The Role of Plant Growth Promoting Rhizosphere Microbiome as Alternative Biofertilizer in Boosting Solanum melongena L. Adaptation to Salinity Stress
Abstract
:1. Introduction
2. Results
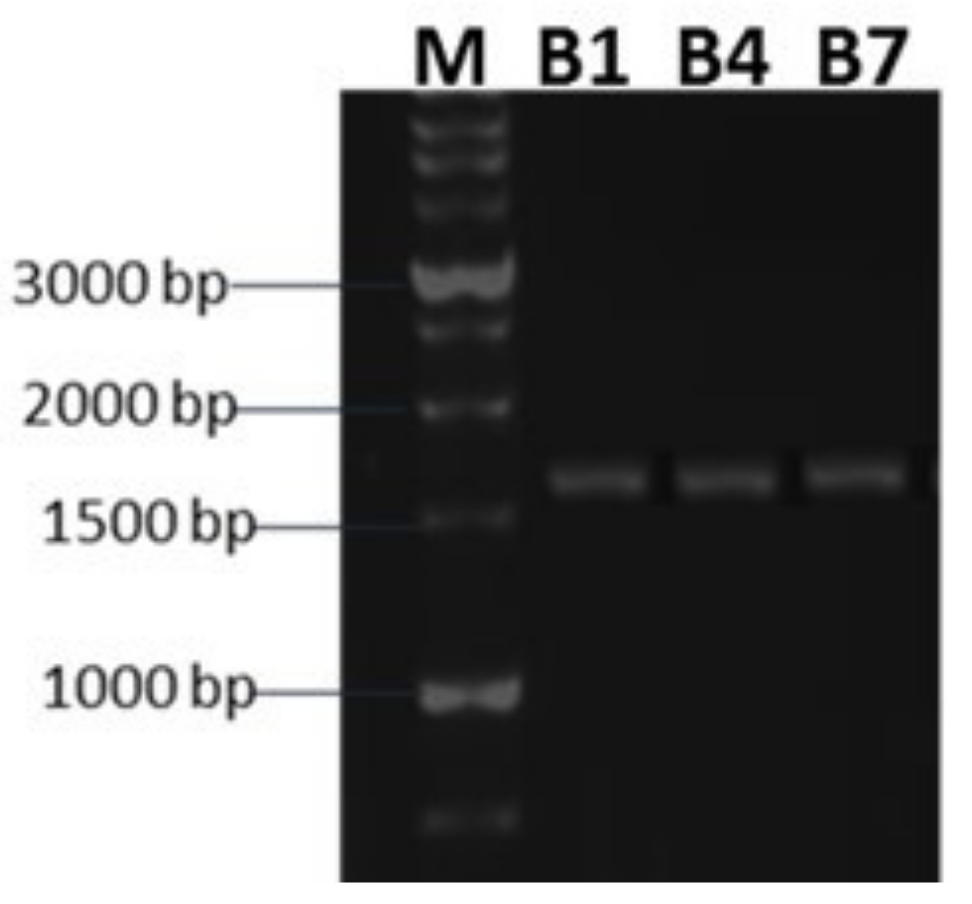
2.1. Molecular Identification and Phylogeny of Bacterial Isolates in the Selected Bioinoculum
2.2. Solanum melongena L. Salt Tolerance Indices Percentage as Affected by Salt Stress and/or Inoculum Treatments
2.3. Solanum melongena L. Leaves Photosynthetic Pigments and Chlorophyll Fluorescence as Affected by Salt Stress and/or Inoculum Treatments
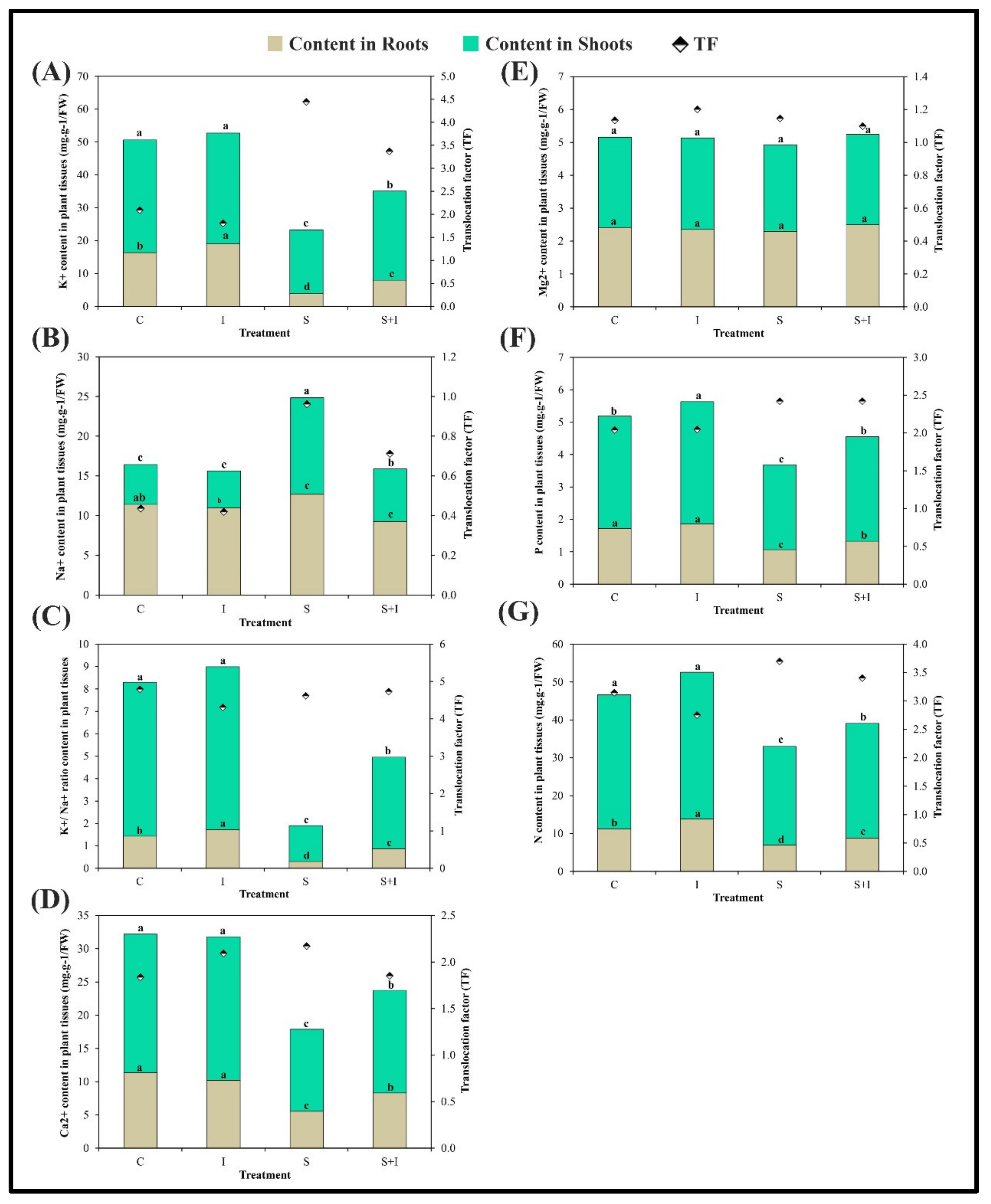
2.4. Element Homeostasis in Solanum melongena L. as Affected by Salt Stress and/or Inoculum Treatments
2.5. Hormonal Status in Salt-Stressed Solanum melongena L. as Affected by Salt Stress and/or Inoculum Treatments
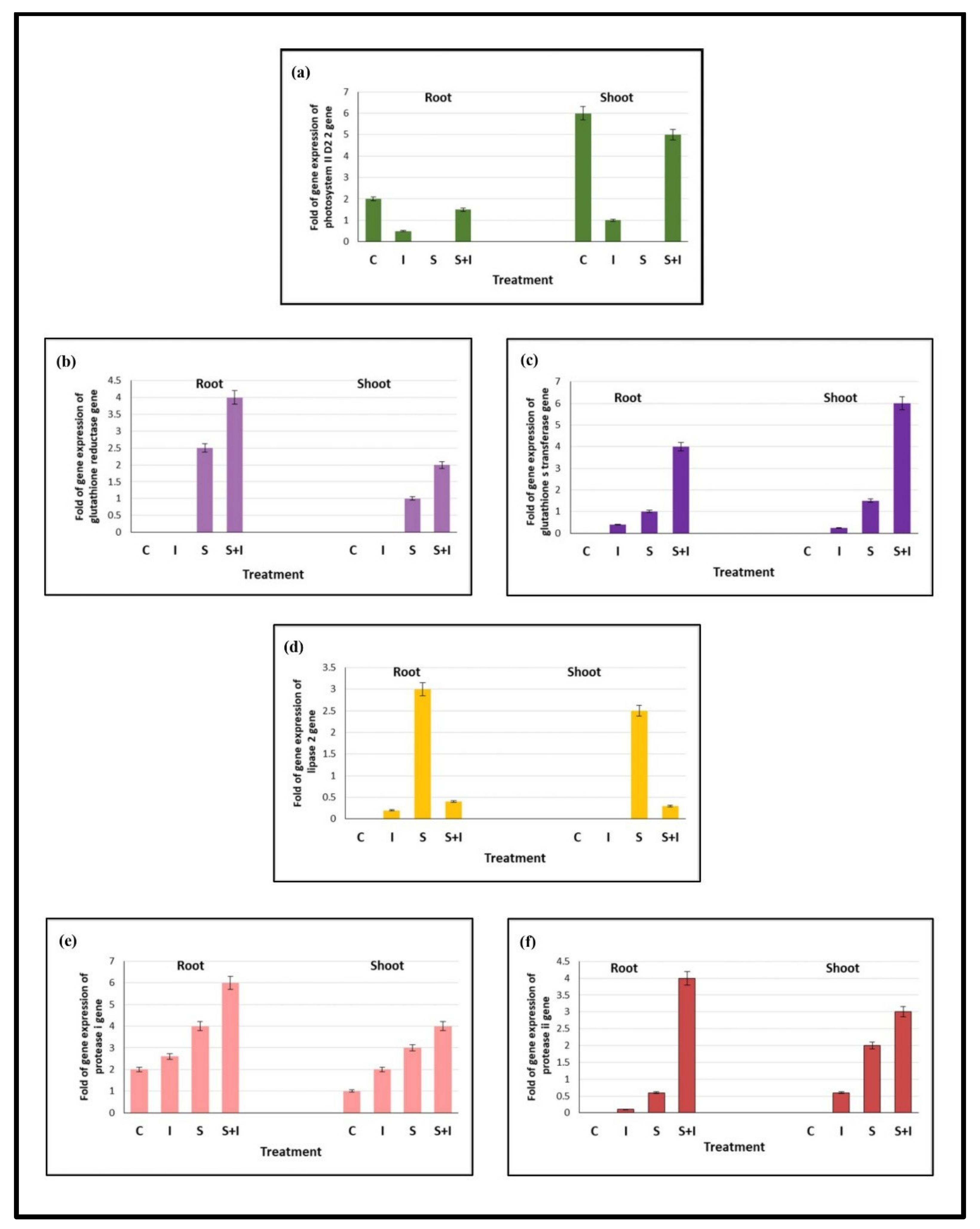
2.6. Selected Inoculum Modulates Some Genes Related to Defense System of Salt-Stressed Solanum melongena L. Roots and Shoots
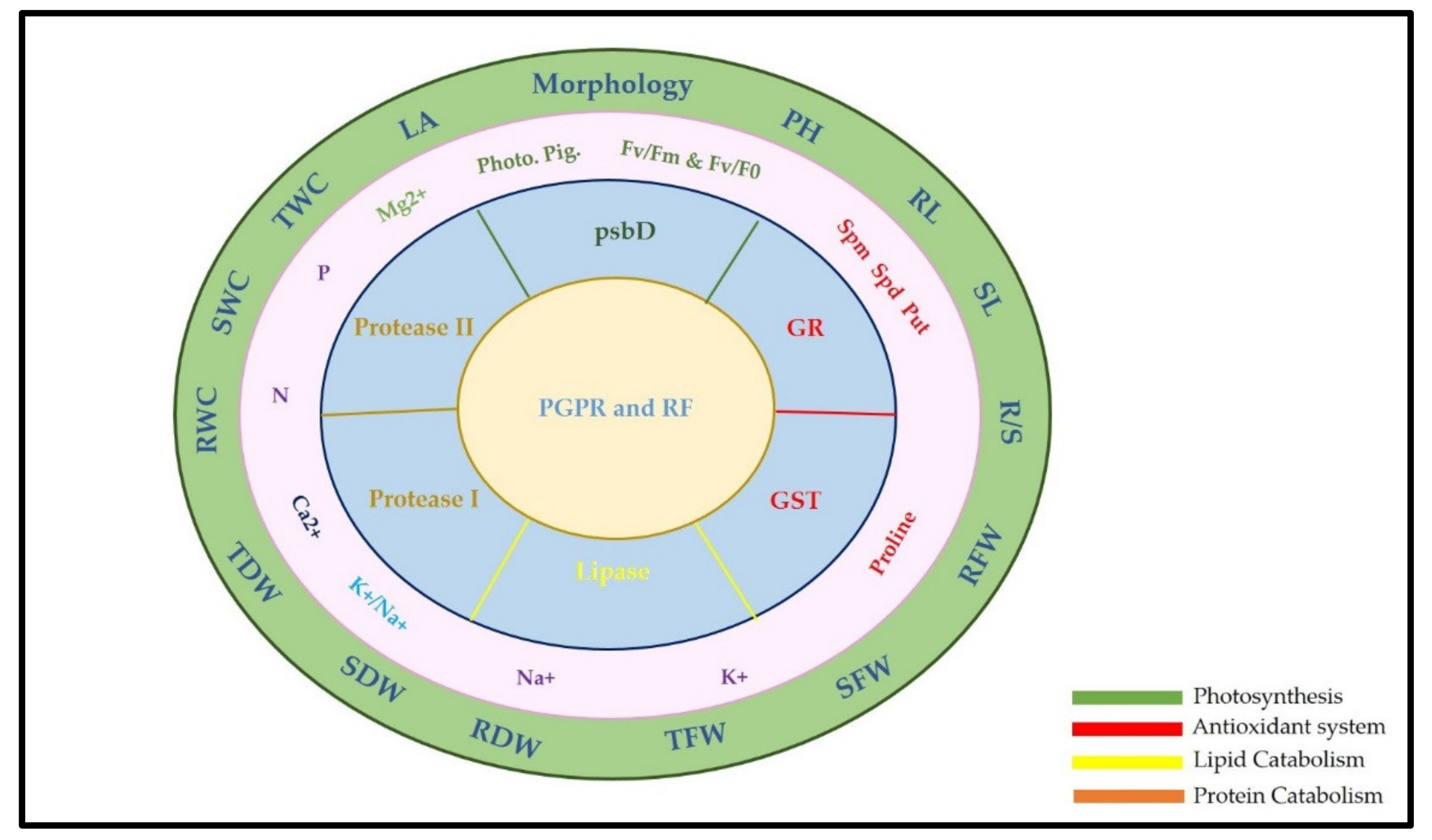
3. Discussion
4. Materials and Methods
4.1. Experimental Materials and Growth Conditions
4.2. Isolation and Identification of Plant Growth Promoting Rhizobacteria and Rhizofungi
4.2.1. Differentiation between Bacterial Isolates
