Less Virulent Leptosphaeria biglobosa Immunizes the Canola Plant to Resist Highly Virulent L. maculans, the Blackleg Pathogen
Abstract
1. Introduction
2. Results
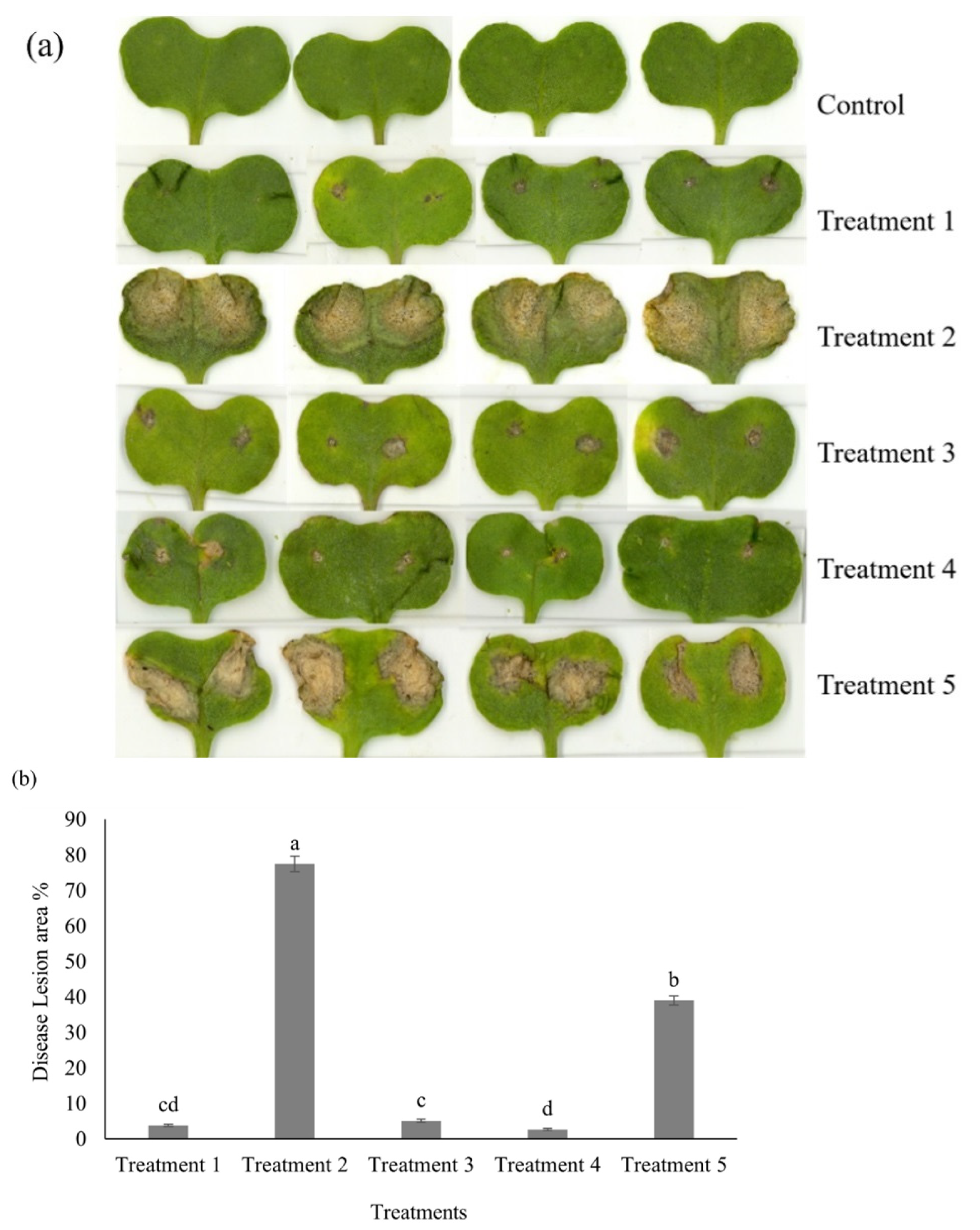
2.1. Disease Development in Cotyledons
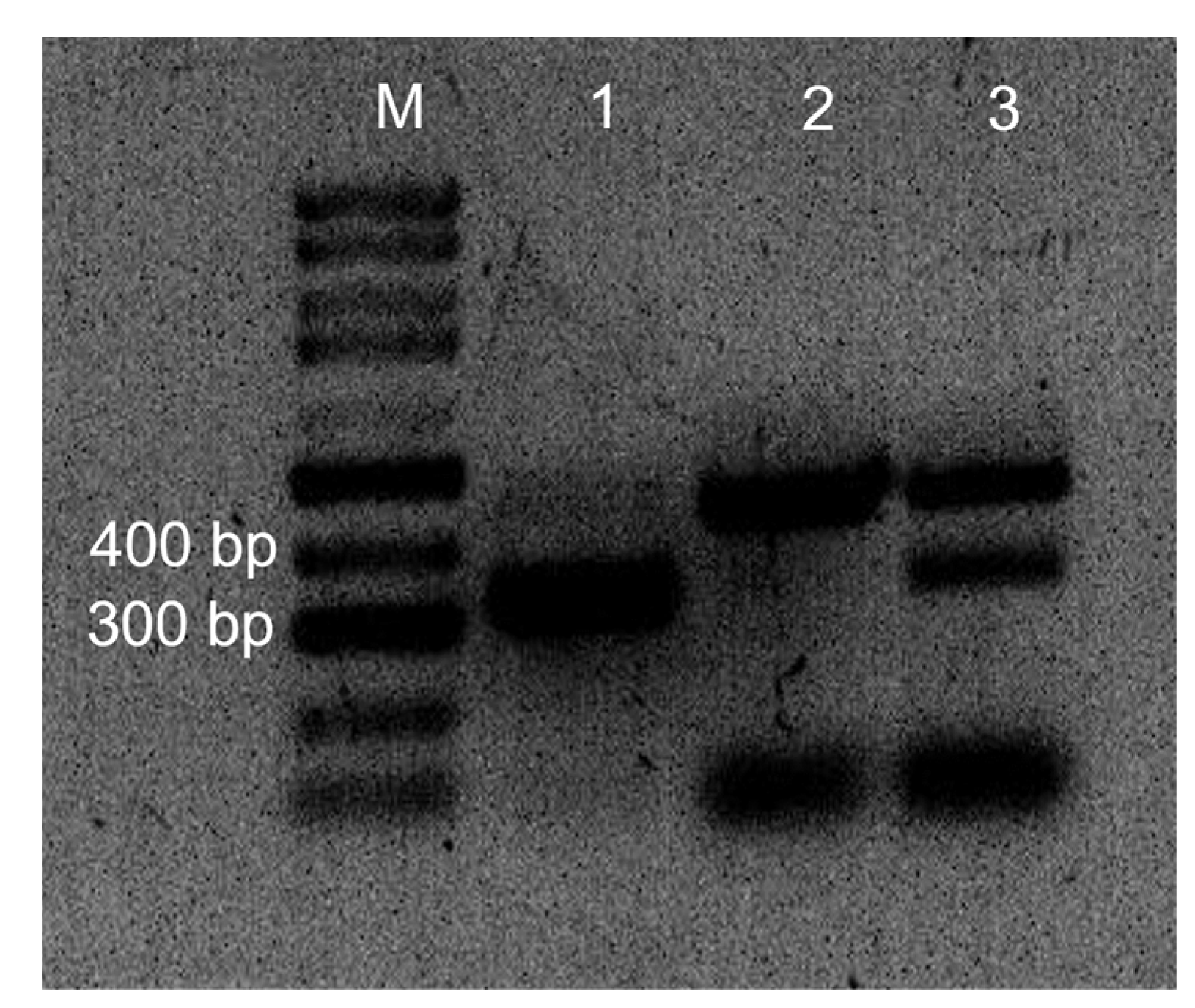
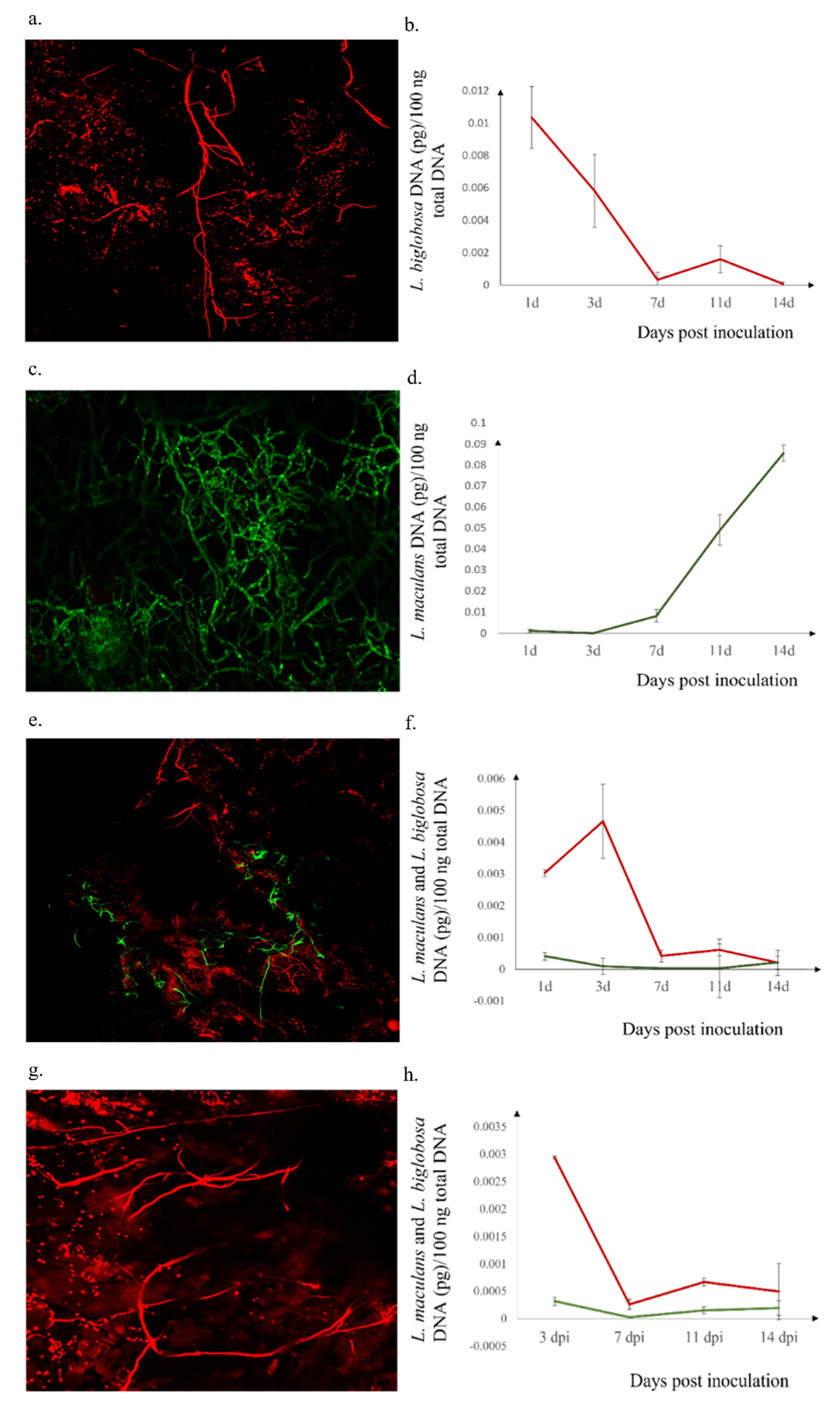
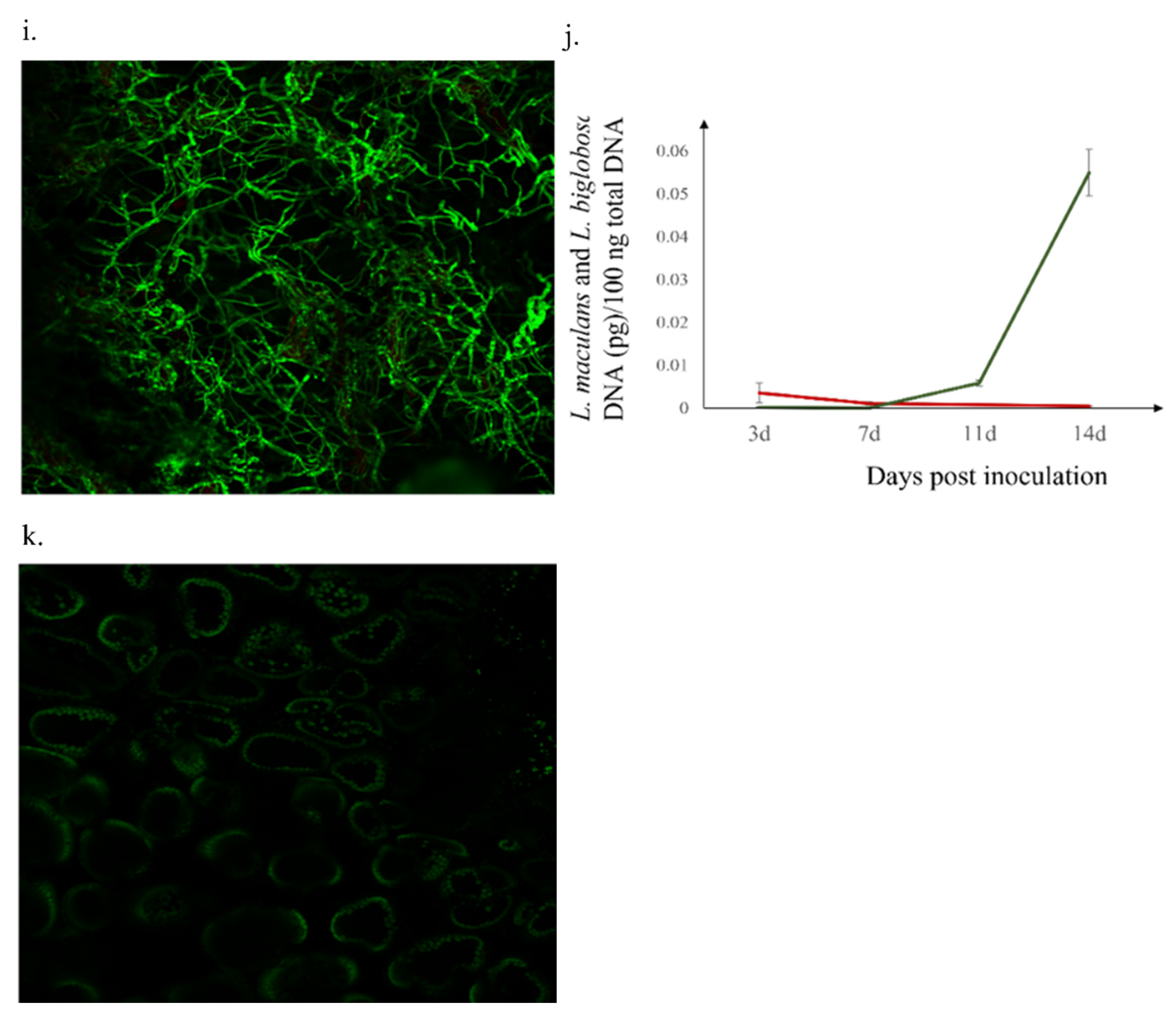
2.2. Presence of Leptosphaeria spp. in Inoculated Westar Cotyledons
2.3. L. biglobosa Inhibits the L. maculans Growth in B. napus Cotyledons
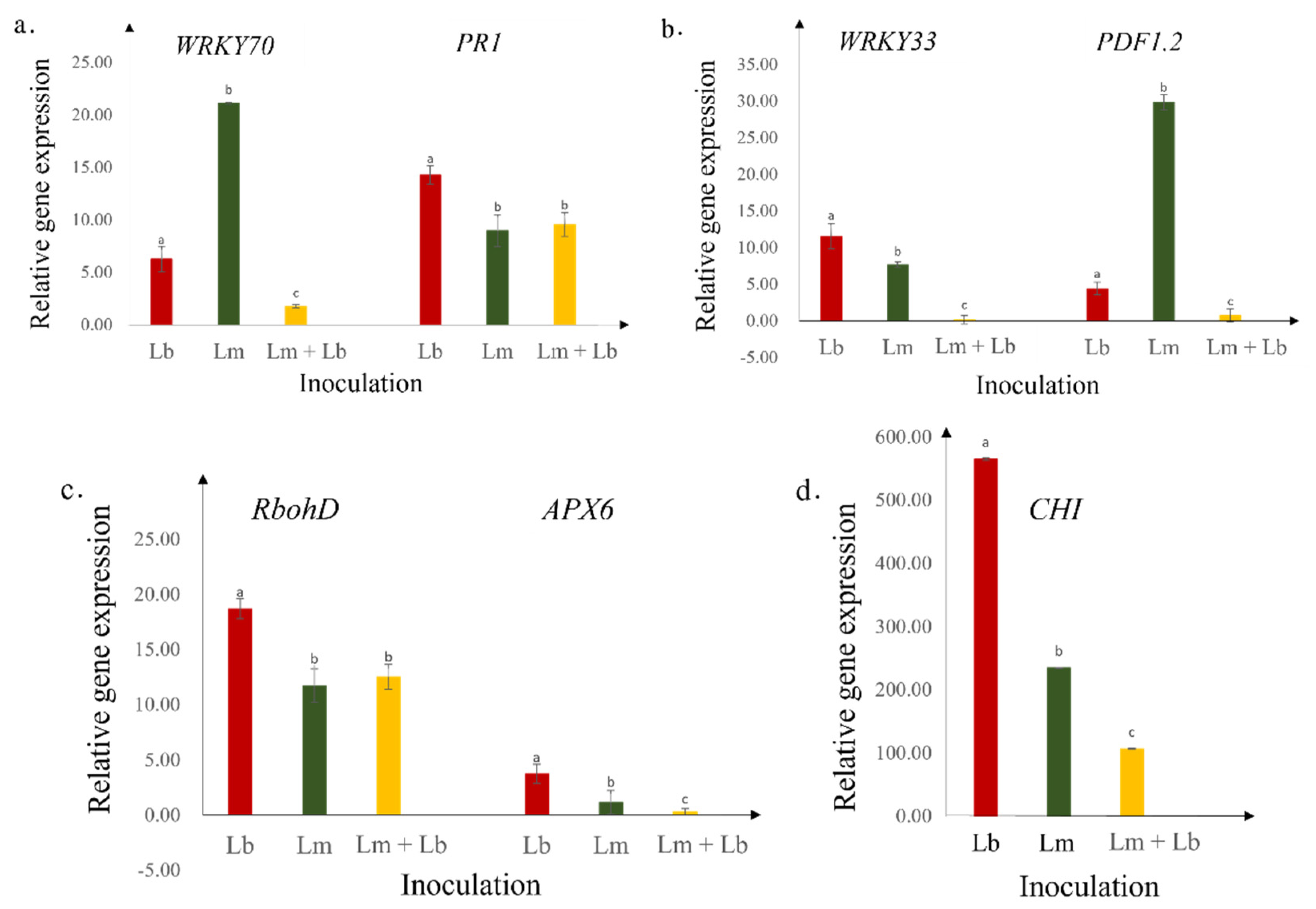
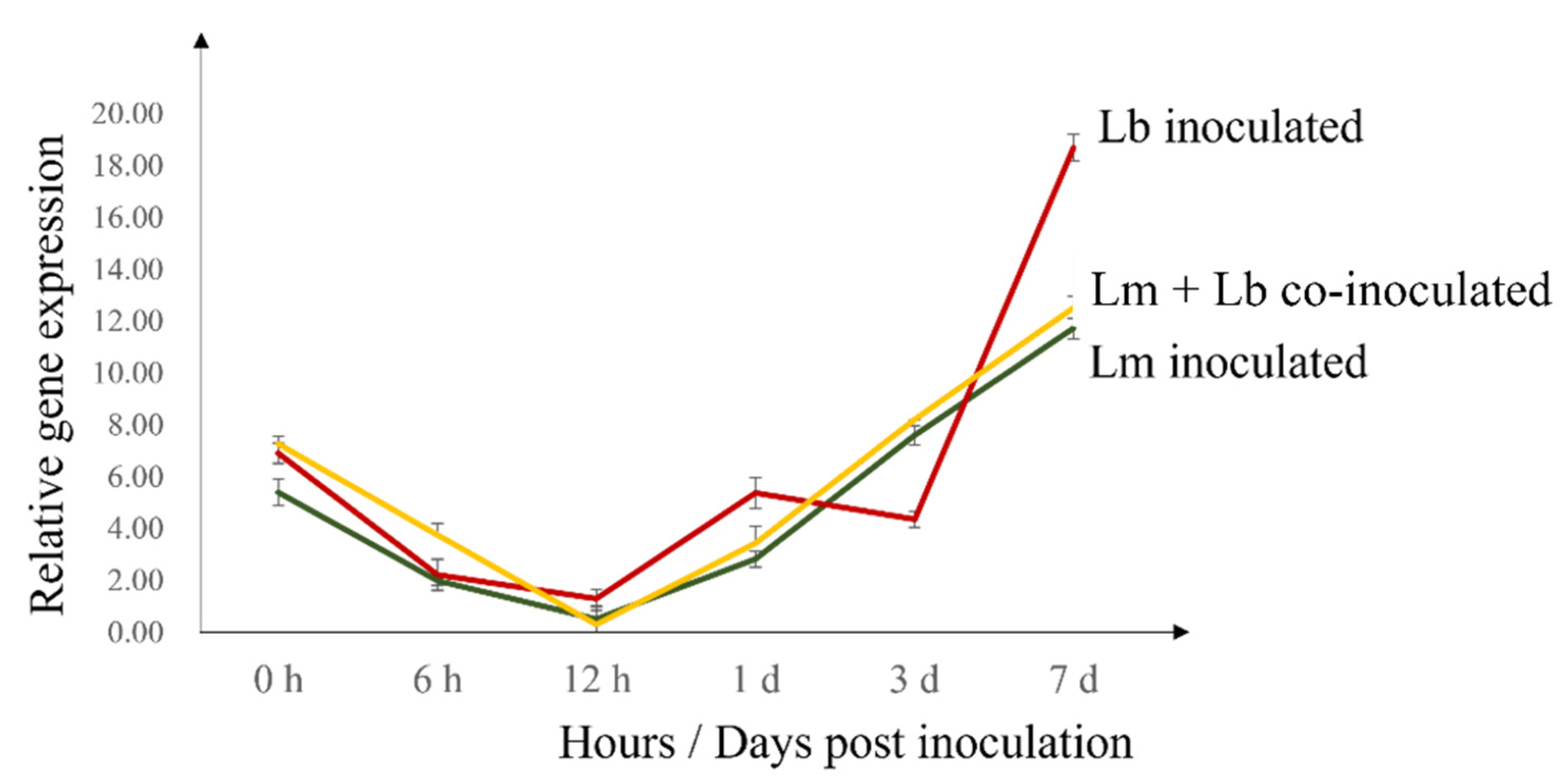
2.4. Expression of Defense Genes
3. Discussion
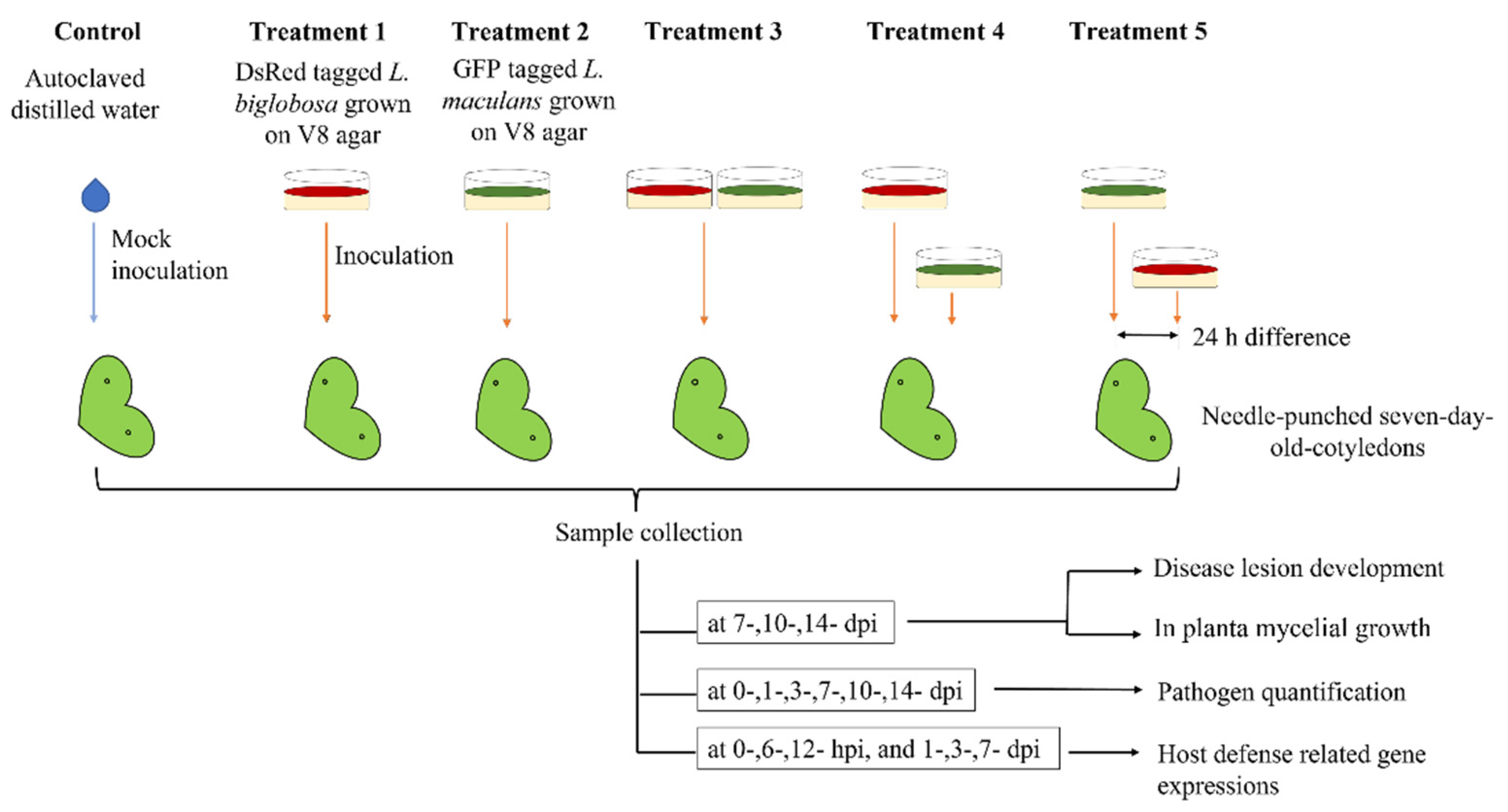
4. Materials and Methods
4.1. Pathogen Isolates
4.2. Plant Growth and Inoculation
4.3. Disease Lesion Development on Cotyledons
4.4. Confocal Observation of in Planta Mycelial Development
4.5. PCR Identification of Leptosphaeria maculans ‘brassicae’ and L. biglobosa ‘brassicae’ Isolates in Inoculated Cotyledons
4.6. Quantification of Pathogen Isolates Using Quantitative PCR
4.7. Defense-Related Gene Expression
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, X.; Fernando, W.G.D. Insights into fighting against blackleg disease of Brassica napus in Canada. Crop Pasture Sci. 2018, 69, 40–47. [Google Scholar] [CrossRef]
- Mendes-Pereira, E.; Balesdent, M.H.; Hortense, B.; Rouxel, T. Molecular phylogeny of the Leptosphaeria maculans–L. biglobosa species complex. Mycol. Res. 2003, 107, 1287–1304. [Google Scholar] [CrossRef]
