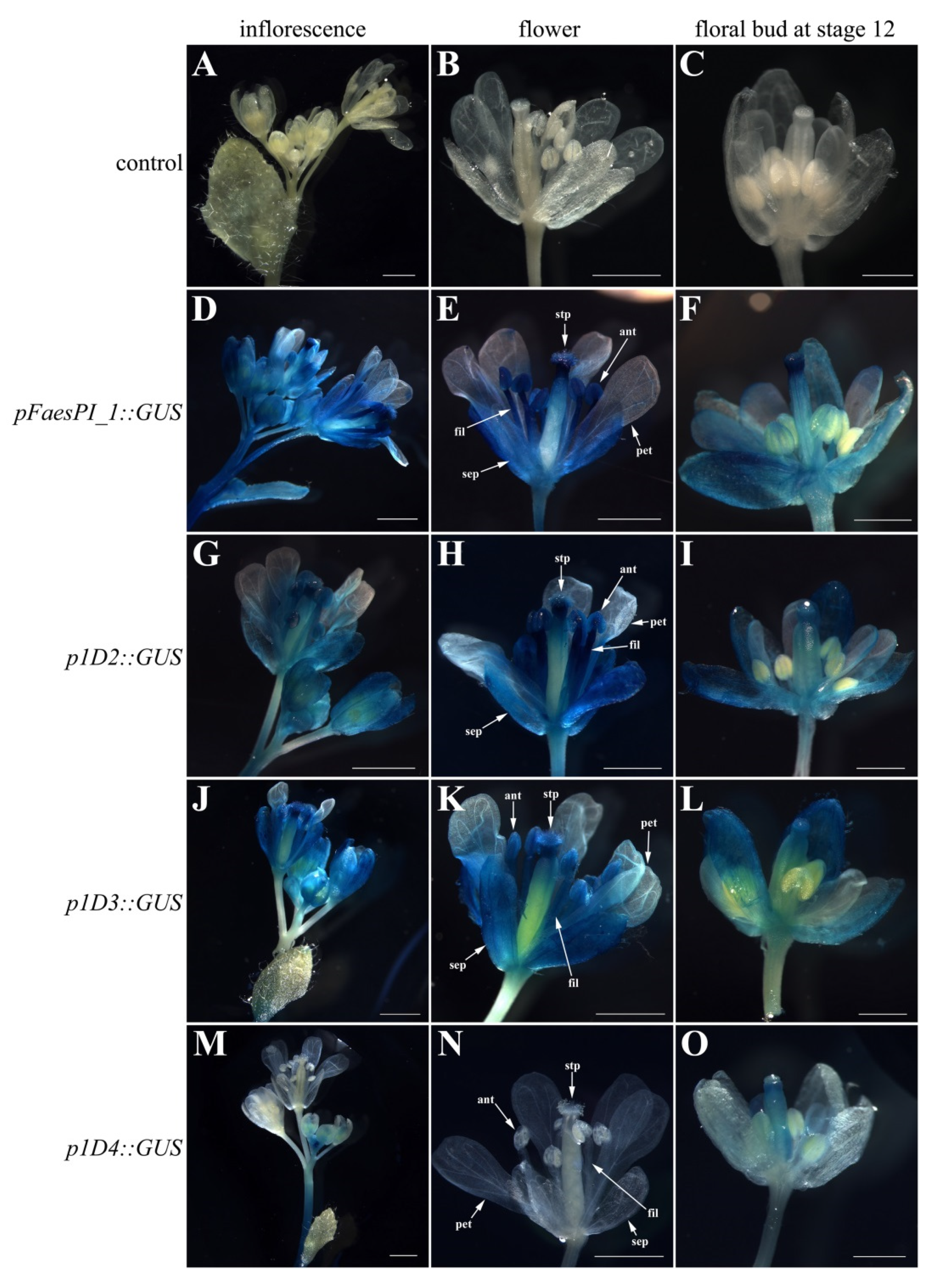
A beta-glucuronidase (GUS) reporter gene separately driven by pFaesPI_1 and pFaesPI_2 was examined in transgenic Arabidopsis (Figure 6 and Figure 7). GUS staining was separately examined in the T1 generation of pFaesPI_1::GUS and pFaesPI_2::GUS independent transgenic lines. GUS expression was found in the inflorescence and flower where sepal, filament, anther, stigma, and stigmatic papillae were high, but was absent in a petal of pFaesPI_1::GUS transgenic Arabidopsis (Figure 6D,E). Moreover, GUS expression was observed in the stage 12 floral bud where sepal, filament, stigma, and stigmatic papillae were intensive, but was absent in petal and anther of pFaesPI_1::GUS transgenic Arabidopsis (Figure 6F) [25]. However, GUS expression was obviously observed in the filament of a mature flower but was almost absent in the sepal, petal, anther, and gynoecium of pFaesPI_2::GUS transgenic Arabidopsis (Figure 7D,E). Moreover, weak GUS expression was observed only at the anther-filament junction in the stage 12 floral bud of pFaesPI_2::GUS transgenic Arabidopsis (Figure 7F).
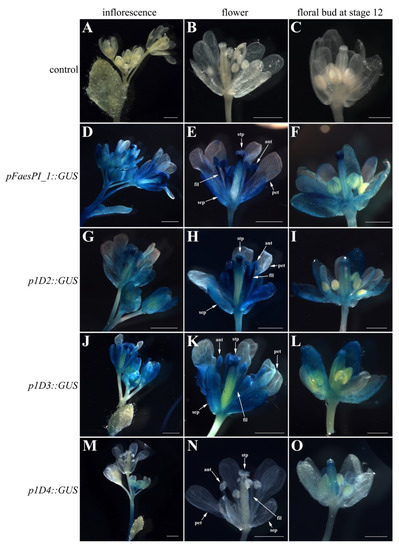
Figure 6.