4.2.2. Molecular Identification and Phylogeny of Bacterial Isolates
4.2.3. Identification of Fungal Strains
4.3. Characterization of the Selected Plant Growth Promoting Bacteria and Rhizofungi
4.4. Experimental Design
4.5. Measurement of Growth Traits
4.6. Extraction and Estimation of Chlorophyll and Carotenoid Contents
4.7. Measurement of Chlorophyll Fluorescence
4.8. Elements Analysis
4.9. Polyamines Detection
4.10. Differential-Display Reverse Transcription-PCR (DDRT-PCR) and Semi Quantitative Gene Expression
4.10.1. RNA Extraction
4.10.2. RAPD-PCR
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Umesha, S.; Manukumar, H.M.G.; Chandrasekhar, B. Sustainable Agriculture and Food Security. In Biotechnology for Sustainable Agriculture; Singh, R.L., Mondal, S., Eds.; Elsevier Inc.: Gurgaon, India, 2018; pp. 67–92. ISBN 978-0-12-812160-3. [Google Scholar] [CrossRef]
- Kumar, M.; Giri, V.P.; Pandey, S.; Gupta, A.; Patel, M.K.; Bajpai, A.B.; Jenkins, S.; Siddique, K.H.M. Plant-Growth-Promoting Rhizobacteria Emerging as an Effective Bioinoculant to Improve the Growth, Production, and Stress Tolerance of Vegetable Crops. Int. J. Mol. Sci. 2021, 22, 12245. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef] [PubMed]
