Plastid Genome of Equisetum xylochaetum from the Atacama Desert, Chile and the Relationships of Equisetum Based on Frequently Used Plastid Genes and Network Analysis
Abstract
:1. Introduction
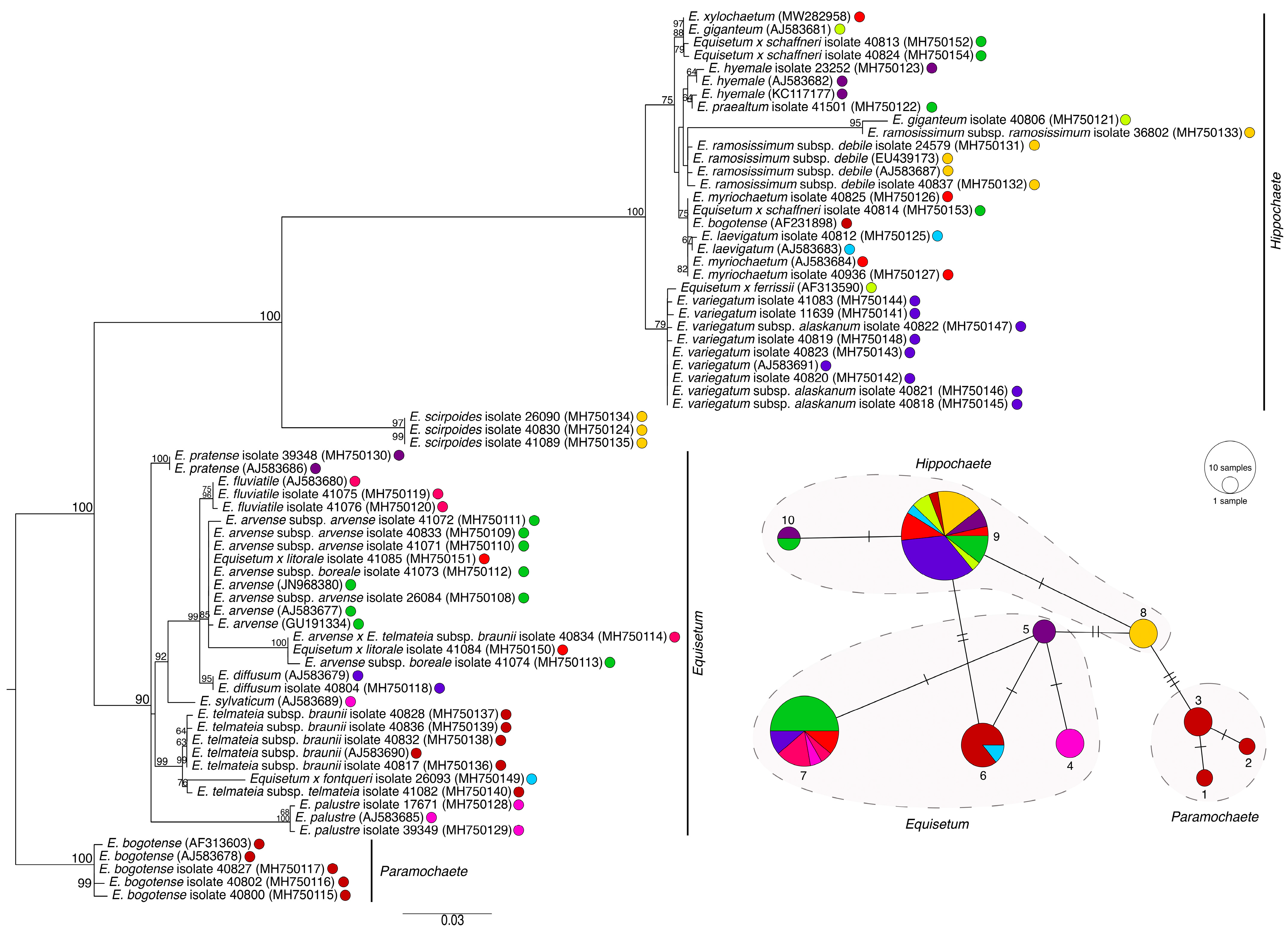
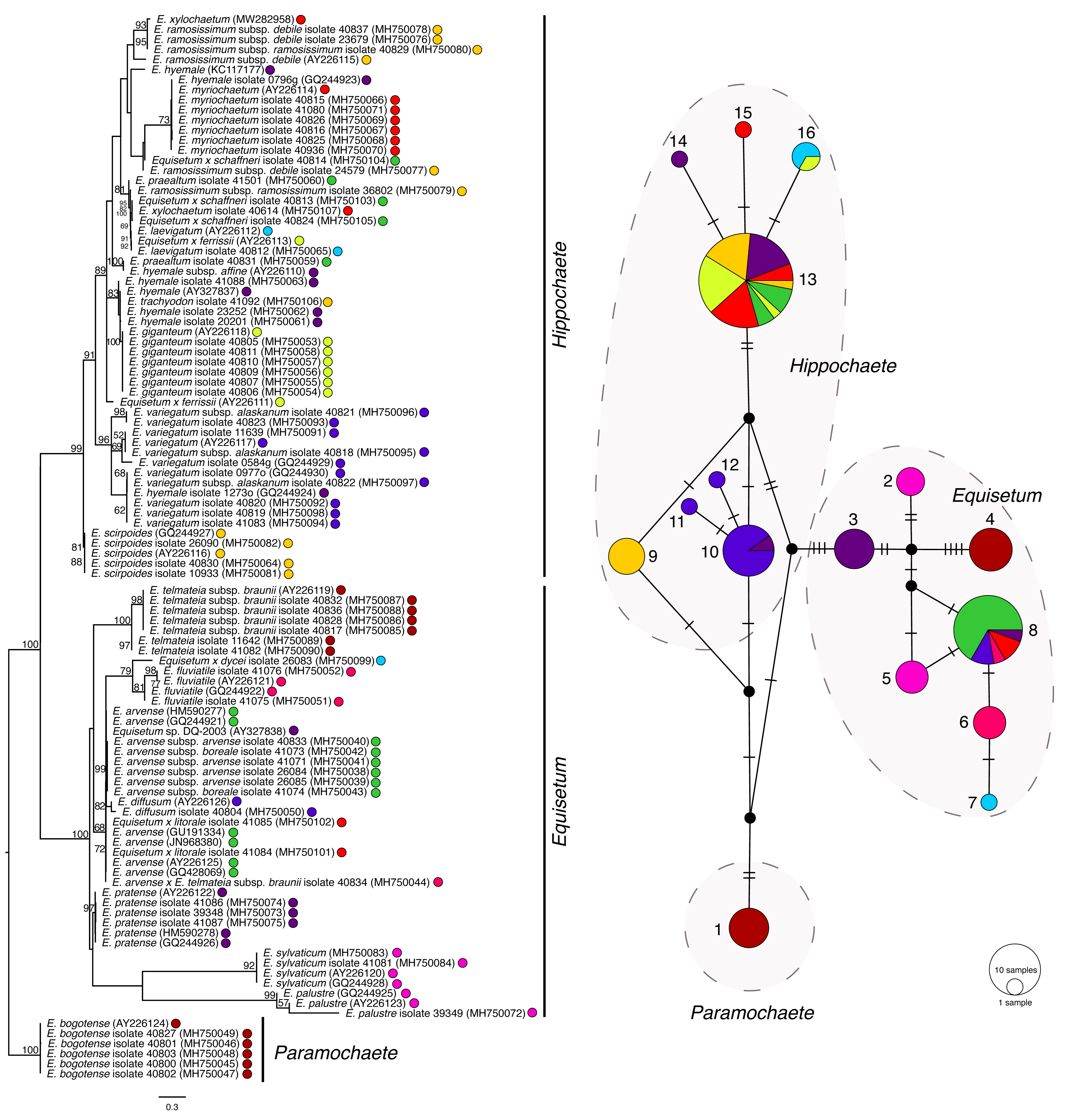
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hauke, R.L. A taxonomical monograph of the genus Equisetum subgenus. Nova Hedwig. 1963, 8, 1–123. [Google Scholar]
- Husby, C. Biology and functional ecology of Equisetum with emphasis on the giant horsetails. Bot. Rev. 2013, 79, 147–177. [Google Scholar] [CrossRef]
- Guillon, J.M. Phylogeny of horsetails (Equisetum) based on the chloroplast rps4 gene and adjacent noncoding sequences. Syst. Bot. 2004, 29, 251–259. [Google Scholar] [CrossRef]
- Elgorriaga, A.; Escapa, I.H.; Rothwell, G.W.; Tomescu, A.M.; Rubén Cúneo, N. Origin of Equisetum: Evolution of horsetails (Equisetales) within the major euphyllophyte clade Sphenopsida. Am. J. Bot. 2018, 105, 1286–1303. [Google Scholar] [CrossRef]
- Christenhusz, M.J.; Bangiolo, L.; Chase, M.W.; Fay, M.F.; Husby, C.; Witkus, M.; Viruel, J. Phylogenetics, classification and typification of extant horsetails (Equisetum, Equisetaceae). Bot. J. Linn. Soc. 2019, 189, 311–352. [Google Scholar] [CrossRef]
- Christenhusz, M.J.; Chase, M.W.; Fay, M.F.; Hidalgo, O.; Leitch, I.J.; Pellicer, J.; Viruel, J. Biogeography and genome size evolution of the oldest extant vascular plant genus, Equisetum (Equisetaceae). Ann. Bot. 2021, 127, 681–695. [Google Scholar] [CrossRef]
- Grewe, F.; Guo, W.; Gubbels, E.A.; Hansen, A.K.; Mower, J.P. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol. Biol. 2013, 13, 8. [Google Scholar] [CrossRef] [Green Version]
- Karol, K.G.; Arumuganathan, K.; Boore, J.L.; Duffy, A.M.; Everett, K.D.; Hall, J.D.; Hansen, S.K.; Kuehl, J.V.; Mandoli, D.F.; Mishler, B.D.; et al. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: Implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol. Biol. 2010, 10, 321. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.T.; Kim, K.J. Chloroplast genome differences between Asian and American Equisetum arvense (Equisetaceae) and the origin of the hypervariable trnY-trnE intergenic spacer. PLoS ONE 2014, 9, e103898. [Google Scholar]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [Green Version]
- Guillon, J.M. Molecular phylogeny of horsetails (Equisetum) including chloroplast atpB sequences. J. Plant Res. 2007, 120, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Satjarak, A.; Piotrowski, M.J.; Graham, L.E.; Trest, M.T.; Wilcox, L.W.; Knack, J.J.; Cook, M.E.; Arancibia-Avila, P. Microscopic and metagenomic evidence for eukaryotic microorganisms associated with Atacama Desert populations of giant Equisetum. Am. Fern. J. 2021, 111, 86–109. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satjarak, A.; Graham, L.E. Comparative DNA sequence analyses of Pyramimonas parkeae (Prasinophyceae) chloroplast genomes. J. Phycol. 2017, 53, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Mekvipad, N.; Satjarak, A. Evolution of organellar genes of chlorophyte algae: Relevance to phylogenetic inference. PLoS ONE 2019, 14, e0216608. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Crandall, M.C.D.P.K.; Clement, M.; Posada, D. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Murdock, A.G. Phylogeny of marattioid ferns (Marattiaceae): Inferring a root in the absence of a closely related outgroup. Am. J. Bot. 2008, 95, 626–641. [Google Scholar] [CrossRef] [PubMed]