Histochemical GUS staining in the T1 generation of pFaesPI_1::GUS transgenic Arabidopsis and deletion analysis of the pFaesPI_1 promoter. (A) wild-type Arabidopsis Inflorescence; (B) wild-type Arabidopsis flower; (C) stage 12 floral bud of wild-type Arabidopsis [25]; (D) inflorescence of pFaesPI_1::GUS transgenic Arabidopsis; (E) mature flower of pFaesPI_1::GUS transgenic Arabidopsis; (F) stage 12 floral bud of pFaesPI_1::GUS transgenic Arabidopsis; (G) inflorescence of p1D2::GUS transgenic Arabidopsis; (H) mature flower of p1D2::GUS transgenic Arabidopsis; (I) stage 12 floral bud of p1D2::GUS transgenic Arabidopsis; (J) inflorescence of p1D3::GUS transgenic Arabidopsis; (K) mature flower of p1D3::GUS transgenic Arabidopsis; (L) stage 12 floral bud of p1D3::GUS transgenic Arabidopsis; (M) inflorescence of p1D4::GUS transgenic Arabidopsis; (N) mature flower of p1D4::GUS transgenic Arabidopsis; (O) stage 12 floral bud of p1D4::GUS transgenic Arabidopsis. sepal (sep), petal (pet), anther (ant), filament (fil), stigmatic papillae (stp); Scale Bars: (A,B,D,E,G,H,J,K,M,N) 1 mm; (C,F,I,L,O) 500 μm.

Figure 7.
Histochemical GUS staining in the T1 generation of pFaesPI_2::GUS transgenic Arabidopsis and deletion analysis of the pFaesPI_2 promoter. (A) wild-type Arabidopsis Inflorescence; (B) wild-type Arabidopsis flower; (C) stage 12 floral bud of Wild-type Arabidopsis; (D) inflorescence of pFaesPI_2::GUS transgenic Arabidopsis; (E) mature flower of pFaesPI_2::GUS transgenic Arabidopsis; (F) stage 12 floral bud of pFaesPI_2::GUS transgenic Arabidopsis; (G) inflorescence of p2D2::GUS transgenic Arabidopsis; (H) mature flower of p2D2::GUS transgenic Arabidopsis; (I) stage 12 floral bud of p2D2::GUS transgenic Arabidopsis; (J) inflorescence of p2D3::GUS transgenic Arabidopsis; (K) mature flower of p2D3::GUS transgenic Arabidopsis; (L) stage 12 floral bud of p2D3::GUS transgenic Arabidopsis; (M) inflorescence of p2D4::GUS transgenic Arabidopsis; (N) mature flower of p2D4::GUS transgenic Arabidopsis; (O) stage 12 floral bud of p2D4::GUS transgenic Arabidopsis. sepal (sep), petal (pet), anther (ant), filament (fil), stigmatic papillae (stp); Scale Bars: (A,B,D,E,G,H,J,K,M,N) 1 mm; (C,F,I,L,O) 500 μm.
2.4. Deletion Analysis of the pFaesPI_1 and pFaesPI_2 in Transgenic Arabidopsis
A series of 5′ deletions fragments of the pFaesPI_1 and pFaesPI_2 were separately fused to the GUS gene and transformed into Arabidopsis to analyze the regulatory effect of different regions of the corresponding promoter. GUS staining suggested that p1D2 (-1402/+68) and p1D3 (-817/+68) constructs presented similar expression patterns with the pFaesPI_1::GUS transgenic Arabidopsis, which was high in the inflorescence, sepal, stamen, stigma, and stigmatic papillae of a mature flower (Figure 6). In addition, further deletion of the -817/-366 fragment from p1D3 to produce p1D4 (-365/+68) caused obviously decreased GUS activity in transgenic Arabidopsis, and weak GUS staining was only observed in the gynoecium of early development floral buds from initiation until stage 12 (Figure 6). These results suggested that the -1402/-366 regions are capable of inducing pFaesPI_1 promoter activity in stamen, stigma, and stigmatic papillae, and an 885 bp region (-817/+68) of pFaesPI_1 was sufficient for driving FaesPI_1 gene to regulate stamen development. However, GUS staining suggested that p2D2 (-1532/+68), p2D3 (-1032/+68), and p2D4 (-250/+68) constructs presented similar expression zones, which were only in sepal, filament, and the gynoecium of transgenic Arabidopsis, and showed different expression zones with the pFaesPI_2::GUS transgenic Arabidopsis (Figure 7). Moreover, intensive GUS staining was found only in the filament of pFaesPI_2::GUS transgenic Arabidopsis. The results also indicated that the -2057/-1532 regions contained regulatory elements critical for restricting FaesPI_2 expression to the filament (Figure 7E).
2.5. Phenotypic Analyses of pFaesPI_1::FaesPI_1 and pFaesPI_2::FaesPI_2 Transgenic pi-1 Arabidopsis
To uncover the roles of FaesPI_1 and FaesPI_2 involved in floral development, pFaesPI_1::FaesPI_1, and pFaesPI_2::FaesPI_2 constructs have been separately transformed into PI/pi-1 heterozygote Arabidopsis to create phenotype complementation lines. All transgenic plants were verified by qRT-PCR. In addition, the independent transgenic lines of pFaesPI_1::FaesPI_1 or pFaesPI_2::FaesPI_2 Arabidopsis under wild-type, heterozygote and homozygous background were verified by using dCAPS method with BspHI (TaKaRa Bio, Otsu, Japan) restriction enzymes, respectively (Supplementary Figure S3). Moreover, FaesPI_1 and FaesPI_2 expression in transgenic lines under homozygous backgrounds were separately detected. In addition, 11 independent pFaesPI_1::FaesPI_1 lines under homozygous pi-1 mutant background and 21 independent pFaesPI_2::FaesPI_2 lines under homozygous pi-1 mutant background were obtained, respectively. Flower phenotypes of each transgenic line after flowering were assessed to evaluate whether FaesPI_1 or FaesPI_2 could replace the endogenous PI gene to control petal and stamen development in Arabidopsis pi-1 mutant.