- Vincenot, L.; Balesdent, M.H.; Li, H.; Barbetti, M.J.; Sivasithamparam, K.; Gout, L.; Rouxel, T. Occurrence of a new subclade of Leptosphaeria biglobosa in Western Australia. Phytopathology 2008, 98, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Zhang, X.; Parks, P.; du Toit, L.J.; Van de Wouw, A.P.; Fernando, W.G.D. A New Subclade of Leptosphaeria biglobosa Identified from Brassica rapa. Int. J. Mol. Sci. 2019, 20, 1668. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Latunde-Dada, A.O.; Hall, A.M.; Fitt, B.D.L. Phoma stem canker disease on oilseed rape (Brassica napus) in China is caused by Leptosphaeria biglobosa ‘brassicae’. Eur. J. Plant Pathol. 2014, 140, 841–857. [Google Scholar] [CrossRef]
- West, J.S.; Biddulph, J.E.; Fitt, B.D.L.; Gladders, P. Epidemiology of Leptosphaeria maculans in relation to forecasting stem canker severity on winter oilseed rape in the UK. Ann. Appl. Biol. 1999, 135, 535–546. [Google Scholar] [CrossRef]
- West, J.S.; Kharbanda, P.D.; Barbetti, M.J.; Fitt, B.D.L. Epidemiology and management of Leptosphaeria maculans (Phoma stem canker) on oilseed rape in Australia, Canada and Europe. Plant Pathol. 2001, 50, 10–27. [Google Scholar] [CrossRef]
- Paul, V.H.; Rawlinson, C.J. Diseases and Pests of Rape; Verlag Th. Mann: Gelsenkirchen-Buer, Germany, 1992. [Google Scholar]
- Fitt, B.D.L.; Brun, H.; Barbetti, M.J.; Rimmer, S.R. World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur. J. Plant Pathol. 2006, 114, 3–15. [Google Scholar] [CrossRef]
- Lowe, R.G.T.; Cassin, A.; Grandaubert, J.; Clark, B.L.; Van de Wouw, A.P.; Rouxel, T.; Howlett, B.J. Genomes and Transcriptomes of Partners in Plant-Fungal- Interactions between Canola (Brassica napus) and Two Leptosphaeria Species. PLoS ONE 2014, 9, e103098. [Google Scholar] [CrossRef]