- Pryer, K.M.; Schneider, H.; Smith, A.R.; Cranfill, R.; Wolf, P.G.; Hunt, J.S.; Sipes, S.D. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 2001, 409, 618–622. [Google Scholar] [CrossRef]
- Cook, R.; Hennell, J.R.; Lee, S.; Khoo, C.S.; Carles, M.C.; Higgins, V.J.; Govindaraghavan, S.; Sucher, N.J. The Saccharomyces cerevisiae transcriptome as a mirror of phytochemical variation in complex extracts of Equisetum arvense from America, China, Europe and India. BMC Genom. 2013, 14, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Hausner, G.; Olson, R.; Simon, D.; Johnson, I.; Sanders, E.R.; Karol, K.G.; McCourt, R.M.; Zimmerly, S. Origin and evolution of the chloroplast trnK (matK) intron: A model for evolution of group II intron RNA structures. Mol. Biol. Evol. 2006, 23, 380–391. [Google Scholar] [CrossRef]
- Knie, N.; Fischer, S.; Grewe, F.; Polsakiewicz, M.; Knoop, V. Horsetails are the sister group to all other monilophytes and Marattiales are sister to leptosporangiate ferns. Mol. Phylogenet. Evol. 2015, 90, 140–149. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Burgess, K.S.; Kesanakurti, P.R.; Graham, S.W.; Newmaster, S.G.; Husband, B.C.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 2008, 3, e2802. [Google Scholar] [CrossRef] [Green Version]
- Magallón, S.; Hilu, K.W.; Quandt, D. Land plant evolutionary timeline: Gene effects are secondary to fossil constraints in relaxed clock estimation of age and substitution rates. Am. J. Bot. 2013, 100, 556–573. [Google Scholar] [CrossRef] [Green Version]
- Scharn, R.; Little, C.J.; Bacon, C.D.; Alatalo, J.M.; Antonelli, A.; Björkman, M.P.; Molau, U.; Nilsson, R.H.; Björk, R.G. Decreased soil moisture due to warming drives phylogenetic diversity and community transitions in the tundra. Environ. Res. Lett. 2021, 16, 064031. [Google Scholar] [CrossRef]
- Kuo, L.Y.; Li, F.W.; Chiou, W.L.; Wang, C.N. First insights into fern matK phylogeny. Mol. Phylogenet. Evol. 2011, 59, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhou, Y.; Wang, Z.W.; Su, Y.J.; Wang, T. Evolution of the rpoB-psbZ region in fern plastid genomes: Notable structural rearrangements and highly variable intergenic spacers. BMC Plant Biol. 2011, 11, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newton, A.E.; Cox, C.J.; Duckett, J.G.; Wheeler, J.A.; Goffinet, B.; Hedderson, T.A.; Mishler, B.D. Evolution of the major moss lineages: Phylogenetic analyses based on multiple gene sequences and morphology. Bryologist 2000, 103, 187–211. [Google Scholar] [CrossRef]
- Des Marais, D.L.; Smith, A.R.; Britton, D.M.; Pryer, K.M. Phylogenetic relationships and evolution of extant horsetails, Equisetum, based on chloroplast DNA sequence data (rbcL and trnL-F). Int. J. Plant Sci. 2003, 164, 737–751. [Google Scholar] [CrossRef] [Green Version]
- Lang, A.; Naciri, Y. New chloroplast primers for intraspecific variation in Dicranum scoparium Hedw. (Dicranaceae) and amplification success in other bryophyte species. Mol. Ecol. Resour. 2010, 10, 735–737. [Google Scholar] [CrossRef]
- Hiiesalu, I.; Oepik, M.; Metsis, M.; Lilje, L.; Davison, J.; Vasar, M.; Moora, M.; Zobel, M.; Wilson, S.D.; Paertel, M. Plant species richness belowground: Higher richness and new patterns revealed by next-generation sequencing. Mol. Ecol. 2012, 21, 2004–2016. [Google Scholar] [CrossRef]
- Soininen, E.M.; Valentini, A.; Coissac, E.; Miquel, C.; Gielly, L.; Brochmann, C.; Brysting, A.K.; Sønstebø, J.H.; Ims, R.A.; Yoccoz, N.G.; et al. Analysing diet of small herbivores: The efficiency of DNA barcoding coupled with high-throughput pyrosequencing for deciphering the composition of complex plant mixtures. Front Zool. 2009, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