- Hashem, A.; Tabassum, B.; Fathi Abd Allah, E. Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 2019, 26, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Riaz, U.; Murtaza, D.G.; Anum, W.; Samreen, T.; Sarfraz, M. Plant Growth-Promoting Rhizobacteria (PGPR) as Biofertilizers and Biopesticides. In Microbiota and Biofertilizers, 1st ed.; Hakeem, K.R., Dar, G.H., Mehmood, M.A., Bhat, R.A., Eds.; Springer Nature: Cham, Switzerland, 2020; pp. 181–196. ISBN 978-3-030-48771-3. [Google Scholar] [CrossRef]
- Mastouri, F.; Björkman, T.; Harman, G.E. Seed Treatment with Trichoderma harzianum Alleviates Biotic, Abiotic, and Physiological Stresses in Germinating Seeds and Seedlings. Phytopathology 2010, 100, 1213–1221. [Google Scholar] [CrossRef] [Green Version]
- Khushdil, F.; Jan, F.G.; Jan, G.; Hamayun, M.; Iqbal, A.; Hussain, A.; Bibi, N. Salt stress alleviation in Pennisetum glaucum through secondary metabolites modulation by Aspergillus terreus L. Plant Physiol. Biochem. 2019, 144, 127–134. [Google Scholar] [CrossRef]
- Khan, S.A.; Hamayun, M.; Yoon, H.; Kim, H.-Y.; Suh, S.-J.; Hwang, S.-K.; Kim, J.-M.; Lee, I.-J.; Choo, Y.-S.; Yoon, U.-H.; et al. Plant growth promotion and Penicillium citrinum. BMC Microbiol. 2008, 8, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 15 June 2020).