- Mahuku, G.S.; Hall, R.; Goodwin, P.H. Co-infection and induction of systemic acquired resistance by weakly and highly virulent isolates of Leptosphaeria maculans in oilseed rape. Physiol. Mol. Plant Pathol. 1996, 49, 61–72. [Google Scholar] [CrossRef]
- Chen, Y.; Fernando, W.G.D. Induced resistance to blackleg (Leptosphaeria maculans) disease of canola (Brassica napus) caused by a weakly virulent isolate of Leptosphaeria biglobosa. Plant Dis. 2006, 90, 1059–1064. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choudhary, D.K.; Prakash, A.; Johri, B.N. Induced systemic resistance (ISR) in plants: Mechanism of action. Indian J. Microbiol. 2007, 47, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; De Zutter, N.; De Saeger, S.; De Boevre, M.; Tran, T.M.; Van der Lee, T.; Waalwijk, C.; Willems, A.; Vandamme, P.; Ameye, M.; et al. Presence of the weakly pathogenic Fusarium poae in the Fusarium Head Blight disease complex hampers biocontrol and chemical control of the virulent Fusarium graminearum pathogen. Front. Plant Sci. 2021, 12, 216. [Google Scholar] [CrossRef] [PubMed]
- De Lamo, F.J.; Spijkers, S.B.; Takken, F.L.W. Protection to Tomato Wilt Disease Conferred by the Nonpathogen Fusarium oxysporum Fo47 is More Effective Than that Conferred by Avirulent Strains. Phytopathology 2021, 111, 253–257. [Google Scholar] [CrossRef]
- Heath, M.C. Apoptosis programmed cell death and the hypersensitive cell death. Eur. J. Plant Pathol. 1998, 104, 117–124. [Google Scholar] [CrossRef]
- Hammerschmidt, R. Induced disease resistance: How do induced plants stop pathogens? Physiol. Mol. Plant Pathol. 1999, 55, 77–84. [Google Scholar] [CrossRef]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef]