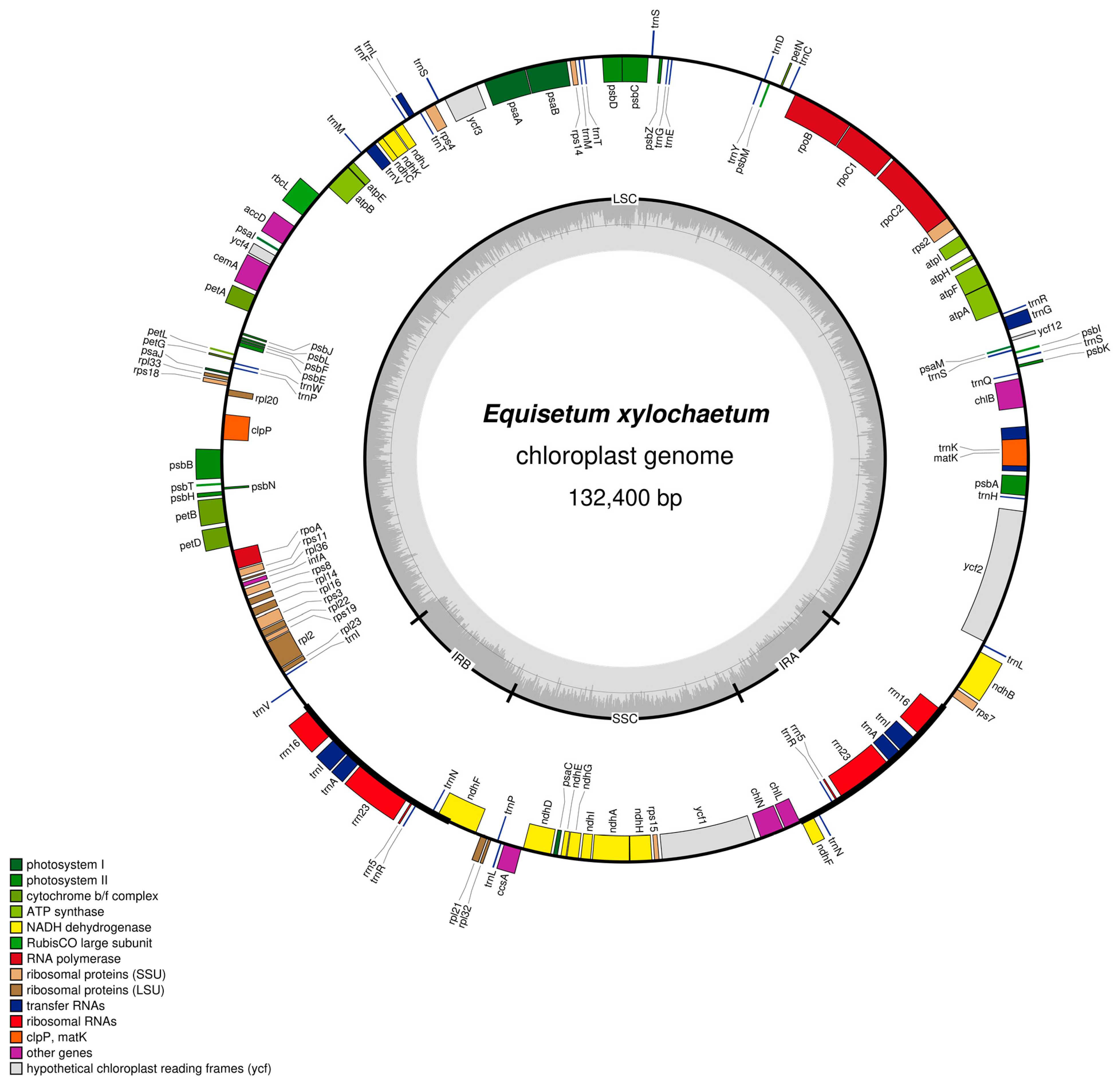

| E. xylochaetum | E. hyemale | E. arvense (US) | E. arvense (Korea) | |
|---|---|---|---|---|
| Accession | MW282958 | KC117177 | GU191334 | JN968380 |
| genome size | 132,400 | 131,760 | 133,309 | 132,726 |
| LSC | 93,902 | 92,580 | 93,542 | 92,961 |
| SSC | 9,726 | 18,994 | 19,469 | 19,477 |
| IRs | 14,386 | 10,093 | 10,149 | 10,144 |
| %GC | 33.9 | 33.7 | 33.4 | 33.4 |
| DNA | Protein | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Gene (# of Intron) | Identical Site (%) | Mean (bp) | SD (bp) | min (bp) | max (bp) | Identical Site (%) | Mean (aa) | SD (aa) | min (aa) | max (aa) |
| 1. | accD | 92.8 | 1021.5 | 81 | 948 | 1143 | 89.9 | 340 | 26.5 | 316 | 380 |
| 2. | atpA | 95.5 | 1539 | 20.8 | 1527 | 1575 | 98 | 512 | 6.9 | 508 | 524 |
| 3. | atpB | 94.4 | 1470 | 0 | 1470 | 1470 | 96.3 | 489 | 0 | 489 | 489 |
| 4. | atpE | 93.4 | 396 | 0 | 396 | 396 | 93.1 | 131 | 0 | 131 | 131 |
| 5. | atpF (1) | 97.5 | 555 | 0 | 555 | 555 | 73.9 | 184 | 0 | 184 | 184 |
| 6. | atpH | 96.3 | 246 | 0 | 246 | 246 | 100 | 81 | 0 | 81 | 81 |
| 7. | atpI | 95.4 | 747 | 0 | 747 | 747 | 99.1 | 248 | 0 | 284 | 248 |
| 8. | ccsA | 93.3 | 943.5 | 1.5 | 942 | 945 | 91.1 | 313.5 | 0.5 | 313 | 314 |
| 9. | cemA | 89.5 | 1438.5 | 21.7 | 1425 | 1476 | 82.7 | 478.5 | 7.2 | 474 | 491 |
| 10. | chlB | 94.3 | 1549.5 | 4.5 | 1545 | 1554 | 93.2 | 515.5 | 1.5 | 514 | 517 |
| 11. | chlL | 91.6 | 879 | 6 | 873 | 885 | 93.9 | 292 | 2 | 290 | 294 |
| 12. | chlN | 92.8 | 1300.5 | 10.5 | 1290 | 1311 | 90.4 | 432.5 | 3.5 | 429 | 436 |
| 13. | clpP (1) | 95.1 | 615 | 0 | 615 | 615 | 98.5 | 204 | 0 | 204 | 204 |
| 14. | infA | 94.7 | 243 | 0 | 243 | 243 | 96.3 | 80 | 0 | 80 | 80 |
| 15. | matK | 88.7 | 1470 | 3 | 1467 | 1473 | 81.4 | 489 | 1 | 488 | 490 |
| 16. | ndhA (1) | 92.7 | 1101.8 | 1.3 | 1101 | 1104 | 91.8 | 366.3 | 0.4 | 366 | 367 |
| 17. | ndhB (1) | 94.8 | 1473 | 0 | 1473 | 1473 | 94.3 | 490 | 0 | 490 | 490 |
| 18. | ndhC | 95.9 | 363 | 0 | 363 | 363 | 98.3 | 120 | 0 | 120 | 120 |
| 19. | ndhD | 95.1 | 1497 | 0 | 1497 | 1497 | 95.2 | 498 | 0 | 498 | 498 |
| 20. | ndhE | 98.3 | 303 | 0 | 303 | 303 | 100 | 100 | 0 | 100 | 100 |
| 21. | ndhF | 92.8 | 2221.5 | 1.5 | 2220 | 2223 | 92.2 | 739.5 | 0.5 | 739 | 740 |
| 22. | ndhG | 91.4 | 606 | 17.2 | 585 | 633 | 85.7 | 201 | 5.7 | 194 | 210 |
| 23. | ndhH | 95.3 | 1182 | 0 | 1182 | 1182 | 97.2 | 393 | 0 | 393 | 393 |
| 24. | ndhI | 97.3 | 549 | 0 | 549 | 549 | 98.4 | 182 | 0 | 182 | 182 |
| 25. | ndhJ | 93.9 | 520.5 | 4.5 | 516 | 525 | 94.8 | 172.5 | 1.5 | 171 | 174 |
| 26. | ndhK | 90.6 | 747.8 | 9.1 | 732 | 753 | 86.8 | 248.3 | 3 | 243 | 250 |
| 27. | petA | 92.4 | 955.5 | 7.5 | 948 | 963 | 93.8 | 317.5 | 2.5 | 315 | 320 |
| 28. | petB (1) | 96 | 648 | 0 | 648 | 648 | 100 | 215 | 0 | 215 | 215 |
| 29. | petD (1) | 96.9 | 483 | 0 | 483 | 483 | 100 | 160 | 0 | 160 | 160 |
| 30. | petG | 97.4 | 114 | 0 | 114 | 114 | 100 | 37 | 0 | 37 | 37 |
| 31. | petL | 93.8 | 96 | 0 | 96 | 96 | 93.5 | 31 | 0 | 31 | 31 |
| 32. | petN | 99 | 96 | 0 | 96 | 96 | 100 | 31 | 0 | 31 | 31 |
| 33. | psaA | 96.2 | 2253 | 0 | 2253 | 2253 | 99.6 | 750 | 0 | 750 | 750 |
| 34. | psaB | 95.7 | 2205 | 0 | 2205 | 2205 | 99.2 | 734 | 0 | 734 | 734 |
| 35. | psaC | 95.1 | 246 | 0 | 246 | 246 | 98.8 | 81 | 0 | 81 | 81 |
| 36. | psaI | 91.9 | 111 | 0 | 111 | 111 | 94.4 | 36 | 0 | 36 | 36 |
| 37. | psaJ | 97.7 | 129 | 0 | 129 | 129 | 100 | 42 | 0 | 42 | 42 |