- Frary, A.; Doganlar, S.; Daunay, M.C. Vegetables. In Genome Mapping and Molecular Breeding in Plants, 1st ed.; Kole, C., Ed.; Springer: Berlin, Germany, 2007; Volume 5, pp. 231–257. [Google Scholar]
- Medakker, A.; Vijayaraghavan, V. Successful commercialization of insect-resistant eggplant by a public-private partnership: Reaching and benefiting resource-poor farmers. In Intellectual Property Management in Health and Agricultural Innovation A Handbook of Best Practices; Krattiger, A., Mahoney, R.T., Eds.; MIHR: Oxford, UK; PIPRA: Davis, CA, USA; Oswaldo Cruz Foundation: Rio de Janeiro, Brazil; Bio Developments-International Institute: Ithaca, NY, USA, 2010; pp. 1829–1831. [Google Scholar]
- Singh, A. Soil salinity: A global threat to sustainable development. Soil Use Manag. 2021, 38, 39–67. [Google Scholar] [CrossRef]
- Brevik, E.C.; Cerdà, A.; Mataix-Solera, J.; Pereg, L.; Quinton, J.N.; Six, J.; Van Oost, K. The interdisciplinary nature of SOIL. SOIL 2015, 1, 117–129. [Google Scholar] [CrossRef] [Green Version]
- Ünlükara, A.; Kurunç, A.; Kesmez, G.D.; Yurtseven, E.; Suarez, D.L. Effects of salinity on eggplant (Solanum melongena L.) growth and evapotranspiration. Irrig. Drain. 2008, 59, 203–214. [Google Scholar]
- Hannachi, S.; Van Labeke, M.-C. Salt stress affects germination, seedling growth and physiological responses differentially in eggplant cultivars (Solanum melongena L.). Sci. Hortic. 2018, 228, 56–65. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safdarian, M.; Askari, H.; Shariati, J.V.; Nematzadeh, G. Transcriptional responses of wheat roots inoculated with Arthrobacter nitroguajacolicus to salt stress. Sci. Rep. 2019, 9, 1792. [Google Scholar] [CrossRef]
- Ćurković-Perica, M.; Vršek, I.; Mitić, B. In vitro propagation of Inula verbascifolia (Willd.) Hausskn. subsp. verbascifolia. Plant Biosyst. 2008, 142, 1–4. [Google Scholar] [CrossRef]
- Khayyat, M.; Rajaee, S.; Abdoreza, S.; Eshghi, S.; Tafazoli, E. Calcium effects on changes in chlorophyll contents, dry weight and micronutrients of strawberry (Fragaria × ananassa Duch.) plants under salt-stress conditions. Fruits 2009, 64, 53–59. [Google Scholar] [CrossRef] [Green Version]
- Ha-Tran, D.M.; Nguyen, T.T.M.; Hung, S.-H.; Huang, E.; Huang, C.-C. Roles of Plant Growth-Promoting Rhizobacteria (PGPR) in Stimulating Salinity Stress Defense in Plants: A Review. Int. J. Mol. Sci. 2021, 22, 3154. [Google Scholar] [CrossRef] [PubMed]
- Bharti, N.; Barnawal, D.; Awasthi, A.; Yadav, A.; Kalra, A. Plant growth promoting rhizobacteria alleviate salinity induced negative effects on growth, oil content and physiological status in Mentha arvensis. Acta Physiol. Plant 2014, 36, 45–60. [Google Scholar] [CrossRef]
- Abd_Allah, E.F.; Alqarawi, A.A.; Hashem, A.; Radhakrishnan, R.; Al-Huqail, A.A.; Al-Otibi, F.O.N.; Malik, J.A.; Alharbi, R.I.; Egamberdieva, D. Endophytic bacterium Bacillus subtilis (BERA 71) improves salt tolerance in chickpea plants by regulating the plant defense mechanisms. J. Plant Interact. 2018, 13, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Khosravi, K.; Sartaj, M.; Tsai, F.T.-C.; Singh, V.P.; Kazakis, N.; Melesse, A.M.; Prakash, I.; Bui, D.T.; Pham, B.T. A comparison study of DRASTIC methods with various objective methods for groundwater vulnerability assessment. Sci. Total Environ. 2018, 642, 1032–1049. [Google Scholar] [CrossRef]
- Taïbia, K.; Taïbia, F.; Abderrahima, L.A.; Ennajahb, A.; Belkhodjac, M.; Mulet, J.M. Effect of salt stress on growth, chlorophyll content, lipid peroxidation and antioxidant defense systems in Phaseolus vulgaris L. S. Afr. J. Bot. 2016, 105, 306–312. [Google Scholar] [CrossRef]
- Taffouo, V.D.; Wamba, O.F.; Yombi, E.; Nono, G.V.; Akoa, A. Growth, yield, water status and ionic distribution response of three bambara groundnut (Vigna subterranean L. verdc.) landraces grown under saline conditions. Int. J. Bot. 2010, 6, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Ashraf, M. Relationships between leaf gas exchange characteristics and growth of differently adapted populations of Blue panicgrass (Panicum antidotale Retz.) under salinity or waterlogging. Plant Sci. 2003, 165, 69–75. [Google Scholar] [CrossRef]
- Saleem, S.; Iqbal, A.; Ahmed, F.; Ahmad, M. Phytobeneficial and salt stress mitigating efficacy of IAA producing salt tolerant strains in Gossypium hirsutum. Saudi J. Biol. Sci. 2021, 28, 5317–5324. [Google Scholar] [CrossRef]
- Abdelmoteleb, A.; Gonzalez-Mendoza, D. Isolation and Identification of Phosphate Solubilizing Bacillus spp. from Tamarix ramosissima Rhizosphere and Their Effect on Growth of Phaseolus vulgaris Under Salinity Stress. Geomicrobiol. J. 2020, 37, 901–908. [Google Scholar] [CrossRef]
- Azarmi-Atajan, F.; Sayyari-Zohan, M.H. Alleviation of salt stress in lettuce (Lactuca sativa L.) by plant growth-promoting rhizobacteria. J. Hortic. Postharvest. Res. 2020, 3, 67–78. [Google Scholar]
- Mishra, M.; Kumar, U.; Mishra, P.K.; Prakash, P. Efficiency of plant growth promoting rhizobacteria for the enhancement of Cicer arietinum L. growth and germination under salinity. Adv. Biol. Res. 2010, 4, 92–96. [Google Scholar]
- Hannachi, S.; Werbrouck, S.; Bahrini, I.; Abdelgadir, A.; Siddiqui, H.A. Agronomical, Physiological and Biochemical Characterization of In Vitro Selected Eggplant Somaclonal Variants under NaCl Stress. Plants 2021, 10, 2544. [Google Scholar] [CrossRef]