- Yang, C.; Fernando, W.G.D. Hormonal responses to susceptible, intermediate, and resistant interactions in the Brassica napus–Leptosphaeria maculans pathosystem. Int. J. Mol. Sci. 2021, 22, 4714. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Valverde, R.A. The Respiratory Burst Oxidase Homolog D (RbohD) Cell and Tissue Distribution in Potato-Potato Virus Y (PVYNTN) Hypersensitive and Susceptible Reactions. Int. J. Mol. Sci. 2019, 20, 2741. [Google Scholar] [CrossRef]
- Sharma, R.K.; Patel, S.; Pargaien, K.C. Synthesis, characterization and properties of Mn-doped ZnO nanocrystals. Adv. Nat. Sci. Nanosci. Nanotechnol. 2012, 3, 035005. [Google Scholar] [CrossRef][Green Version]
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012, 159, 266–285. [Google Scholar] [CrossRef]
- Suzuki, N.; Miller, G.; Morales, J.; Shulaev, V.; Torres, M.A.; Mittler, R. Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant Biol. 2011, 14, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Keller, H.; Blein, J.P.; Bonnet, P.; Ricci, P. Physiological and Molecular Characteristics of Elicitin-Induced Systemic Acquired Resistance in Tobacco. Plant Physiol. 1996, 110, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Bestwick, C.S.; Brown, I.R.; Bennett, M.H.R.; Mansfield, J.W. Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola. Plant Cell 1997, 9, 209–221. [Google Scholar] [PubMed]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.G.; Zhang, X.; Walker, P.L.; Wan, J.C.; Millar, J.L.; Khan, D.; Granger, M.J.; Cavers, J.D.; Chan, A.C.; Fernando, D.W.; et al. Transcriptome analysis of the Brassica napus–Leptosphaeria maculans pathosystem identifies receptor, signaling and structural genes underlying plant resistance. Plant J. 2017, 90, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Farmer, E.E.; Alméras, E.; Krishnamurthy, V. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 2003, 6, 372–378. [Google Scholar] [CrossRef]