| 38. | psaM | 96 | 99 | 0 | 99 | 99 | 96.9 | 32 | 0 | 32 | 32 |
| 39. | psbA | 98.1 | 1062 | 0 | 1062 | 1062 | 100 | 353 | 0 | 353 | 353 |
| 40. | psbB | 96.1 | 1527 | 0 | 1527 | 1527 | 99 | 508 | 0 | 508 | 508 |
| 41. | psbC | 95 | 1422 | 0 | 1422 | 1422 | 99.4 | 473 | 0 | 473 | 473 |
| 42. | psbD | 95.6 | 1062 | 0 | 1062 | 1062 | 87.3 | 353 | 0 | 353 | 353 |
| 43. | psbE | 97.2 | 246 | 0 | 246 | 246 | 100 | 81 | 0 | 81 | 81 |
| 44. | psbF | 98.3 | 120 | 0 | 120 | 120 | 100 | 39 | 0 | 39 | 39 |
| 45. | psbH | 94.7 | 225 | 0 | 225 | 225 | 89.2 | 74 | 0 | 74 | 74 |
| 46. | psbI | 97.3 | 111 | 0 | 111 | 111 | 100 | 36 | 0 | 36 | 36 |
| 47. | psbJ | 99.2 | 123 | 0 | 123 | 123 | 100 | 40 | 0 | 40 | 40 |
| 48. | psbK | 97 | 168 | 0 | 168 | 168 | 96.4 | 55 | 0 | 55 | 55 |
| 49. | psbL | 98.3 | 117 | 0 | 117 | 117 | 100 | 38 | 0 | 38 | 38 |
| 50. | psbM | 98.2 | 111 | 0 | 111 | 111 | 94.4 | 36 | 0 | 36 | 36 |
| 51. | psbN | 95.5 | 132 | 0 | 132 | 132 | 93 | 43 | 0 | 43 | 43 |
| 52. | psbT | 97.4 | 112.5 | 1.5 | 111 | 114 | 97.3 | 36.5 | 0.5 | 36 | 37 |
| 53. | psbZ | 94.2 | 189 | 0 | 189 | 189 | 100 | 62 | 0 | 62 | 62 |
| 54. | rbcL | 96.1 | 1428 | 0 | 1428 | 1428 | 99.2 | 475 | 0 | 475 | 475 |
| 55. | rpl14 | 97.6 | 369 | 0 | 369 | 369 | 99.2 | 122 | 0 | 122 | 122 |
| 56. | rpl16 (1 in E. arvense) | 93.1 | 423 | 0 | 423 | 423 | 95 | 140 | 0 | 140 | 140 |
| 57. | rpl2 (1) | 94.3 | 834.8 | 1.3 | 834 | 837 | 95.3 | 277.3 | 0.4 | 277 | 278 |
| 58. | rpl20 | 89.7 | 347.3 | 1.3 | 345 | 348 | 85.2 | 114.8 | 0.4 | 114 | 115 |
| 59. | rpl21 | 91.3 | 364.5 | 1.5 | 363 | 366 | 86 | 120.5 | 0.5 | 120 | 121 |
| 60. | rpl22 | 94.1 | 372 | 0 | 372 | 372 | 96.6 | 123 | 0 | 123 | 123 |
| 61. | rpl23 | 94.9 | 273 | 0 | 273 | 273 | 93.3 | 90 | 0 | 90 | 90 |
| 62. | rpl32 | 95.3 | 171 | 0 | 171 | 171 | 98.2 | 56 | 0 | 56 | 56 |
| 63. | rpl33 | 95.5 | 201 | 0 | 201 | 201 | 90.9 | 66 | 0 | 66 | 66 |
| 64. | rpl36 | 93.9 | 114 | 0 | 114 | 114 | 100 | 37 | 0 | 37 | 37 |
| 65. | rpoA | 93.5 | 1.18.5 | 4.5 | 1014 | 1023 | 93.5 | 338.5 | 1.5 | 337 | 340 |
| 66. | rpoB | 93.9 | 3235.5 | 33.8 | 3216 | 3294 | 92.9 | 1.00.5 | 11.3 | 1071 | 1097 |
| 67. | rpoC1 (1) | 93.3 | 2060.3 | 3.9 | 2058 | 2067 | 91.3 | 685.8 | 1.3 | 685 | 688 |
| 68. | rpoC2 | 92.3 | 4143 | 21 | 4122 | 4164 | 87.2 | 1380 | 7 | 1373 | 1387 |
| 69. | rps11 | 94.7 | 396 | 0 | 396 | 396 | 95.4 | 131 | 0 | 131 | 131 |
| 70. | rps12 | 98.1 | 372 | 0 | 372 | 372 | 100 | 123 | 0 | 123 | 123 |
| 71. | rps14 | 92.2 | 306 | 0 | 306 | 306 | 93.1 | 101 | 0 | 101 | 101 |
| 72. | rps15 | 95.2 | 270 | 0 | 270 | 270 | 92.1 | 89 | 0 | 89 | 89 |
| 73. | rps18 | 96.5 | 228 | 0 | 228 | 228 | 98.7 | 75 | 0 | 75 | 75 |
| 74. | rps19 | 95.7 | 279 | 0 | 279 | 279 | 98.9 | 92 | 0 | 92 | 92 |
| 75. | rps2 | 95.2 | 708 | 0 | 708 | 708 | 97 | 235 | 0 | 235 | 235 |
| 76. | rps3 | 95.4 | 657 | 0 | 657 | 657 | 96.3 | 218 | 0 | 218 | 218 |
| 77. | rps4 | 94.1 | 624 | 0 | 624 | 624 | 92.3 | 207 | 0 | 207 | 207 |
| 78. | rps7 | 94.9 | 468 | 0 | 468 | 468 | 94.8 | 155 | 0 | 155 | 155 |
| 79. | rps8 | 95.7 | 399 | 0 | 399 | 399 | 95.5 | 132 | 0 | 132 | 132 |
| No. | Name | GenBank Accession | Locality | References |
|---|---|---|---|---|
| atpB | ||||
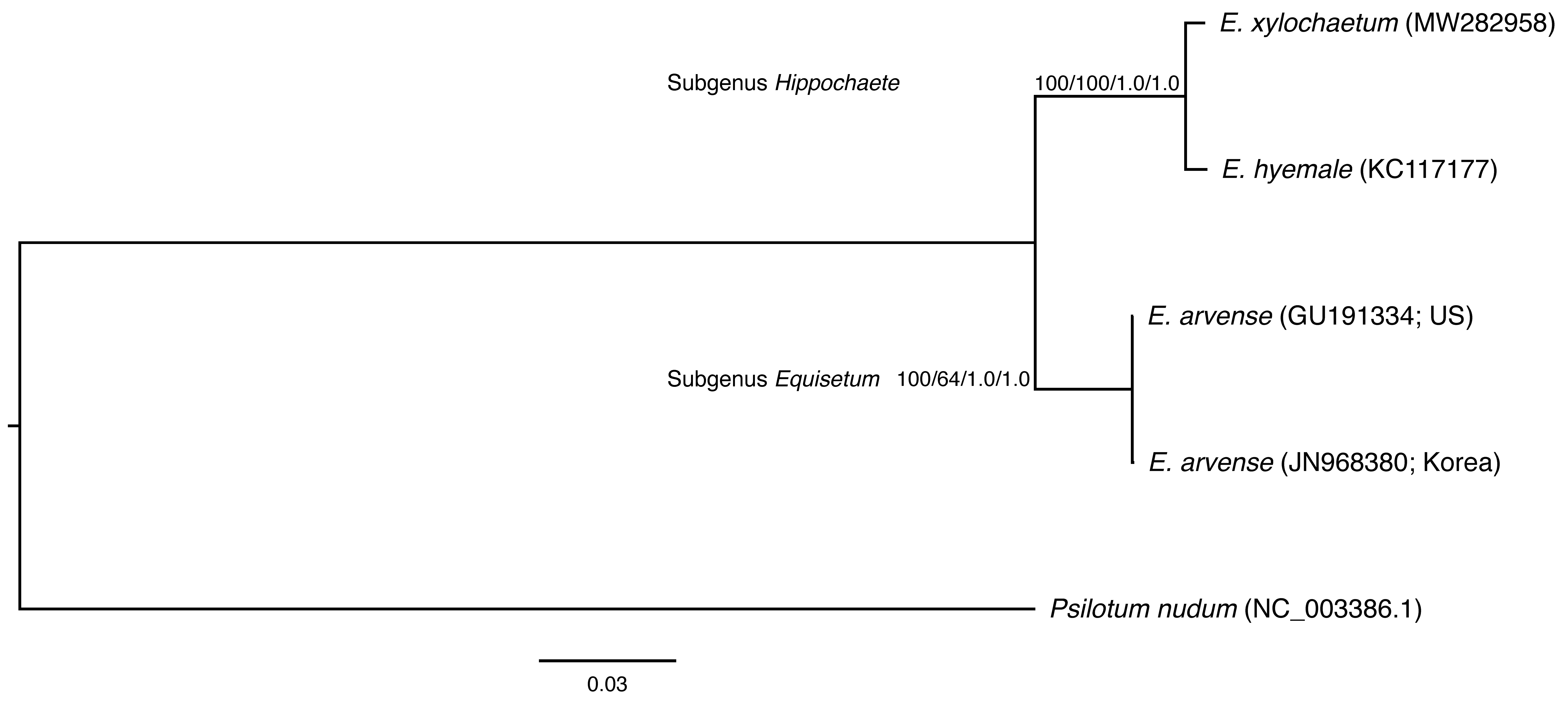
| 1. | E. arvense | GU191334 | USA | [8] |
| 2. | E. arvense | JN968380 | Korea | [9] |
| 3. | E. hyemale | KC117177 | unknown | [7] |
| 4. | E. ramosissimum subsp. debile | EU439074 | unknown | [23] |
| 5. | E. telmateia | AF313542 | unknown | [24] |
| 6. | E. xylochaetum | MW282958 | Chile | This study |