- Zribi, L.; Fatma, G.; Fatma, R.; Salwa, R.; Hassan, N.; Néjib, R.M. Application of chlorophyll fluorescence for the diagnosis of salt stress in tomato “Solanum lycopersicum (variety Rio Grande). Sci. Hortic. 2009, 120, 367–372. [Google Scholar] [CrossRef]
- Gallé, Ă.; Csiszăr, J.; Tari, I.; Erdei, L. Changes in water relation and chlorophyll fluorescence parameters under osmotic stress in wheat cultivars. Acta Biol. Szeged. 2002, 46, 85–86. [Google Scholar]
- Yaghoubian, I.; Ghassemi, S.; Nazari, M.; Raei, Y.; Smith, D.L. Response of physiological traits, antioxidant enzymes and nutrient uptake of soybean to Azotobacter Chroococcum and zinc sulfate under salinity. S. Afr. J. Bot. 2021, 143, 42–51. [Google Scholar] [CrossRef]
- Osman, H.; Gowayed, S.; Elbagory, M.; Omara, A.; El-Monem, A.; El-Razek, U.A.; Hafez, E. Interactive Impacts of Beneficial Microbes and Si-Zn Nanocomposite on Growth and Productivity of Soybean Subjected to Water Deficit under Salt-Affected Soil Conditions. Plants 2021, 10, 1396. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M. Some important physiological selection criteria for salt tolerance in plants. Flora Morphol. Distrib. Funct. Ecol. Plants 2004, 199, 361–376. [Google Scholar] [CrossRef]
- El-Dakak, R.; El-Aggan, W.; Badr, G.; Helaly, A.; Tammam, A. Positive Salt Tolerance Modulation via Vermicompost Regulation of SOS1 Gene Expression and Antioxidant Homeostasis in Viciafaba Plant. Plants 2021, 10, 2477. [Google Scholar] [CrossRef]
- Halfter, U.; Ishitani, M.; Zhu, J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef]
- Zhu, J.-K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Hauer-Jákli, M.; Tränkner, M. Critical Leaf Magnesium Thresholds and the Impact of Magnesium on Plant Growth and Photo-Oxidative Defense: A Systematic Review and Meta-Analysis From 70 Years of Research. Front. Plant Sci. 2019, 10, 766. [Google Scholar] [CrossRef] [PubMed]
- Ceylan, B.; Khorshid, L.; Güneş, Ü.Y.; Zaybak, A. Evaluation of oxygen saturation values in different body positions in healthy individuals. J. Clin. Nurs. 2016, 25, 1095–1100. [Google Scholar] [CrossRef]
- Desnoues, N.; Lin, M.; Guo, X.; Ma, L.; Carreño-Lopez, R.; Elmerich, C. Nitrogen fixation genetics and regulation in a Pseudomonas stutzeri strain associated with rice. Microbiology 2003, 149, 2251–2262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Ghany, T.M.; AlAwlaqi, M.M. Molecular Identification of Rhizospheric Thermo-halotolerant Aspergillus terreus and its Correlation to Sustainable Agriculture. Bioresources 2018, 13, 8012–8023. [Google Scholar] [CrossRef]
- Brenes, M.; Pérez, J.; González-Orenga, S.; Solana, A.; Boscaiu, M.; Prohens, J.; Plazas, M.; Fita, A.; Vicente, O. Comparative Studies on the Physiological and Biochemical Responses to Salt Stress of Eggplant (Solanum melongena) and Its Rootstock S. torvum. Agriculture 2020, 10, 328. [Google Scholar] [CrossRef]
- Ilangumaran, G.; Smith, D.L. Plant Growth Promoting Rhizobacteria in Amelioration of Salinity Stress: A Systems Biology Perspective. Front. Plant Sci. 2017, 8, 1768. [Google Scholar] [CrossRef]
- Zhou, C.; Ma, Z.; Zhu, L.; Xiao, X.; Xie, Y.; Zhu, J.; Wang, J. Rhizobacterial Strain Bacillus megaterium BOFC15 Induces Cellular Polyamine Changes that Improve Plant Growth and Drought Resistance. Int. J. Mol. Sci. 2016, 17, 976. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-W.; Xu, S.-C.; Hu, Q.-Z.; Mao, W.-H.; Gong, Y.-M. Putrescine Plays a Positive Role in Salt-Tolerance Mechanisms by Reducing Oxidative Damage in Roots of Vegetable Soybean. J. Integr. Agric. 2014, 13, 349–357. [Google Scholar] [CrossRef] [Green Version]
- Shi, K.; Huang, Y.Y.; Xia, X.J.; Zhang, Y.L.; Zhou, Y.H.; Yu, J.Q. Protective Role of Putrescine Against Salt Stress is Partially Related to the Improvement of Water Relation and Nutritional Imbalance in Cucumber. J. Plant Nutr. 2008, 31, 1820–1831. [Google Scholar] [CrossRef]
- Zapata, P.J.; Serrano, M.; Pretel, M.; Amorós, A.; Botella, M. Ángeles Polyamines and ethylene changes during germination of different plant species under salinity. Plant Sci. 2004, 167, 781–788. [Google Scholar] [CrossRef]
- El-Shintinawy, F. Photosynthesis in Two Wheat Cultivars Differing in Salt Susceptibility. Photosynthetica 2000, 38, 615–620. [Google Scholar] [CrossRef]
- Xie, S.-S.; Wu, H.-J.; Zang, H.-Y.; Wu, L.-M.; Zhu, Q.-Q.; Gao, X.-W. Plant Growth Promotion by Spermidine-Producing Bacillus subtilis OKB105. MPMI 2014, 27, 655–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Effects of salinity stress on growth and yield of two varieties of eggplant under greenhouse conditions. Res. Crop. 2018, 19, 436–440. [CrossRef]
- Prittesh, P.; Avnika, P.; Kinjal, P.; Jinal, H.N.; Sakthivel, K.; Amaresan, N. Amelioration effect of salt-tolerant plant growth-promoting bacteria on growth and physiological properties of rice (Oryza sativa) under salt-stressed conditions. Arch. Microbiol. 2020, 202, 2419–2428. [Google Scholar] [CrossRef]
- Komenda, J.; Reisinger, V.; Müller, B.C.; Dobáková, M.; Granvogl, B.; Eichacker, L.A. Accumulation of the D2 Protein Is a Key Regulatory Step for Assembly of the Photosystem II Reaction Center Complex in Synechocystis PCC 6803. J. Biol. Chem. 2004, 279, 48620–48629. [Google Scholar] [CrossRef] [Green Version]
- Minai, L.; Wostrikoff, K.; Wollman, F.-A.; Choquet, Y. Chloroplast Biogenesis of Photosystem II Cores Involves a Series of Assembly-Controlled Steps That Regulate Translation. Plant Cell 2006, 18, 159–175. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, H.; Zhang, X.; Tang, M. Arbuscular Mycorrhizal Symbiosis Alleviates Salt Stress in Black Locust through Improved Photosynthesis, Water Status, and K+/Na+ Homeostasis. Front. Plant Sci. 2017, 8, 1739. [Google Scholar] [CrossRef]