Among eleven pFaesPI_1::FaesPI_1 transgenic pi-1 Arabidopsis, eight (72.73%) showed stamen complementation phenotypes with carpelloid stamen, filament-like organs, or filament with carpelloid anther or stigmatic papillae at the top in the third whorl of the flower (Figure 8B,C), the remaining three (27.27%) lines showed similar flower phenotypes with homozygous pi-1 Arabidopsis. Among twenty-one pFaesPI_2::FaesPI_2 transgenic pi-1 Arabidopsis, nineteen (90.48%) showed a graded complementation phenotype with carpelloid stamen, filament-like organs, or filament with carpelloid anther at the top in the third whorl of the flower (Figure 8E,F), other three (9.52%) lines showed similar flower phenotype with homozygous pi-1 Arabidopsis.

Figure 8.
Flower phenotypes of wild-type, Arabidopsis pi-1 mutant, pFaesPI_1::FaesPI_1 transgenic pi-1 Arabidopsis, and pFaesPI_2::FaesPI_2 transgenic pi-1 Arabidopsis. (A) wild-type Arabidopsis flower with perfect flower (sepal in the first floral whorl, petal in the second floral whorl, stamen in the third floral whorl and fused carpel in the fourth floral whorl); (B) flower of pFaesPI_1::FaesPI_1 transgenic homozygous pi-1 Arabidopsis with carpelloid stamen or filament with stigmatic papillae at the top in the third floral whorl; (C) flower of pFaesPI_1::FaesPI_1 transgenic homozygous pi-1 Arabidopsis with filament and filament with carpelloid anther at the top in the third floral whorl; (D) flower of Arabidopsis pi-1 mutant exhibits homeotic transformations of the second floral whorl petal to sepal, and stamen deficiency in the third floral whorl; (E) flower of pFaesPI_2::FaesPI_2 transgenic homozygous pi-1 Arabidopsis with carpelloid stamen and filament-like organ in the third floral whorl; (F) flower of pFaesPI_2::FaesPI_2 transgenic homozygous pi-1 Arabidopsis with carpelloid stamen and filament with carpelloid anther at the top in the third floral whorl. sepal (sep), petal (pet), anther (ant), filament (fil), filament-like organ (fio), stamen-like organ (sto), gynoecia (gyn), stigmatic papillae (stp). Scale Bars: 500 μm.
3. Discussion
In core eudicots, most GLO-/PI-like genes, such as EjPI from Eriobotrya japonica [26], GGLO1 from Gerbera hybrida [27], and AsPI from Argania spinosa [28], were expressed in the petal and stamen and were mainly required in controlling perfect petal and stamen identities during flower development. The data indicate that the functions of GLO-/PI-like genes are highly correlaTaKaRa, Shigated with their expression zones in core eudicots. However, many GLO-/PI-like genes usually displayed broader expression zones and versatile functions in basal angiosperms, basal eudicots, and monocots. For example, Hedyosmum orientale PI-like genes HoPI were broadly expressed in all floral organs but were involved only in specifying petal and stamen identities [29]. Magnolia wufengensis PI orthologous gene, MAwuPI, was expressed in petaloid tepal and stamen but was required only for stamen identity [30]. Lily PI orthologous genes, LMADS8/9, were expressed in tepal and stamen and were required for tepal and stamen formation [31,32]. Some orchid PI-like genes, such as OMADS8 from Oncidium and OPI from Phalaenopsis, were found expressed in all floral organs but were required only for specifying perianths (sepal/petal and lip) and androecium formation [33,34,35]. The data indicate that stamen-specific function obtained from PI orthologs antedate their petal-specific function during angiosperm evolution. F. esculentum is a member of the family Polygonaceae in the order Caryophyllales, a member of an early-diverging clade of higher eudicots, and has distylous flowers without the petal whorl [3,9]. In addition, F. esculentum PI-like genes, FaesPI_1 and FaesPI_2, were expressed only in the stamen. The expression absence of FaesPI_1 and FaesPI_2 in showy tepals suggested that the PI-like gene-dependent petal identity program was not observed in F. esculentum. The findings provided new clues for understanding flower variation and interpretation of petal evolution across core eudicots. Moreover, intensive GUS staining was observed in the whole stamen (filament and anther) of pFaesPI_1::GUS transgenic Arabidopsis, while intensive GUS staining was found only in the filament of pFaesPI_2::GUS transgenic Arabidopsis. Phenotype complementation analysis suggested that pFaesPI_1::FaesPI_1/pFaesPI_2::FaesPI_2 transgenic pi-1 Arabidopsis showed similar flower structure with stamen-like organs or filament-like organs in the third whorl. The data suggest that FaesPI_2 may be involved only in filament development, and FaesPI_1 may specify stamen development in common buckwheat.