| 7. | Equisetum x ferrissii | AF313541 | unknown | [24] |
| matK | ||||
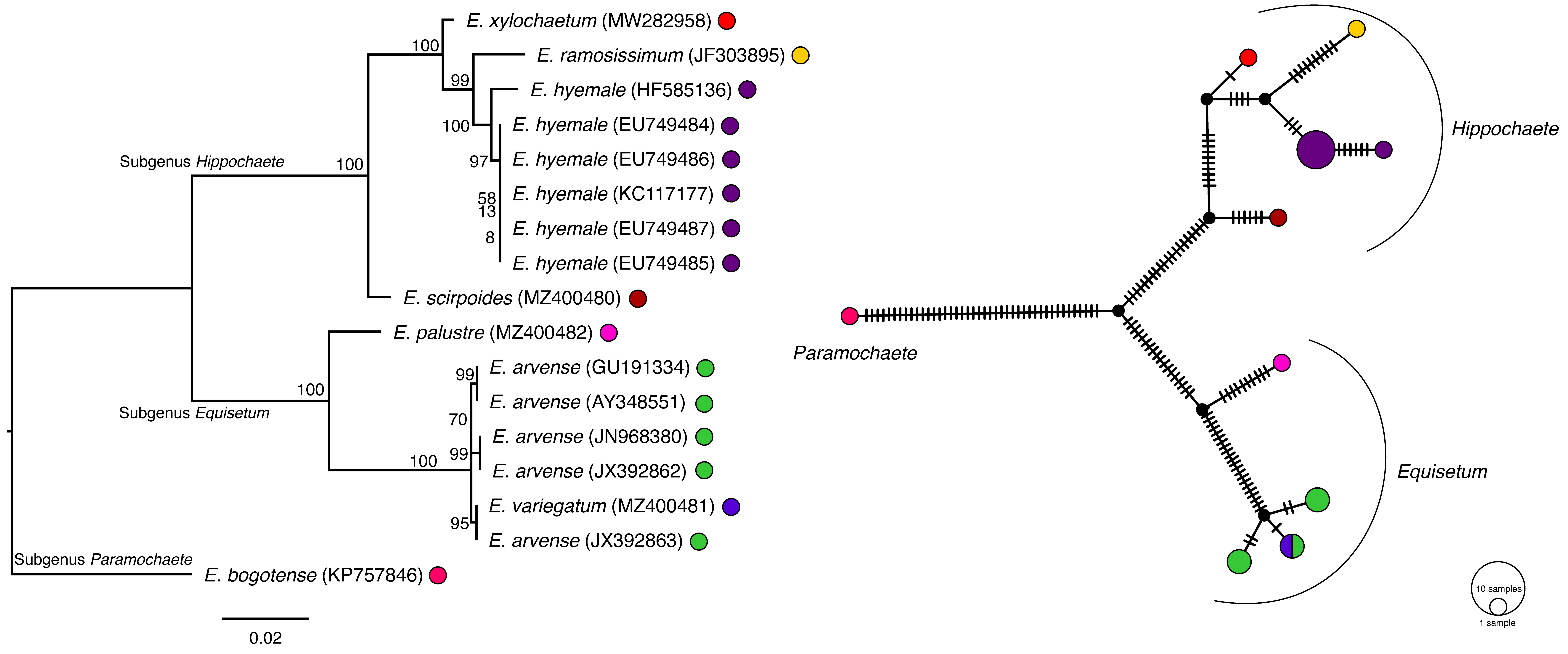
| 1. | E. arvense | JX392862 | China | [25] |
| 2. | E. arvense | JX392863 | Europe | [25] |
| 3. | E. arvense | AY348551 | unknown | [26] |
| 4. | E. arvense | GU191334 | USA | [8] |
| 5. | E. arvense | JN968380 | Korea | [9] |
| 6. | E. bogotense | KP757846 | unknown | [27] |
| 7. | E. hyemale | EU749486 | unknown | [28] |
| 8. | E. hyemale | EU749485 | unknown | [28] |
| 9. | E. hyemale | EU749484 | unknown | [28] |
| 10. | E. hyemale | EU749487 | unknown | [28] |
| 11. | E. hyemale | HF585136 | unknown | [29] |
| 12. | E. hyemale | KC117177 | unknown | [7] |
| 13. | E. palustre | MZ400482 | Sweden | [30] |
| 14. | E. ramosissimum | JF303895 | unknown | [31] |
| 15. | E. scirpoides | MZ400480 | Sweden | [30] |
| 16. | E. variegatum | MZ400481 | Sweden | [30] |
| 17. | E. xylochaetum | MW282958 | Chile | This study |
| rpoB | ||||
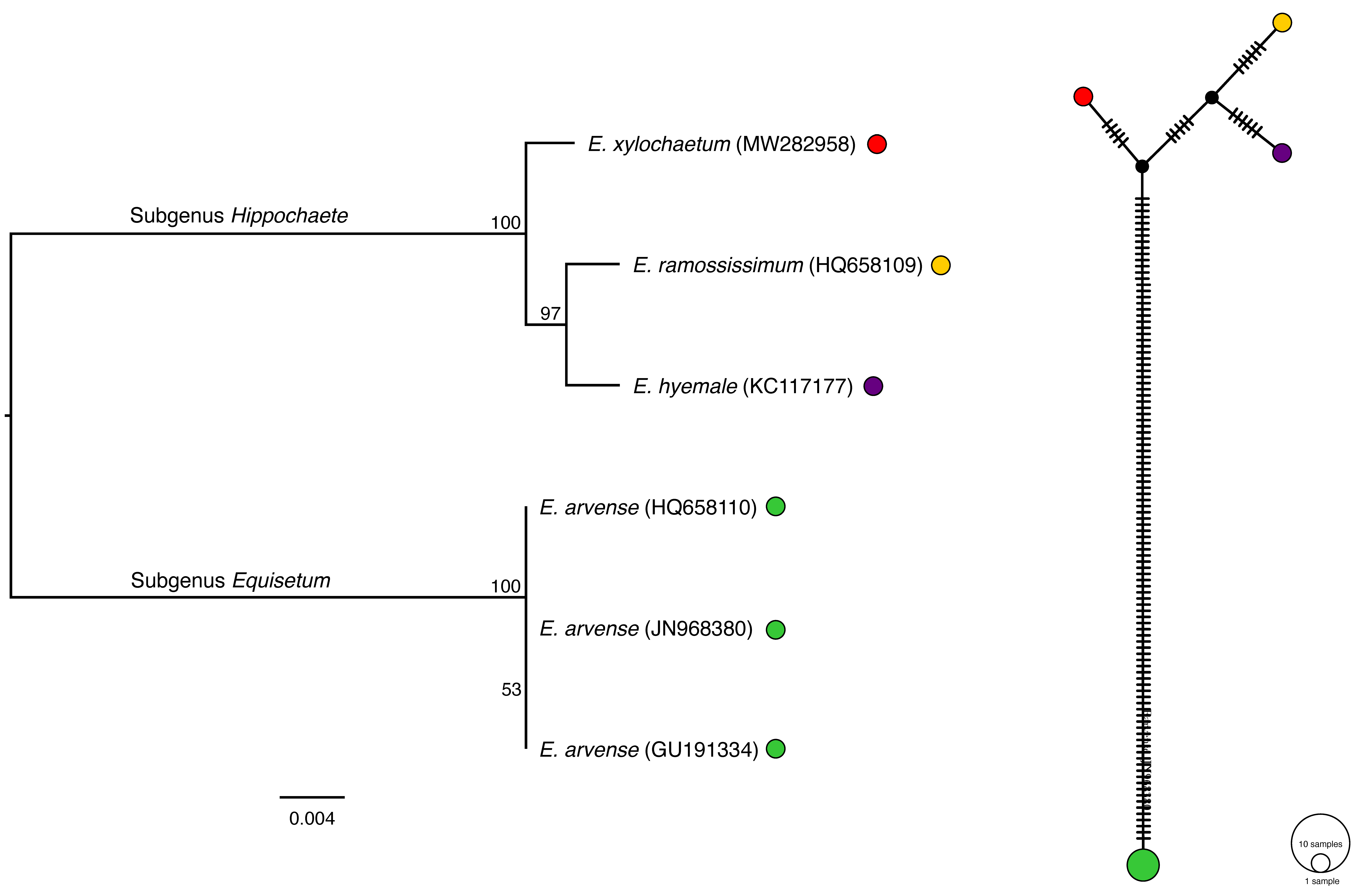
| 1. | E. arvense | HQ658110 | China | [32] |
| 2. | E. arvense | GU191334 | USA | [8] |
| 3. | E. arvense | JN968380 | Korea | [9] |
| 4. | E. hyemale | KC117177 | Unknown | [7] |
| 5. | E. ramossissimum | HQ658109 | China | [32] |
| 6. | E. xylochaetum | MW282958 | Chile | This study |
| rps4 | ||||
| 1. | E. arvense subsp. arvense isolate 41072 | MH750111 | Finland | [5] |
| 2. | E. arvense | AJ583677 | unknown | [3] |
| 3. | E. arvense | JN968380 | Korea | [9] |
| 4. | E. arvense | GU191334 | USA | [8] |
| 5. | E. arvense subsp. arvense isolate 26084 | MH750108 | India (Himachal Pradesh) | [5] |
| 6. | E. arvense subsp. arvense isolate 40833 | MH750109 | USA (California) | [5] |
| 7. | E. arvense subsp. arvense isolate 41071 | MH750110 | Finland | [5] |
| 8. | E. arvense subsp. boreale isolate 41073 | MH750112 | Finland/Norway (border) | [5] |
| 9. | E. arvense subsp. boreale isolate 41074 | MH750113 | Finland/Norway (border) | [5] |
| 10. | E. arvense x E. telmateia subsp. braunii isolate 40834 | MH750114 | USA (California) | [5] |
| 11. | E. bogotense | AF231898 | unknown | [33] |
| 12. | E. bogotense | AF313603 | unknown | [24] |
| 13. | E. bogotense | AJ583678 | unknown | [3] |
| 14. | E. bogotense isolate 40800 | MH750115 | Argentina | [5] |
| 15. | E. bogotense isolate 40802 | MH750116 | Ecuador | [5] |
| 16. | E. bogotense isolate 40827 | MH750117 | Colombia | [5] |
| 17. | E. diffusum | AJ583679 | unknown | [3] |
| 18. | E. diffusum isolate 40804 | MH750118 | India | [5] |
| 19. | E. fluviatile | AJ583680 | unknown | [3] |
| 20. | E. fluviatile isolate 41075 | MH750119 | Finland | [5] |
| 21. | E. fluviatile isolate 41076 | MH750120 | Finland | [5] |
| 22. | E. giganteum | AJ583681 | unknown | [3] |
| 23. | E. giganteum isolate 40806 | MH750121 | Chile | [5] |
| 24. | E. hyemale | AJ583682 | unknown | [3] |
| 25. | E. hyemale | KC117177 | unknown | [7] |
| 26. | E. hyemale isolate 23252 | MH750123 | Norway | [5] |
| 27. | E. laevigatum | AJ583683 | unknown | [3] |
| 28. | E. laevigatum isolate 40812 | MH750125 | USA (California) | [5] |
| 29. | E. myriochaetum | AJ583684 | unknown | [3] |
| 30. | E. myriochaetum isolate 40825 | MH750126 | Mexico | [5] |
| 31. | E. myriochaetum isolate 40936 | MH750127 | El Salvador | [5] |
| 32. | E. palustre | AJ583685 | unknown | [3] |
| 33. | E. palustre isolate 17671 | MH750128 | UK (England, Norfolk) | [5] |
| 34. | E. palustre isolate 39349 | MH750129 | UK (England, Surrey) | [5] |
| 35. | E. praealtum isolate 41501 | MH750122 | USA (Ohio) | [5] |
| 36. | E. pratense | AJ583686 | unknown | [3] |