- Henry, E.; Yadeta, K.A.; Coaker, G. Recognition of bacterial plant pathogens: Local, systemic and transgenerational immunity. New Phytol. 2013, 199, 908–915. [Google Scholar] [CrossRef]
- Kumar, M.; Brar, A.; Yadav, M.; Chawade, A.; Vivekanand, V.; Pareek, N. Chitinases—Potential Candidates for Enhanced Plant Resistance towards Fungal Pathogens. Agriculture 2018, 8, 88. [Google Scholar] [CrossRef]
- Li, C.; Barker, S.J.; Gilchrist, D.G.; Lincoln, J.E.; Cowling, W.A. Leptosphaeria maculans elicits apoptosis coincident with leaf lesion formation and hyphal advance in Brassica napus. Mol. Plant. Microbe Interact. 2008, 21, 1143–1153. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.G.; Haddadi, P.; Wan, J.; Adam, L.; Walker, P.; Larkan, N.J.; Daayf, F.; Borhan, M.H.; Belmonte, M.F. Transcriptome analysis of Rlm2-mediated host immunity in the Brassica napus–Leptosphaeria maculans pathosystem. Mol. Plant-Microbe Interact. 2019, 32, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Dilmaghani, A.; Balesdent, M.H.; Didier, J.P.; Wu, C.; Davey, J.; Barbetti, M.J.; Li, H.; Moreno-Rico, O.; Phillips, D.; Despeghel, J.P.; et al. The Leptosphaeria maculans–Leptosphaeria biglobosa species complex in the American continent. Plant Pathol. 2009, 58, 1044–1058. [Google Scholar] [CrossRef]