Small-scale gene duplication events of GLO-/PI-like genes happened throughout angiosperms. In addition, most PI-like paralogs have undergone functional overlap or subfunctionalization after gene duplication. For example, NdPI1 and NdPI2 were two PI-like genes from basal eudicots Nigella damascene (Ranunculaceae). NdPI1 and NdPI2 were mainly expressed in sepal, petal, and stamen, and have functioned redundantly in specifying petal and stamen identity [36]. Two GLO/PI orthologous genes, PLPI1 and PLPI2 from basal eudicots Paeonia lactiflora, were strongly expressed in petal, stamen, and carpel; PLPI1 was required for petal and stamen identity, while PLPI2 was sufficient to guarantee stamen identity [37]. However, few paralogs acquire new functions after duplication. For example, Medicago truncatula PI-like gene MtPI maintained the overall ancestral function for specifying petal and stamen identity, while another PI-like gene MtNGL9, may be required to maintain the critical dosage for the B-function in M. truncatula [38]. In distylous Primula, GLO/PI orthologous gene GLO1 maintained the ancestral B-class function in specifying petal and stamen identity after duplication, while GLO2 underwent neofunctionalization for promoting the cell expansion in the fused tube of petals and stamen filaments and determining anther position. Moreover, the GLO2 gene at the S locus worked together with the style-length-determining gene CYP734A50 for the development of heterostyly [4,39]. In our study, F. esculentum pFaesPI_2 could drive the GUS gene to be expressed only in the filament of transgenic Arabidopsis, while pFaesPI_1 could drive the GUS gene to be expressed in the whole stamen (filament and anther) of transgenic Arabidopsis. A previous study also proved that the GLO/PI-like orthologs and AP3/DEF usually work together to specify perfect petal and stamen identity during flower development in almost all core eudicots [7,8]. Our previous study also suggested that two AP3-like paralogs, FaesAP3_1 and FaesAP3_2, had functioned redundantly in controlling filament identity, and FaesAP3_2 had a key role in regulating anther development [40]. The data suggest that FaesPI_2 may interact with FaesAP3_1 to filament development during common buckwheat flower development. In addition, a future challenge was to explore whether the FaesPI_2 works together with the candidate S locus gene to regulate heterostyly in F. esculentum. Our data suggest the functional divergences of PI-like paralogs after duplication in an early-diverging clade of core eudicots, and also provide an idea candidate gene for the potential application in bioengineering to develop a common buckwheat male sterile line.
4. Materials and Methods
4.1. Plant Material
Floral buds at various developmental stages were sampled from thrum and pin plants of buckwheat ‘Beizaosheng’ planted under natural conditions in Jingzhou, Hubei Provence, China, respectively. Moreover, each sample was divided in half; one was immediately frozen in liquid nitrogen and then stored at −80 °C, and another was incubated in FAA [38% formaldehyde: acetic acid: 70% ethanol = 1:1:18 (V/V)]. The roots, stems, juvenile leaves, tepals, stamens, gynoecia and 6-day-old fruits (achenes) of thrum and pin plants were dissected, respectively, and were immediately frozen and stored in liquid nitrogen. The Arabidopsis pi-1 mutant (CS77) seeds were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University, USA.
4.2. Characterization of Genomic DNA FaesPI_1 and FaesPI_2 from F. esculentum
Buckwheat genomic DNA was extracted from juvenile leaves of thrum and pin plants using the CTAB Plant Genomic DNA Rapid Extraction Kit (Aidlab, Beijing, China) referring to the manufacturer’s protocol. The full-length genomic DNA sequences of FaesPI_1 and FaesPI_2 were separately isolated from F. esculentum genomic DNA with a forward primer DFaesPIF and reverse primer DFaesPIR (Supplementary Table S1), and then were cloned into the pTOPO-TA vector (Aidlab, Beijing, China) for sequencing, respectively. The PCR primers were designed based on the buckwheat PI-like gene (Genbank accession numbers: JN605356.1) identified before. The PCR amplification of FaesPI_1 or FaesPI_2 genomic DNA was carried out in a 25 µL reaction volumes containing 4 µL dNTP Mixture (2.5 mM each) (TaKaRa Bio, Otsu, Japan), 2.5 µL 10 × LA PCR Buffer II (Mg2+ plus), and 0.3 µL LA Taq DNA Polymerase (5 U/µL) (TaKaRa Bio, Otsu, Japan). PCR was carried out with denaturation at 94 °C (3 min), followed by 30 cycles of 30 s at 94 °C, annealing at 58 °C (30 s), extension at 72 °C (90 s), with a final extension period (10 min). Phylogenetic tree construction was performed by using the neighbor-joining (NJ) method in MEGA version 5.05. The NJ tree was built with the Poisson model and 1000 bootstrap replications. All the B-class MADS-box transcription factors (TF) sequences containing whole M, I, K, and C domains were obtained from NCBI Genbank (Supplementary Table S2). In order to characterize FaesPI_1 and FaesPI_2 in detail, the TF sequence alignment was aligned by the ClustalW algorithm with 1000 bootstrap replications in BioEdit version 7.0.9. Pairwise alignment was carried out with a gap opening penalty of 10 and a gap extension penalty of 0.1, and multiple alignments were performed with a gap opening penalty of 10 and a gap extension penalty of 0.2.