- Trivedi, D.K.; Gill, S.S.; Yadav, S.; Tuteja, N. Genome-wide analysis of glutathione reductase (GR) genes from rice and Arabidopsis. Plant Signal. Behav. 2013, 8, e23021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gururani, M.A.; Venkatesh, J.; Upadhyaya, C.P.; Nookaraju, A.; Pandey, S.K.; Park, S.W. Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant Pathol. 2012, 78, 51–65. [Google Scholar] [CrossRef]
- Habib, S.H.; Kausar, H.; Saud, H.M. Plant Growth-Promoting Rhizobacteria Enhance Salinity Stress Tolerance in Okra through ROS-Scavenging Enzymes. BioMed Res. Int. 2016, 2016, 6284547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, I.; Chiu, M.; Cheng, S.; Hsu, Y.; Tsai, C. The glutathione transferase of Nicotiana benthamiana Nb GSTU 4 plays a role in regulating the early replication of Bamboo mosaic virus. New Phytol. 2013, 199, 749–757. [Google Scholar] [CrossRef] [Green Version]
- Jia, B.; Sun, M.; Sun, X.; Li, R.; Wang, Z.; Wu, J.; Wei, Z.; Duanmu, H.; Xiao, J.; Zhu, Y. Overexpression of GsGSTU13 and SCMRP in Medicago sativa confers increased salt-alkaline tolerance and methionine content. Physiol. Plant 2015, 156, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xing, X.-J.; Tian, Y.-S.; Peng, R.-H.; Xue, Y.; Zhao, W.; Yao, Q.-H. Transgenic Arabidopsis Plants Expressing Tomato Glutathione S-Transferase Showed Enhanced Resistance to Salt and Drought Stress. PLoS ONE 2015, 10, e0136960. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Hossain, M.A.; Fujita, M. Trehalose pretreatment induces salt tolerance in rice (Oryza sativa L.) seedlings: Oxidative damage and co-induction of antioxidant defense and glyoxalase systems. Protoplasma 2015, 252, 461–475. [Google Scholar] [CrossRef]
- Sappl, P.G.; Carroll, A.J.; Clifton, R.; Lister, R.; Whelan, J.; Millar, A.H.; Singh, K.B. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009, 58, 53–68. [Google Scholar] [CrossRef]
- Kissoudis, C.; Kalloniati, C.; Flemetakis, E.; Madesis, P.; Labrou, N.; Tsaftaris, A.; Nianiou-Obeidat, I. Stress-inducible GmGSTU4 shapes transgenic tobacco plants metabolome towards increased salinity tolerance. Acta Physiol. Plant 2015, 37, e102. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Durrett, T.P.; Benning, C. Functional diversity of glycerolipid acylhydrolases in plant metabolism and physiology. Prog. Lipid Res. 2019, 75, 100987. [Google Scholar] [CrossRef]
- Lu, J.; Xu, Y.; Wang, J.; Singer, S.D.; Chen, G. The Role of Triacylglycerol in Plant Stress Response. Plants 2020, 9, 472. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.P.; Jha, P.N. The Multifarious PGPR Serratia marcescens CDP-13 Augments Induced Systemic Resistance and Enhanced Salinity Tolerance of Wheat (Triticum aestivum L.). PLoS ONE 2016, 11, e0155026. [Google Scholar] [CrossRef] [Green Version]
- Cook, R.; Lupette, J.; Benning, C. The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses. Cells 2021, 10, 706. [Google Scholar] [CrossRef]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aminisarte, M. Biological Control of Meloidogyne incognita on Eggplant (Solanum melongena). Asian J. Plant Sci. 2021, 21, 1–8. [Google Scholar] [CrossRef]
- Siddiqui, I.A.; Ehetshamul-Haque, S.; Shaukat, S.S. Use of Rhizobacteria in the Control of Root Rot-Root Knot Disease Complex of Mungbean. J. Phytopathol. 2001, 149, 337–346. [Google Scholar] [CrossRef]
- Schönbeck, F.; Dehne, H.W.; Beicht, W. Untersuchungen zur Aktivierung unspezifischer Resistenzmechanismen in Pflanzen. Z. Pflk. Pflschutz 1980, 87, 654–666. [Google Scholar]
- Behera, B.C.; Sethi, B.K.; Mohapatra, S.; Thatoi, H.; Mishra, R.R. Bio-production of alkaline protease by Trichoderma longibrachiatum and Penicillium rubidurum using different agro-industrial products. Nov. Res. Microbiol. J. 2021, 5, 1241–1255. [Google Scholar] [CrossRef]
- Abu-Tahon, M.A.; Arafat, H.H.; Isaac, G.S. Laundry Detergent Compatibility and Dehairing Efficiency of Alkaline Thermostable Protease Produced from Aspergillus terreus under Solid-state Fermentation. J. Oleo Sci. 2020, 69, 241–254. [Google Scholar] [CrossRef] [Green Version]
- Alkhafaje, W.K.; Ali, S.M.; Olama, Z.A. Isolation and molecular differentiation of MDR bacteria isolated from dairy products. Pollut. Res. 2019, 38, 109–115. [Google Scholar]
- Hassan, S.W.M.; El-Latif, H.H.A.; Ali, S.M. Production of Cold-Active Lipase by Free and Immobilized Marine Bacillus cereus HSS: Application in Wastewater Treatment. Front. Microbiol. 2018, 9, 2377. [Google Scholar] [CrossRef] [Green Version]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage, 3rd ed.; Springer: Boston, MA, USA, 2009; p. 519. [Google Scholar] [CrossRef]
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi, 2nd ed.; IHW-Verlg Eching: Eching, Germany, 2007; p. 672. [Google Scholar]
- Harman, G.E.; Kubicek, C.P. Trichoderma And Gliocladium. Volume 1: Basic Biology, Taxonomy and Genetics, 1st ed.; CRC Press: London, UK, 1998; p. 300. [Google Scholar]
- Houbraken, J.; Frisvad, J.; Samson, R. Taxonomy of Penicillium section Citrina. Stud. Mycol. 2011, 70, 53–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeuchi, T.; Ishida, A.; Tajifi, M.; Nagata, S. Induction of salt tolerance in Bacillus subtilis IFO 3025. J. Biosci. Bioeng. 2003, 96, 184–186. [Google Scholar] [CrossRef]