- Fitt, B.D.L.; Hu, B.C.; Li, Z.Q.; Liu, S.Y.; Lange, R.M.; Kharbanda, P.D.; Butterworth, M.H.; White, R.P. Strategies to prevent spread of Leptosphaeria maculans (phoma stem canker) onto oilseed rape crops in China; costs and benefits. Plant Pathol. 2008, 57, 652–664. [Google Scholar] [CrossRef]
- Jacques, N.; Balesdent, M.; Rouxel, T.; Laval, V. New specific quantitative real-time PCR assays shed light on the epidemiology of two species of the Leptosphaeria maculans–Leptosphaeria biglobosa species complex. Plant Pathol. 2021, 70, 643–654. [Google Scholar] [CrossRef]
- West, J.S.; Balesdent, M.H.; Rouxel, T.; Nancy, J.P.; Huang, Y.J.; Roux, J.; Steed, J.M.; Fitt, B.D.L.; Schmit, J. Colonisation of winter oilseed rape tissues by A/Tox+ and B/Tox0 Leptosphaeria maculans (phoma stem canker) in France and England. Plant Pathol. 2002, 5, 311–321. [Google Scholar] [CrossRef]
- Mazáková, J.; Urban, J.; Zouhar, M.; Ryšánek, P. Analysis of Leptosphaeria species complex causing phoma leaf spot and stem canker of winter oilseed rape (Brassica napus) in the Czech Republic. Crop Pasture Sci. 2017, 68, 254–264. [Google Scholar] [CrossRef]
- Liu, S.Y.; Liu, Z.; Fitt, B.D.L.; Evans, N.; Foster, S.J.; Huang, Y.J.; Latunde-Dada, A.O.; Lucas, J.A. Resistance to Leptosphaeria maculans (phoma stem canker) in Brassica napus (oilseed rape) induced by L. biglobosa and chemical defence activators in field and controlled environments. Plant Pathol. 2006, 55, 401–412. [Google Scholar] [CrossRef]
- Martyn, J.D. Induced resistance to fusarium wilt of watermelon under simulated field conditions. Plant Dis. 1991, 75, 874–877. [Google Scholar] [CrossRef]
- Eckert, M.; Maguire, K.; Urban, M.; Foster, S.; Fitt, B.; Lucas, J.; Hammond-Kosack, K. Agrobacterium tumefaciens-mediated transformation of Leptosphaeria spp. and Oculimacula spp. with the reef coral gene DsRed and the jellyfish gene gfp. FEMS Microbiol. Lett. 2005, 253, 67–74. [Google Scholar] [CrossRef]