4.3. Isolation and Sequence Analysis of FaesPI_1 and FaesPI_2 Promoters from F. esculentum
The FaesPI_1 5′ flanking regions were cloned using the method suggested by Liu et al. [41], but with three reverse primers D1pPISP1, D1pPISP2, and D1pPISP3 for the walking sequencing. The FaesPI_2 5′ flanking regions were coloned following the above method, but with three reverse primers D1pPISP1, D1pPISP2, and D1pPISP3 for the first walking sequencing, and with three reverse primers D2pPI_2SP1, D2pPI_2SP2, and D2pPI_2SP3 for the second walking sequencing. Moreover, the full-length FaesPI_1 promoter (pFaesPI_1) was amplified via PCR and cloned into the pTOPO-TA vector (Aidlab, Beijing, China) with the forward primer TpFaesPI_1F and the reverse primer TpFaesPI_1R for sequencing. The full-length FaesPI_2 promoter (pFaesPI_2) was amplified via PCR and cloned into the pTOPO-TA vector (Aidlab, Beijing, China) with the forward primer TpFaesPI_2F and the reverse primer TpFaesPI_2R for sequencing. In addition, the putative transcription start site of FaesPI_1 or FaesPI_2 was searched by using the 5′RACE method with the 5′RACE System for Rapid Amplification of cDNA Ends (Invitrogen, Carlsbad, CA, USA) referring to the manufacturer’s protocol, and three gene-specific reverse primers 5RPI_1GSP1, 5 RPI_1GSP2 and 5 RPI_1GSP3 for FaesPI_1, but three gene-specific reverse primers 5RPI_2GSP1, 5 RPI_2GSP2 and 5 RPI_2GSP3 for FaesPI_2. The cis-acting regulatory elements of the pFaesPI_1 and pFaesPI_2 promoters were found in the PLACE database [42].
4.4. Characterization of pFaesPI_1 and pFaesPI_2 Activity from the 5′ Deleted Promoter Fragments in Transgenic Arabidopsis
Four forward primers (TpFaesPI_1F, TpFaesPI_1F1, TpFaesPI_1F2, and TpFaesPI_1F3) and a reverse primer TpFaesPI_1/2R were designed to obtain 5′-deletion fragments of pFaesPI_1. Moreover, four forward primers (TpFaesPI_2F, TpFaesPI_2F1, TpFaesPI_2F2, and TpFaesPI_2F3) and reverse primer TpFaesPI_1/2R were designed to obtain 5′-deletion fragments of pFaesPI_2. Four 5′-deletion fragments of pFaesPI_1 were designated as p1D1 (-2186/+68),p1D2 (-1402/+68),p1D3 (-817/+68), and p1D4 (-365/+68), and were separately cloned into the pCAMBIA1300 with Xba I (TaKaRa Bio, Otsu, Japan) and Sac I (TaKaRa Bio, Otsu, Japan) restriction enzymes using the ClonExpress® Ultra One Step Cloning Kit (Vazyme, Nanjing, China) following the manufacturer’s protocol. Moreover, four 5′-deletion fragments of pFaesPI_2 were designated as p2D1 (-2057/+68), p2D2 (-1532/+68), p2D3 (-1032/+68), and p2D4 (-250/+68), and were separately cloned into the pCAMBIA1300 vector using the above method. All the constructs were separately transformed into A. thaliana Col-0 plants using the floral-dip method according to Clough and Bent [43]. Transgenic Arabidopsis seedlings were selected, cultivated, and prepared for histochemical GUS staining using the method suggested by Liu et al. [41].
For GUS staining, the inflorescences of transgenic Arabidopsis were incubated in 90% acetone (4 °C for 20 min) and then rinsed with GUS assay buffer [50 mM sodium phosphate (pH 7.0), 1 mM K3Fe(CN)6, 1 mM K4Fe(CN)6·3H2O, 10 mM EDTA (pH 8.0), 0.2 % Triton X-100 (V/V)] 2–3 times, followed by vacuum infiltrated in a mixture of GUS assay buffer and 2 mM X-Gluc for 30 min at room temperature, and then incubated for 6 h at 37 °C, discarding the liquids and later cleared in an ethanol series (75, 85, 95 and 100%). The samples were observed with a Leica 165C microscope (Leica Microsystems, Wetzlar, Germany), and the photomicrographs were taken.
4.5. Cytomorphological Observation and Expression Analysis of FaesPI_1 and FaesPI_2
The floral bud samples of thrum and pin plants fixed in FAA above were separately dehydrated using an ethanol series, cleared twice in xylene, infiltrated three times in molten paraffin, embedded into paraffin block, serially sectioned, and then sections were separately stained according to the method described by Liu et al. [41]. Each section was observed under a CAIKON RCK-40C microscope (CAIKON, Shanghai, China) and the Photomicrographs were taken.
Total RNA and the first-strand cDNA of each sample were prepared for quantitative real-time PCR (qRT-PCR) according to Zeng et al. [40]. The expressions of FaesPI_1 and FaesPI_2 were separately detected in seven organs (root, stem, juvenile leaf, tepal, stamen, gynoecium, and 6-day-old fruit) of different flower phenotype plants according to Zeng et al. [40], but with the primers qFaesPI_1F and qFaesPI_1R for FaesPI_1, and the primers qFaesPI_2F and qFaesPI_2R for FaesPI_2. In addition, FaesPI_1 and FaesPI_2 expressions were separately detected in thrum and pin floral buds at sequential developmental stages using qRT-PCR suggested above. The amplicons of F. esculentum actin gene (Genbank accession number: HQ398855.1) were selected as the internal control with the forward primer qFaesactinF and the reversed primer qFaesactinR. qRT-PCR was performed with three biological replicates and the relative expression levels were measured according to Liu et al. [41], but with annealing at 58 °C.