| 37. | E. pratense isolate 39348 | MH750130 | Finland | [5] |
| 38. | E. ramosissimum subsp. debile | AJ583687 | unknown | [3] |
| 39. | E. ramosissimum subsp. debile | EU439173 | unknown | [23] |
| 40. | E. ramosissimum subsp. debile isolate 24579 | MH750131 | Sri Lanka | [5] |
| 41. | E. ramosissimum subsp. debile isolate 40837 | MH750132 | New Caledonia | [5] |
| 42. | E. ramosissimum subsp. ramosissimum isolate 36802 | MH750133 | Spain (Andalucia) | [5] |
| 43. | E. scirpoides | AJ583688 | unknown | [3] |
| 44. | E. scirpoides isolate 26090 | MH750134 | Greenland | [5] |
| 45. | E. scirpoides isolate 40830 | MH750124 | Russia (Kamtschatka) | [5] |
| 46. | E. sylvaticum | AJ583689 | unknown | [3] |
| 47. | E. telmateia subsp. braunii | AJ583690 | unknown | [3] |
| 48. | E. telmateia subsp. braunii isolate 40817 | MH750136 | USA (California) | [5] |
| 49. | E. telmateia subsp. braunii isolate 40828 | MH750137 | Canada (British Columbia) | [5] |
| 50. | E. telmateia subsp. braunii isolate 40832 | MH750138 | USA (California) | [5] |
| 51. | E. telmateia subsp. braunii isolate 40836 | MH750139 | USA (California) | [5] |
| 52. | E. telmateia subsp. telmateia isolate 41082 | MH750140 | Ireland | [5] |
| 53. | E. variegatum | AJ583691 | unknown | [3] |
| 54. | E. variegatum isolate 11639 | MH750141 | UK (Wales) | [5] |
| 55. | E. variegatum isolate 40819 | MH750148 | Ireland | [5] |
| 56. | E. variegatum isolate 40820 | MH750142 | France (Pyrenees) | [5] |
| 57. | E. variegatum isolate 40823 | MH750143 | USA (Keweenaw, Michigan) | [5] |
| 58. | E. variegatum isolate 41083 | MH750144 | Ireland | [5] |
| 59. | E. variegatum subsp. alaskanum isolate 40818 | MH750145 | USA (Alaska) | [5] |
| 60. | E. variegatum subsp. alaskanum isolate 40821 | MH750146 | Canada (British Columbia) | [5] |
| 61. | E. variegatum subsp. alaskanum isolate 40822 | MH750147 | Canada (Banff) | [5] |
| 62. | E. xylochaetum | MW282958 | Chile | This study |
| 63. | Equisetum scirpoides isolate 41089 | MH750135 | Finland | [5] |
| 64. | Equisetum x ferrissii | AF313590 | unknown | [24] |
| 65. | Equisetum x fontqueri isolate 26093 (E. telmateia x E. palustre) | MH750149 | UK (Scotland) | [5] |
| 66. | Equisetum x litorale isolate 41084 (E. arvense x E. fluviatile) | MH750150 | Ireland | [5] |
| 67. | Equisetum x litorale isolate 41085 (E. arvense x E. fluviatile) | MH750151 | Ireland | [5] |
| 68. | Equisetum x schaffneri isolate 40813 (E. giganteum x E. myriochaetum) | MH750152 | Mexico | [5] |
| 69. | Equisetum x schaffneri isolate 40814 (E. myriochaetum x E. giganteum) | MH750153 | Peru (cult RBG Edinburgh) | [5] |
| 70. | Equisetum x schaffneri isolate 40824 (E. giganteum x E. myriochaetum) | MH750154 | Mexico | [5] |
| trnL-trnF | ||||
| 1. | E. arvense | JN968380 | Korea | [9] |
| 2. | E. arvense | GU191334 | USA | [8] |
| 3. | E. arvense | AY226125 | Franc | [34] |
| 4. | E. arvense | GQ428069 | unknown | [35] |
| 5. | E. arvense | HM590277 | Estonia | [36] |
| 6. | E. arvense | GQ244921 | unknown | [37] |
| 7. | E. arvense subsp boreale isolate 41074 | MH750043 | Finland/Norway | [5] |
| 8. | E. arvense subsp. arvense isolate 26084 | MH750038 | India | [5] |
| 9. | E. arvense subsp. arvense isolate 26085 | MH750039 | UK | [5] |
| 10. | E. arvense subsp. arvense isolate 40833 | MH750040 | USA | [5] |
| 11. | E. arvense subsp. arvense isolate 41071 | MH750041 | Finland | [5] |
| 12. | E. arvense subsp. boreale isolate 41073 | MH750042 | Finland/Norway | [5] |
| 13. | E. arvense x E. telmateia subsp. braunii isolate 40834 | MH750044 | USA | [5] |