- Hubbard, M.; Peng, G. Quantitative resistance against an isolate of Leptosphaeria maculans (blackleg) in selected Canadian canola cultivars remains effective under increased temperatures. Plant Pathol. 2018, 67, 1329–1338. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Iwase, A.; Rymen, B.; Lambolez, A.; Kojima, M.; Takebayashi, Y.; Heyman, J.; Watanabe, S.; Seo, M.; De Veylder, L.; et al. Wounding Triggers Callus Formation via Dynamic Hormonal and Transcriptional Changes. Plant Physiol. 2017, 175, 1158–1174. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Liban, S.H.; Cross, D.J.; Kutcher, H.R.; Peng, G.; Fernando, W.G.D. Race structure and frequency of avirulence genes in the western Canadian Leptosphaeria maculans pathogen population, the causal agent of blackleg in Brassica species. Plant Pathol. 2016, 65, 1161–1169. [Google Scholar] [CrossRef]
- Huang, Y.J.; Karandeni-Dewage, C.S.; Fitt, B.D.L. Importance of Leptosphaeria biglobosa as a cause of phoma stem canker on winter oilseed rape in the UK. Asp. Appl. Biol. 2014, 127, 117–122. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Gene | Full Name | Defense Signaling Pathway | Forward Primer (5′3′) | Reverse Primer (5′3′) |
|---|---|---|---|---|
| WRKY70 | WRKY transcription factor 70 | Salicyclic acid signaling pathway | ACATACATAGGAAACCACACG | ACTTGGACTATCTTCAGAATGC |
| PR1 | Pathogenesis-related protein 1 | Salicyclic acid pathway | GGCTAACTATAACCACGATTC | GTTCCACCATTGTTACACC |
| WRKY33 | WRKY transcription factor 33 | Jasmonic acid signaling pathway | TGTCGGACAGCTTGGGAAAG | AGAGGACGGTTACAACTGGAGAAA |