4.6. Phenotypic Analyses of pFaesPI_1::FaesPI_1 and pFaesPI_2::FaesPI_2 Transgenic pi-1 Arabidopsis
Full-length pFaesPI_1::FaesPI_1 genomic DNA was cloned into pCAMBIA1300 with Xba I (TaKaRa Bio, Otsu, Japan) and Sac I (TaKaRa Bio, Otsu, Japan) restriction enzymes, and the primer pairs Tp1DFaesPI_1F and TpDFaesPIR using the ClonExpress® Ultra One Step Cloning Kit (Vazyme, Nanjing, China) following the manufacturer’s protocol. Meanwhile, full-length pFaesPI_2::FaesPI_2 genomic DNAs were cloned into the pCAMBIA1300 vector with the above method, but the forward primer Tp2DFaesPI_2F and the reverse primer TpDFaesPIR. The pFaesPI_1::FaesPI_1 and pFaesPI_2::FaesPI_2 constructs were separately transformed into PI/pi-1 heterozygote Arabidopsis through the floral-dip suggested by Clough and Bent [43]. Transgenic Arabidopsis seedlings were screened, cultivated, and identified referring to Fang et al. [10]. Homozygous pi-1 transgenic Arabidopsis lines were obtained using the dCAPS genotyping method described by Lamb and Irish [44]. Phenotypes of all transgenic Arabidopsis lines were separately assessed after flowering.
Moreover, the phenotype complementation degrees of independent transgenic lines of pFaesPI_1::FaesPI_1 or pFaesPI_2::FaesPI_2 homozygous pi-1 Arabidopsis were classified as ‘no complementation’, ‘medium complementation’, and ‘strong complementation’, respectively. In addition, independent transgenic lines of each complementation degree were verified by qRT-PCR according to the above method, but with the primers qTFaesPI_1F and qTFaesPI_1R for FaesPI_1, and with the primers qTFaesPI_2F and qTFaesPI_2F for FaesPI_2. Amplification fragment of A. thaliana Actin (Genbank accession numbers: AY114679.1) with the primers qActinF and qActinF was the internal control.
5. Conclusions
F. esculentum (Polygonaceae) belongs to the order Caryophyllales (an early-diverging core eudicots clade) and produces distylous flowers with single-whorl undifferentiated showy tepals, representing an obvious difference with flowers of most core eudicots, which makes it an ideal model for exploring floral organ development and evolution. The Arabidopsis floral homeotic B-class MADS-box gene PISTILLATA (PI) is expressed in petal and stamen and works together with another B-function gene APETALA3 (AP3) to specify petal and stamen identity. However, a small-scale gene duplication (GD) event was found in the common buckwheat PI ortholog and resulted in FaesPI_1 and FaesPI_2. Furthermore, FaesPI_1/2 were expressed only in the stamen of the distylous flower. The expression absence of FaesPI_1/2 in showy tepals suggested that the PI-like gene-dependent petal identity program was not observed in F. esculentum. In addition, GUS driven by pFaesPI_1 promoter was expressed in the whole stamen of pFaesPI_1::GUS transgenic Arabidopsis, while GUS driven by pFaesPI_2 promoter was expressed only in the filament of stamen in pFaesPI_2::GUS transgenic Arabidopsis. Moreover, pFaesPI_1::FaesPI_1/pFaesPI_2::FaesPI_2 transgenic pi-1 Arabidopsis produced a similar flower with stamen-/filament-like organs in the third whorl. All these suggested that FaesPI_2 may only specify filament development, but FaesPI_1 may specify stamen development. Meanwhile, FaesPI_1 and FaesPI_2 had overlapping functions in specifying stamen filament identity and working together to regulate normal stamen development.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants11081047/s1, Table S1: Primers used in this study. Table S2: Information on Sequences selected for alignments and phylogenetic analyses from NCBI GenBank. Figure S1: FaesPI_1 promoter sequence. Figure S2: FaesPI_2 promoter sequence. Figure S3: Genotyping of wildtype, heterozygous and homozygous pi-1 mutant A. thaliana by dCAPS.
Author Contributions
W.Y.; writing—original draft preparation; W.Y., X.C., L.Z. and Z.M.; methodology; Z.L. writing-review and edition, supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31771867).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Huda, M.N.; Lu, S.; Jahan, T.; Ding, M.; Jha, R.; Zhang, K.; Zhang, W.; Georgiev, M.I.; Park, S.U.; Zhou, M. Treasure from Garden: Bioactive Compounds of Buckwheat. Food Chem. 2021, 335, 127653. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Yasui, Y. Buckwheat Heteromorphic Self-Incompatibility: Genetics, Genomics and Application to Breeding. Breed. Sci. 2020, 70, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-Y.; Fang, Z.-W.; Li, X.-F.; Liu, Z.-X. Isolation and Characterization of the C-class MADS-box Gene from the Distylous Pseudo-cereal Fagopyrum sculentum. J. Plant Biol. 2017, 60, 189–198. [Google Scholar] [CrossRef]
- Burrows, B.A.; McCubbin, A.G. Sequencing the Genomic Regions Flanking S-linked PvGLO Sequences Confirms the Presence of Two GLO Loci, One of which Lies Adjacent to the Style-Length Determinant Gene CYP734A50. Plant Reprod. 2017, 30, 53–67. [Google Scholar] [CrossRef]