| 14. | E. bogotense | AY226124 | Colombia | [34] |
| 15. | E. bogotense isolate 40800 | MH750045 | Argentina | [5] |
| 16. | E. bogotense isolate 40801 | MH750046 | Chile | [5] |
| 17. | E. bogotense isolate 40802 | MH750047 | Ecuador | [5] |
| 18. | E. bogotense isolate 40803 | MH750048 | Chile | [5] |
| 19. | E. bogotense isolate 40827 | MH750049 | Colombia | [5] |
| 20. | E. diffusum | AY226126 | India | [34] |
| 21. | E. diffusum isolate 40804 | MH750050 | India | [5] |
| 22. | E. fluviatile | AY226121 | Canada | [34] |
| 23. | E. fluviatile | GQ244922 | unknown | [37] |
| 24. | E. fluviatile isolate 41075 | MH750051 | Finland | [5] |
| 25. | E. fluviatile isolate 41076 | MH750052 | Finland | [5] |
| 26. | E. giganteum | AY226118 | Ecuador | [34] |
| 27. | E. giganteum isolate 40805 | MH750053 | Jamaica | [5] |
| 28. | E. giganteum isolate 40806 | MH750054 | Chile | [5] |
| 29. | E. giganteum isolate 40807 | MH750055 | Peru | [5] |
| 30. | E. giganteum isolate 40810 | MH750057 | Argentina | [5] |
| 31. | E. giganteum isolate 40811 | MH750058 | Argentina | [5] |
| 32. | E. hyemale | KC117177 | unknown | [7] |
| 33. | E. hyemale | AY327837 | unknown | [34] |
| 34. | E. hyemale isolate 0796g | GQ244923 | unknown | [37] |
| 35. | E. hyemale isolate 1273o | GQ244924 | unknown | [37] |
| 36. | E. hyemale isolate 20201 | MH750061 | France | [5] |
| 37. | E. hyemale isolate 23252 | MH750062 | Norway | [5] |
| 38. | E. hyemale isolate 41088 | MH750063 | Finland | [5] |
| 39. | E. hyemale subsp. affine | AY226110 | USA | [34] |
| 40. | E. iganteum isolate 40809 | MH750056 | Argentina | [5] |
| 41. | E. laevigatum | AY226112 | USA | [34] |
| 42. | E. laevigatum isolate 40812 | MH750065 | USA | [5] |
| 43. | E. myriochaetum | AY226114 | USA | [34] |
| 44. | E. myriochaetum isolate 40815 | MH750066 | USA | [5] |
| 45. | E. myriochaetum isolate 40816 | MH750067 | USA | [5] |
| 46. | E. myriochaetum isolate 40825 | MH750068 | Mexico | [5] |
| 47. | E. myriochaetum isolate 40826 | MH750069 | Ecuador | [5] |
| 48. | E. myriochaetum isolate 40936 | MH750070 | El Savador | [5] |
| 49. | E. myriochaetum isolate 41080 | MH750071 | Guatemala | [5] |
| 50. | E. palustre | AY226123 | Canada | [34] |
| 51. | E. palustre | GQ244925 | unknown | [37] |
| 52. | E. palustre isolate 39349 | MH750072 | UK | [5] |
| 53. | E. praealtum isolate 40831 | MH750059 | USA | [5] |
| 54. | E. praealtum isolate 41501 | MH750060 | USA | [5] |
| 55. | E. pratense | AY226122 | Canada | [34] |
| 56. | E. pratense | GQ244926 | unknown | [37] |
| 57. | E. pratense | HM590278 | Estonia | [36] |
| 58. | E. pratense isolate 39348 | MH750073 | Finland | [5] |
| 59. | E. pratense isolate 41086 | MH750074 | Finland | [5] |
| 60. | E. pratense isolate 41087 | MH750075 | Finland | [5] |
| 61. | E. ramosissimum subsp. debile | AY226115 | Taiwan | [34] |
| 62. | E. ramosissimum subsp. debile isolate 23679 | MH750076 | Reunion | [5] |
| 63. | E. ramosissimum subsp. debile isolate 24579 | MH750077 | Sri Lanka | [5] |
| 64. | E. ramosissimum subsp. debile isolate 40837 | MH750078 | New Caledonia | [5] |
| 65. | E. ramosissimum subsp. ramosissimum isolate 36802 | MH750079 | Spain | [5] |
| 66. | E. ramosissimum subsp. ramosissimum isolate 40829 | MH750080 | Turkey | [5] |
| 67. | E. scirpoides | AY226116 | Canada | [34] |
| 68. | E. scirpoides | GQ244927 | unknown | [37] |
| 69. | E. scirpoides isolate 26090 | MH750082 | Greenland | [5] |
| 70. | E. scirpoides isolate 40830 | MH750064 | Russia | [5] |
| 71. | E. scirpoides isolate10933 | MH750081 | UK | [5] |