| PDF1.2 | Plant defensin 1.2 | Ethylene and Jasmonic acid pathway | AAATGCTTCCTGCGACAACG | AGTCCACGTCTCCGATCTCT |
| RbohD | Respiratory burst oxidase homolog protein D | Reactive Oxygen Species Production | TATCCTCAAGGACATCATCAG | TATCCTCAAGGACATCATCAG |
| APX6 | Ascorbate peroxidase | Reactive Oxygen Species Scavenging | AGTTCGTAGCTGCTAAATATT | GGAGTTGTTATTACCAAGAAA |
| CHI | Chitinase | Pathogen chitin degradation | TGCTACATAGAAGAAATAAACGG | TTCCATGATAGTTGAATCGG |
| Actin | Actin | Reference gene used in the assay | CTGGAATTGCTGACCGTATGAG | GTTGGAAAGTGCTGAGGGATG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padmathilake, K.R.E.; Fernando, W.G.D. Less Virulent Leptosphaeria biglobosa Immunizes the Canola Plant to Resist Highly Virulent L. maculans, the Blackleg Pathogen. Plants 2022, 11, 996. https://doi.org/10.3390/plants11070996
Padmathilake KRE, Fernando WGD. Less Virulent Leptosphaeria biglobosa Immunizes the Canola Plant to Resist Highly Virulent L. maculans, the Blackleg Pathogen. Plants. 2022; 11(7):996. https://doi.org/10.3390/plants11070996
Chicago/Turabian StylePadmathilake, Kaluhannadige Rasanie Eranka, and Wannakuwattewaduge Gerard Dilantha Fernando. 2022. "Less Virulent Leptosphaeria biglobosa Immunizes the Canola Plant to Resist Highly Virulent L. maculans, the Blackleg Pathogen" Plants 11, no. 7: 996. https://doi.org/10.3390/plants11070996
APA StylePadmathilake, K. R. E., & Fernando, W. G. D. (2022). Less Virulent Leptosphaeria biglobosa Immunizes the Canola Plant to Resist Highly Virulent L. maculans, the Blackleg Pathogen. Plants, 11(7), 996. https://doi.org/10.3390/plants11070996