- Shore, J.S.; Hamam, H.J.; Chafe, P.D.J.; Labonne, J.D.J.; Henning, P.M.; McCubbin, A.G. The Long and Short of the S-locus in Turnera (Passifloraceae). New Phytol. 2019, 224, 1316–1329. [Google Scholar] [CrossRef]
- Kim, S.; Yoo, M.; Albert, V.A.; Farris, J.S.; Soltis, P.S.; Soltis, D.E. Phylogeny and Diversification of B-Function MADS-box Genes in Angiosperms: Evolutionary and Functional Implications of a 260-million-year-old Duplication. Am. J. Bot. 2004, 91, 2102–2118. [Google Scholar] [CrossRef]
- Melzer, R.; Härter, A.; Rümpler, F.; Kim, S.; Soltis, P.S.; Soltis, D.E.; Theißen, G. DEF- and GLO-like Proteins May Have Lost Most of Their Interaction Partners during Angiosperm Evolution. Ann. Bot. 2014, 114, 1431–1443. [Google Scholar] [CrossRef]
- Wuest, S.E.; O’Maoileidigh, D.S.; Rae, L.; Kwasniewska, K.; Raganelli, A.; Hanczaryk, K.; Lohan, A.J.; Loftus, B.; Graciet, E.; Wellmer, F. Molecular Basis for the Specification of Floral Organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 2012, 109, 13452–13457. [Google Scholar] [CrossRef]
- Brockington, S.F.; Rudall, P.J.; Frohlich, M.W.; Oppenheimer, D.G.; Soltis, P.S.; Soltis, D.E. ‘Living stones’ reveal alternative petal identity programs within the core eudicots. Plant J. 2012, 69, 193–203. [Google Scholar] [CrossRef]
- Fang, Z.-W.; Li, X.-P.; Li, X.-F.; Liu, Z.-X. FaesPI, a Fagopyrum esculentum PISTILLATA Ortholog, Is Involved Only in Stamen Development. J. Plant Biol. 2015, 58, 102–109. [Google Scholar] [CrossRef]
- Yang, Y.; Jack, T. Defining Subdomains of the K Domain Important for Protein-Protein Interactions of Plant MADS Proteins. Plant Mol. Biol. 2004, 55, 45–59. [Google Scholar] [CrossRef] [PubMed]
- De Folter, S.; Angenent, G.C. Trans Meets cis in MADS Science. Trends Plant Sci. 2006, 11, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Leonard, J.M.; Monteros, A.; Liu, P.-P.; Nonogaki, H. A Novel Endo-β-Mannanase Gene in Tomato LeMAN5 Is Associated with Anther and Pollen Development. Plant Physiol. 2004, 134, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Rogers, H.J.; Bate, N.; Combe, J.; Sullivan, J.; Sweetman, J.; Swan, C.; Lonsdale, D.M.; Twell, D. Functional Analysis of cis-Regulatory Elements within the Promoter of the Tobacco Late Pollen Gene g10. Plant Mol. Biol. 2001, 45, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Dinh, T.T.; Girke, T.; Liu, X.; Yant, L.; Schmid, M.; Chen, X. The Floral Homeotic Protein APETALA2 Recognizes and Acts through an AT-Rich Sequence Element. Development 2012, 139, 1978–1986. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.-H.; Fujii, H.; Zheng, X.; Zhu, J.-K. A R2R3 Type MYB Transcription Factor Is Involved in the Cold Regulation of CBF Genes and in Acquired Freezing Tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef]
- Zhang, Z.-L.; Xie, Z.; Zou, X.; Casaretto, J.; Ho, T.D.; Shen, Q.J. A Rice WRKY Gene Encodes a Transcriptional Repressor of the Gibberellin Signaling Pathway in Aleurone Cells. Plant Physiol. 2004, 134, 1500–1513. [Google Scholar] [CrossRef]
- Gubler, F.; Kalla, R.; Roberts, J.K.; Jacobsen, J.V. Gibberellin-Regulated Expression of a Myb Gene in Barley Aleurone Cells: Evidence for Myb Transactivation of a High-PI α-Amylase Gene Promoter. Plant Cell 1995, 7, 1879–1891. [Google Scholar] [CrossRef]
- Mena, M.; Cejudo, F.J.; Isabel-Lamoneda, I.; Carbonero, P. A Role for the DOF Transcription Factor BPBF in the Regulation of Gibberellin-Responsive Genes in Barley Aleurone. Plant Physiol. 2002, 130, 111–119. [Google Scholar] [CrossRef]
- Gowik, U.; Burscheidt, J.; Akyildiz, M.; Schlue, U.; Koczor, M.; Streubel, M.; Westhoff, P. cis -Regulatory Elements for Mesophyll-Specific Gene Expression in the C4 Plant Flaveria Trinervia, the Promoter of the C4 Phosphoenolpyruvate Carboxylase Gene. Plant Cell 2004, 16, 1077–1090. [Google Scholar] [CrossRef]
- Tatematsu, K.; Ward, S.; Leyser, O.; Kamiya, Y.; Nambara, E. Identification of cis-Elements That Regulate Gene Expression during Initiation of Axillary Bud Outgrowth in Arabidopsis. Plant Physiol. 2005, 138, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Le Gourrierec, J.; Samach, A.; Coupland, G. CONSTANS and the CCAAT Box Binding Complex Share a Functionally Important Domain and Interact to Regulate Flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef] [PubMed]