| 72. | E. sylvaticum | MH750083 | UK | [5] |
| 73. | E. sylvaticum | AY226120 | France | [34] |
| 74. | E. sylvaticum | GQ244928 | unknown | [37] |
| 75. | E. sylvaticum isolate 41081 | MH750084 | Finland | [5] |
| 76. | E. telmateia isolate 11642 | MH750089 | China | [5] |
| 77. | E. telmateia isolate 41082 | MH750090 | Ireland | [5] |
| 78. | E. telmateia subsp. braunii | AY226119 | USA | [34] |
| 79. | E. telmateia subsp. braunii isolate 40817 | MH750085 | USA | [5] |
| 80. | E. telmateia subsp. braunii isolate 40828 | MH750086 | Canada | [5] |
| 81. | E. telmateia subsp. braunii isolate 40832 | MH750087 | USA | [5] |
| 82. | E. telmateia subsp. braunii isolate 40836 | MH750088 | USA | [5] |
| 83. | E. trachyodon isolate 41092 | MH750106 | Finland | [5] |
| 84. | E. variegatum | AY226117 | USA | [34] |
| 85. | E. variegatum isolate 0584g | GQ244929 | unknown | [37] |
| 86. | E. variegatum isolate 0977o | GQ244930 | unknown | [37] |
| 87. | E. variegatum isolate 11639 | MH750091 | UK | [5] |
| 88. | E. variegatum isolate 40819 | MH750098 | Ireland | [5] |
| 89. | E. variegatum isolate 40820 | MH750092 | France | [5] |
| 90. | E. variegatum isolate 40823 | MH750093 | USA | [5] |
| 91. | E. variegatum isolate 41083 | MH750094 | Ireland | [5] |
| 92. | E. variegatum subsp. alaskanum isolate 40818 | MH750095 | USA | [5] |
| 93. | E. variegatum subsp. alaskanum isolate 40821 | MH750096 | Canada | [5] |
| 94. | E. variegatum subsp. alaskanum isolate 40822 | MH750097 | Canada | [5] |
| 95. | E. xylochaetum | MW282958 | Chile | This study |
| 96. | E. xylochaetum isolate 40614 | MH750107 | Chile | [5] |
| 97. | Equisetum sp. | AY327838 | unknown | [34] |
| 98. | Equisetum x dycei isolate 26083 | MH750099 | UK | [5] |
| 99. | Equisetum x ferrissii (E. laevigatum x E. hyemale) | AY226113 | USA | [34] |
| 100. | Equisetum x ferrissii (Equisetum hyemale x laevigatum) | AY226111 | Canada | [34] |
| 101. | Equisetum x litorale isolate 41084 | MH750101 | Ireland | [5] |
| 102. | Equisetum x litorale isolate 41085 | MH750102 | Ireland | [5] |
| 103. | Equisetum x schaffneri isolate 40813 | MH750103 | Mexico | [5] |
| 104. | Equisetum x schaffneri isolate 40814 | MH750104 | Peru | [5] |
| 105. | Equisetum x schaffneri isolate 40824 | MH750105 | Mexico | [5] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satjarak, A.; Graham, L.E.; Trest, M.T.; Arancibia-Avila, P. Plastid Genome of Equisetum xylochaetum from the Atacama Desert, Chile and the Relationships of Equisetum Based on Frequently Used Plastid Genes and Network Analysis. Plants 2022, 11, 1001. https://doi.org/10.3390/plants11071001
Satjarak A, Graham LE, Trest MT, Arancibia-Avila P. Plastid Genome of Equisetum xylochaetum from the Atacama Desert, Chile and the Relationships of Equisetum Based on Frequently Used Plastid Genes and Network Analysis. Plants. 2022; 11(7):1001. https://doi.org/10.3390/plants11071001
Chicago/Turabian StyleSatjarak, Anchittha, Linda E. Graham, Marie T. Trest, and Patricia Arancibia-Avila. 2022. "Plastid Genome of Equisetum xylochaetum from the Atacama Desert, Chile and the Relationships of Equisetum Based on Frequently Used Plastid Genes and Network Analysis" Plants 11, no. 7: 1001. https://doi.org/10.3390/plants11071001
APA StyleSatjarak, A., Graham, L. E., Trest, M. T., & Arancibia-Avila, P. (2022). Plastid Genome of Equisetum xylochaetum from the Atacama Desert, Chile and the Relationships of Equisetum Based on Frequently Used Plastid Genes and Network Analysis. Plants, 11(7), 1001. https://doi.org/10.3390/plants11071001