- Latif, S.; Mohamed, A.; Sueyoshi, K.; Mohamed, H.; Saber, N. Effect of Bacillus subtilis on Some Physiological and Biochemical Processes in Barley (Hordeum vulgare L.) Plant Grown under Salt Stress. Egypt. J. Bot. 2020, 61, 141–153. [Google Scholar] [CrossRef]
- Fall, R.; Kinsinger, R.F.; Wheeler, K.A. A Simple Method to Isolate Biofilm-forming Bacillus subtilis and Related Species from Plant Roots. Syst. Appl. Microbiol. 2004, 27, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Lastochkina, O.; Baymiev, A.; Shayahmetova, A.; Garshina, D.; Koryakov, I.; Shpirnaya, I.; Pusenkova, L.; Mardanshin, I.; Kasnak, C.; Palamutoglu, R. Effects of Endophytic Bacillus Subtilis and Salicylic Acid on Postharvest Diseases (Phytophthora infestans, Fusarium oxysporum) Development in Stored Potato Tubers. Plants 2020, 9, 76. [Google Scholar] [CrossRef] [Green Version]
- Preston, G.M. Plant perceptions of plant growth-promoting Pseudomonas. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 907–918. [Google Scholar] [CrossRef] [Green Version]
- Egamberdieva, D.; Li, L.; Lindstrom, K.; Räsänen, L.A. A synergistic interaction between salt-tolerant Pseudomonas and Mesorhizobium strains improves growth and symbiotic performance of liquorice (Glycyrrhiza uralensis Fish.) under salt stress. Appl. Microbiol. Biotechnol. 2016, 100, 2829–2841. [Google Scholar] [CrossRef]
- Vimal, S.R.; Gupta, J.; Singh, J.S. Effect of salt tolerant Bacillus sp. and Pseudomonas sp. on wheat (Triticum aestivum L.) growth under soil salinity: A comparative study. Microbiol. Res. 2018, 9, 26–32. [Google Scholar] [CrossRef]
- Gupta, S.; Smith, P.M.C.; Boughton, B.A.; Rupasinghe, T.W.T.; Natera, S.H.A.; Roessner, U. Inoculation of barley with Trichoderma harzianum T-22 modifies lipids and metabolites to improve salt tolerance. J. Exp. Bot. 2021, 72, 7229–7246. [Google Scholar] [CrossRef] [PubMed]
- Zin, N.A.; Badaluddin, N.A. Biological functions of Trichoderma spp. for agriculture applications. Ann. Agric. Sci. 2020, 65, 168–178. [Google Scholar] [CrossRef]
- Bae, H.; Roberts, D.P.; Lim, H.-S.; Strem, M.D.; Park, S.-C.; Ryu, C.-M.; Melnick, R.L.; Bailey, B.A. Endophytic Trichoderma Isolates from Tropical Environments Delay Disease Onset and Induce Resistance Against Phytophthora capsici in Hot Pepper Using Multiple Mechanisms. MPMI 2011, 24, 336–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, R.; Lee Rutgers, S. Applications of Aspergillus in Plant Growth Promotion. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 223–227. ISBN 9780444635051. [Google Scholar] [CrossRef]
- Vassileva, M.; Malusá, E.; Eichler-Löbermann, B.; Vassilev, N. Aspegillus terreus: From Soil to Industry and Back. Microorganisms 2020, 8, 1655. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.-L.; Chiang, M.W.-L.; Guo, S.-Y.; Shih, C.-Y.; Dahms, H.U.; Hwang, J.-S.; Cha, H.-J. Growth study under combined effects of temperature, pH and salinity and transcriptome analysis revealed adaptations of Aspergillus terreus NTOU4989 to the extreme conditions at Kueishan Island Hydrothermal Vent Field, Taiwan. PLoS ONE 2020, 15, e0233621. [Google Scholar] [CrossRef]
- Dhakar, K.; Sharma, A.; Pandey, A. Cold, pH and salt tolerant Penicillium spp. inhabit the high altitude soils in Himalaya, India. World J. Microbiol. Biotechnol. 2014, 30, 1315–1324. [Google Scholar] [CrossRef]
- Saleem, M.; Arshad, M.; Hussain, S.; Bhatti, A.S. Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J. Ind. Microbiol. Biotechnol. 2007, 34, 635–648. [Google Scholar] [CrossRef]
- Wu, H.; Guo, J.; Wang, C.; Li, K.; Zhang, X.; Yangm, Z.; Li, M.; Wang, B. An Effective Screening method and an eliable screening trait for salt tolerance of Brassica napus at the germination stage. Front. Plant Sci. 2019, 10, 530. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Moustakas, M.; Ouzounidou, G.; Lannoye, R. Rapid Screening for Aluminum Tolerance in Cereals by Use of the Chlorophyll Fluorescence Test. Plant Breed. 1993, 111, 343–346. [Google Scholar] [CrossRef]
- Kimbrough, D.E.; Wakakuwa, J.R. Acid digestion for sediments, sludges, soils, and solid wastes. A proposed alternative to EPA SW 846 Method 3050. Environ. Sci. Technol. 1989, 23, 898–900. [Google Scholar] [CrossRef]
- Gong, X.; Liu, J.H. Detection of Free Polyamines in Plants Subjected to Abiotic Stresses by High-Performance Liquid Chromatography (HPLC). In Plant Stress Tolerance. Methods in Molecular Biology; Sunkar, R., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1631, pp. 305–311. [Google Scholar] [CrossRef]
- IBM Corp. IBM SPSS Statistics for Windows, Version 21.0; IBM Corp.: Armonk, NY, USA, 2012. [Google Scholar]
- Zhang, P.; Ma, Y.; Xie, C.; Guo, Z.; He, X.; Valsami-Jones, E.; Lynch, I.; Luo, W.; Zheng, L.; Zhang, Z. Plant species-dependent transformation and translocation of ceria nanoparticles. Environ. Sci. Nano 2019, 6, 60–67. [Google Scholar] [CrossRef] [Green Version]
- Sokal, R.R.; Rohlf, F. Biometry: The Principles and Practice of Statistics in Biological Research; Correa, J., Ed.; W. H. Freeman and Company: New York, NY, USA, 2012; pp. 177–602. [Google Scholar]

| STI% of Parameters | S | S + I | T | p |
|---|---|---|---|---|
| PH | 76.89 ± 11.62 | 82.41 ± 11.65 | 0.581 | 0.592 |
| RL | 118.61 ± 28.20 | 112.53 ± 24.45 | 0.282 | 0.792 |
| SL | 60.35 ± 12.71 | 70.93 ± 17.23 | 0.856 | 0.440 |
| R/S | 204.65 ± 69.85 | 167.30 ± 59.26 | 0.706 | 0.519 |
| RFW | 4.12 * ± 0.55 | 72.79 ± 39.92 | 2.979 * | 0.041 * |
| SFW | 16.97 * ± 5.20 | 50.56 ± 6.48 | 7.002 * | 0.002 * |
| TFW | 11.03 * ± 1.54 | 54.07 ± 11.72 | 6.310 * | 0.022 * |