- Causier, B.; Bradley, D.; Cook, H.; Davies, B. Conserved Intragenic Elements Were Critical for the Evolution of the Floral C-Function: Intragenic Regulation of the Floral C-Function. Plant J. 2009, 58, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Welchen, E.; Gonzalez, D.H. Differential Expression of the Arabidopsis Cytochrome c Genes Cytc-1 and Cytc-2. Evidence for the Involvement of TCP-Domain Protein-Binding Elements in Anther- and Meristem-Specific Expression of the Cytc-1 Gene. Plant Physiol. 2005, 139, 88–100. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early Flower Development in Arabidopsis. Plant Cell 1990, 2, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shi, M.; Chen, W.; Hu, R.; Jing, D.; Wu, D.; Wang, S.; Li, Q.; Deng, H.; Guo, Q.; et al. Expression Pattern and Functional Characterization of PISTILLATA Ortholog Associated With the Formation of Petaloid Sepals in Double-Flower Eriobotrya Japonica (Rosaceae). Front. Plant Sci. 2020, 10, 1685. [Google Scholar] [CrossRef]
- Broholm, S.K.; Pöllänen, E.; Ruokolainen, S.; Tähtiharju, S.; Kotilainen, M.; Albert, V.A.; Elomaa, P.; Teeri, T.H. Functional Characterization of B Class MADS-Box Transcription Factors in Gerbera hybrida. J. Exp. Bot. 2010, 61, 75–85. [Google Scholar] [CrossRef]
- Louati, M.; Salazar-Sarasua, B.; Roque, E.; Beltrán, J.P.; Salhi Hannachi, A.; Gómez-Mena, C. Isolation and Functional Analysis of a PISTILLATA-like MADS-Box Gene from Argan Tree (Argania Spinosa). Plants 2021, 10, 1665. [Google Scholar] [CrossRef]
- Liu, S.; Sun, Y.; Du, X.; Xu, Q.; Wu, F.; Meng, Z. Analysis of the APETALA3- and PISTILLATA-like Genes in Hedyosmum orientale (Chloranthaceae) Provides Insight into the Evolution of the Floral Homeotic B-Function in Angiosperms. Ann. Bot. 2013, 112, 1239–1251. [Google Scholar] [CrossRef]
- Liu, W.; Shen, X.; Liang, H.; Wang, Y.; He, Z.; Zhang, D.; Chen, F. Isolation and Functional Analysis of PISTILLATA Homolog From Magnolia wufengensis. Front. Plant Sci. 2018, 9, 1743. [Google Scholar] [CrossRef]
- Chen, M.-K.; Hsieh, W.-P.; Yang, C.-H. Functional Analysis Reveals the Possible Role of the C-Terminal Sequences and PI Motif in the Function of Lily (Lilium Longiflorum) PISTILLATA (PI) Orthologues. J. Exp. Bot. 2011, 63, 941–961. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Hsu, W.; Li, J.; Yang, C. Distance-based Measurement Determines the Coexistence of B Protein Hetero- and Homodimers in Lily Tepal and Stamen Tetrameric Complexes. Plant J. 2021, 105, 1357–1373. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.-T.; Hsu, H.-F.; Hsu, W.-H.; Li, J.-Y.; Lee, Y.-I.; Yang, C.-H. The C-Terminal Sequence and PI Motif of the Orchid (Oncidium Gower Ramsey) PISTILLATA (PI) Ortholog Determine Its Ability to Bind AP3 Orthologs and Enter the Nucleus to Regulate Downstream Genes Controlling Petal and Stamen Formation. Plant Cell Physiol. 2015, 56, 2079–2099. [Google Scholar] [CrossRef][Green Version]
- Hsu, H.-F.; Hsu, W.-H.; Lee, Y.-I.; Mao, W.-T.; Yang, J.-Y.; Li, J.-Y.; Yang, C.-H. Model for Perianth Formation in Orchids. Nat. Plants 2015, 1, 15046. [Google Scholar] [CrossRef]
- Hsu, H.-F.; Chen, W.-H.; Shen, Y.-H.; Hsu, W.-H.; Mao, W.-T.; Yang, C.-H. Multifunctional Evolution of B and AGL6 MADS Box Genes in Orchids. Nat. Commun. 2021, 12, 902. [Google Scholar] [CrossRef]
- Wang, P.; Liao, H.; Zhang, W.; Yu, X.; Zhang, R.; Shan, H.; Duan, X.; Yao, X.; Kong, H. Flexibility in the Structure of Spiral Flowers and Its Underlying Mechanisms. Nat. Plants 2016, 2, 15188. [Google Scholar] [CrossRef]
- Gong, P.; Ao, X.; Liu, G.; Cheng, F.; He, C. Duplication and Whorl-Specific Down-Regulation of the Obligate AP3–PI Heterodimer Genes Explain the Origin of Paeonia Lactiflora Plants with Spontaneous Corolla Mutation. Plant Cell Physiol. 2017, 58, 411–425. [Google Scholar] [CrossRef][Green Version]
- Roque, E.; Fares, M.A.; Yenush, L.; Rochina, M.C.; Wen, J.; Mysore, K.S.; Gómez-Mena, C.; Beltrán, J.P.; Cañas, L.A. Evolution by Gene Duplication of Medicago Truncatula PISTILLATA-like Transcription Factors. J. Exp. Bot. 2016, 67, 1805–1817. [Google Scholar] [CrossRef][Green Version]
- Huu, C.N.; Keller, B.; Conti, E.; Kappel, C.; Lenhard, M. Supergene Evolution via Stepwise Duplications and Neofunctionalization of a Floral-Organ Identity Gene. Proc. Natl. Acad. Sci. USA 2020, 117, 23148–23157. [Google Scholar] [CrossRef]
- Zeng, L.; Zhang, J.; Wang, X.; Liu, Z. Isolation and Characterization of APETALA3 Orthologs and Promoters from the Distylous Fagopyrum esculentum. Plants 2021, 10, 1644. [Google Scholar] [CrossRef]
- Liu, Z.; Fei, Y.; Zhang, K.; Fang, Z. Ectopic Expression of a Fagopyrum esculentum APETALA1 Ortholog Only Rescues Sepal Development in Arabidopsis Ap1 Mutant. Int. J. Mol. Sci. 2019, 20, 2021. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-Acting Regulatory DNA Elements (PLACE) Database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method for Agrobacterium-Mediated Transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.S.; Irish, V.F. Functional Divergence within the APETALA3/PISTILLATA Floral Homeotic Gene Lineages. Proc. Natl. Acad. Sci. USA 2003, 100, 6558–6563. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).