| RDW | 3.68 ± 0.33 | 73.14 ± 36.09 | 3.333 * | 0.029 * |
| SDW | 17.35 * ± 2.93 | 65.79 ± 15.08 | 5.459 * | 0.005 * |
| TDW | 9.12 * ± 0.23 | 67.21 ± 20.81 | 4.834 * | 0.040 * |
| RWC | 4.35 * ± 0.71 | 73.15 ± 42.58 | 2.798 * | 0.049 * |
| SWC | 16.90 * ± 5.69 | 47.71 ± 5.13 | 6.964 * | 0.002 * |
| TWC | 11.64 * ± 1.98 | 51.18 ± 9.91 | 6.774 * | 0.002 * |
| LA | 41.24 ± 5.95 | 92.68 ± 35.89 | 2.449 | 0.128 |
| Parameters | Treatments | |||
|---|---|---|---|---|
| C | I | S | S + I | |
| Chl a (μg g−1 FW) | 10.11 a ± 0.78 | 7.38 b ± 0.61 | 5.75 b ± 0.77 | 10.37 a ± 2.39 |
| Chl b (μg g−1 FW) | 3.30 a ± 0.38 | 1.99 c ± 0.11 | 2.14 bc ± 0.09 | 2.98 ab ± 0.82 |
| Chl a + b (μg g−1 FW) | 13.41 a ± 0.74 | 9.36 b ± 0.71 | 7.89 b ± 0.85 | 13.35 a ± 3.20 |
| Chl a/b | 3.06 b ± 0.23 | 3.71 a ± 0.02 | 2.69 c ± 0.08 | 3.48 a ± 0.05 |
| Carot. (μg g−1 FW) | 2.36 a ± 0.24 | 1.78 b ± 0.11 | 1.31 b ± 0.12 | 2.39 a ± 0.49 |
| Fv/Fm | 0.79 a ± 0.01 | 0.79 a ± 0.03 | 0.75 a ± 0.02 | 0.76 a ± 0.02 |
| Fv/F0 | 3.84 a ± 0.27 | 3.76 a ± 0.60 | 2.98 a ± 0.36 | 3.38 a ± 0.31 |
| Parameters | Treatment | |||||||
|---|---|---|---|---|---|---|---|---|
| Roots | Shoots | |||||||
| C | I | S | S + I | C | I | S | S + I | |
| Spm (mg g−1FW) | 0.58 b ± 0.01 | 0.0 c ± 0.0 | 0.0 c ± 0.0 | 0.66 a ± 0.02 | 0.31 d ± 0.02 | 0.80 b ± 0.04 | 0.39 c ± 0.02 | 0.93 a ± 0.02 |
| Spd (mg g−1FW) | 0.31 a ± 0.02 | 0.0 d ± 0.0 | 0.10 c ± 0.01 | 0.18 b ± 0.01 | 0.08 c ± 0.02 | 0.10 c ± 0.01 | 0.17 b ± 0.01 | 0.21 a ± 0.02 |
| Put (mg g−1FW) | 3.37 a ± 0.07 | 0.18 d ± 0.01 | 1.50 c ± 0.03 | 2.37 b ± 0.02 | 0.08 b ± 0.01 | 0.03 c ± 0.01 | 0.08 b ± 0.02 | 0.17 a ± 0.02 |
| Prol (µg g−1 FW) | 10.35 c ± 0.28 | 7.73 d ± 0.67 | 16.22 b ± 0.30 | 19.22 a ± 1.22 | 18.71 c ± 1.43 | 8.87 d ± 0.26 | 24.37 b ± 0.29 | 30.05 a ± 0.54 |
| Organism | Type of Organism | Location of the Organism | Function of the Organism | Reference |
|---|---|---|---|---|
| Bacillus subtilis | Gram + ve non-pathogenic Bacterium | Soil/Colonizing plant roots | * Salt tolerant bacterium * Protect cellular membranes integrity * Increase nitrate reductase and glutamine synthetase activities * Supply IAA to the cultures * Reduce ethylene generation under salt stress through ACC deaminase secretion, thus increase nutrient uptake and growth. * Decrease oxidative and osmotic induced stress * Manifested plant growth improvement, slowing down statolite starch hydrolysis under salinity | [4,83,84,85,86] |
| Pseudomonas sp. | Gram -ve Bacterium | Saprophytic/ parasite on plant surfaces | * Salt tolerant bacteria * Promote plant growth by suppressing pathogenic micro-organisms * Synthesize growth-stimulating plant hormones * Promote increased plant disease resistance | [87,88,89] |
| Trichoderma harizanum | Free-living saprophytic fungi | In most types of soils/mutualistic endophytic with plant species | * Salt tolerant fungi * Significantly suppress the growth of plant pathogenic microorganisms * Regulate the rate of plant growth * Well known for biological control mechanism * Produce secondary metabolites in agroecosystems | [90,91,92] |
| Aspergillus terrus | Saprophytic filamentous fungi | Part of the soil microbiota/can be found in many types of soils/frequently found as endophytic | * May live at pH 3 and 30% salinity * Used with PGPR, induces positive effects on plant growth and development * Induce systemic resistance and reduce plant stress * Attain phosphorous-solubilizing activity * Strong biocontrol activity | [93,94,95] |
| Penicillium citrinum | Mesophilic Fungus | Soil is their natural habitat | * Isolates tolerate salt concentration above 10% * Plant growth promoting ability * Contain ACC deaminase activity which sustains plant growth and development under stress conditions * Produces mycotoxin citrinin, cellulase, endoglucanase, as well as xylulase. * Produces Gibberellins | [8,96,97] |
| Primer Sequence | Length (bp) | Gene |
|---|---|---|
| F: AGGCTGTGGACCGACATCTA | 266 | psbD |
| R: GCTCATGAACACGTCCCTCT | ||
| F: CACATCCTGATCGCCACCG | 200 | GR |
| R: TCCTTCCTGAAGCACAGGTC | ||
| F: GAAGATCCCCGTGCTGATCC, | 390 | GST |
| R: AAGTTGGGGAACTTCTCGCT | ||
| F: GCACATCCTGAGGGTGAACA | 369 | Lip. |
| R: AGCTCGTAGTCCTCCCTGTC | ||
| F: AGGCTGTGGACCGACATCTA, | 489 | Prot. I |
| R: GCTCATGAACACGTCCCTCT | ||
| F: CGACACCATGCAGTACGTGA, | 386 | Prot. II |
| R: TGGCGTAGTTGGCGTACATC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mokabel, S.; Olama, Z.; Ali, S.; El-Dakak, R. The Role of Plant Growth Promoting Rhizosphere Microbiome as Alternative Biofertilizer in Boosting Solanum melongena L. Adaptation to Salinity Stress. Plants 2022, 11, 659. https://doi.org/10.3390/plants11050659
Mokabel S, Olama Z, Ali S, El-Dakak R. The Role of Plant Growth Promoting Rhizosphere Microbiome as Alternative Biofertilizer in Boosting Solanum melongena L. Adaptation to Salinity Stress. Plants. 2022; 11(5):659. https://doi.org/10.3390/plants11050659
Chicago/Turabian StyleMokabel, Souhair, Zakia Olama, Safaa Ali, and Rehab El-Dakak. 2022. "The Role of Plant Growth Promoting Rhizosphere Microbiome as Alternative Biofertilizer in Boosting Solanum melongena L. Adaptation to Salinity Stress" Plants 11, no. 5: 659. https://doi.org/10.3390/plants11050659
APA StyleMokabel, S., Olama, Z., Ali, S., & El-Dakak, R. (2022). The Role of Plant Growth Promoting Rhizosphere Microbiome as Alternative Biofertilizer in Boosting Solanum melongena L. Adaptation to Salinity Stress. Plants, 11(5), 659. https://doi.org/10.3390/plants11050659






