Abstract
Major biotic stresses viz., bacterial blight (BB) and blast and brown plant hopper (BPH) coupled with abiotic stresses like drought stress, significantly affect rice yields. To address this, marker-assisted intercross (IC) breeding involving multiple donors was used to combine three BB resistance genes—xa5, xa13 and Xa21, two blast resistance genes—Pi9 and Pi54, two BPH resistance genes—Bph20 and Bph21, and four drought tolerant quantitative trait loci (QTL)—qDTY1.1, qDTY2.1, qDTY3.1 and qDTY12.1—in the genetic background of the elite Indian rice cultivar ‘Krishna Hamsa’. Three cycles of selective intercrossing followed by selfing coupled with foreground selection and phenotyping for the target traits resulted in the development of 196 introgression lines (ILs) with a myriad of gene/QTL combinations. Based on the phenotypic reaction, the ILs were classified into seven phenotypic classes of resistance/tolerance to the following: (1) BB, blast and drought—5 ILs; (2) BB and blast—10 ILs; (3) BB and drought—9 ILs; (4) blast and drought—42 ILs; (5) BB—3 ILs; (6) blast—84 ILs; and (7) drought—43 ILs; none of the ILs were resistant to BPH. Positive phenotypic response (resistance) was observed to both BB and blast in 2 ILs, BB in 9 ILs and blast in 64 ILs despite the absence of corresponding R genes. Inheritance of resistance to BB and/or blast in such ILs could be due to the unknown genes from other parents used in the breeding scheme. Negative phenotypic response (susceptibility) was observed in 67 ILs possessing BB-R genes, 9 ILs with blast-R genes and 9 ILs harboring QTLs for drought tolerance. Complex genic interactions and recombination events due to the involvement of multiple donors explain susceptibility in some of the marker positive ILs. The present investigation successfully demonstrates the possibility of rapid development of multiple stress-tolerant/resistant ILs in the elite cultivar background involving multiple donors through selective intercrossing and stringent phenotyping. The 196 ILs in seven phenotypic classes with myriad of gene/QTL combinations will serve as a useful genetic resource in combining multiple biotic and abiotic stress resistance in future breeding programs.
1. Introduction
Rice, an important crop for human sustenance, occupies a prominent place in the Indian agriculture. It is the cheapest source of calories for the developing countries and is a staple food crop for more than half of the global population, despite the changing climatic, social and economic scenario. The major biotic stresses like bacterial leaf blight (BB), blast, sheath blight, brown plant hoppers (BPH), stem borer, etc., result in a severe yield reduction in rice. In addition, abiotic stresses viz., drought, salinity, temperature extremities, etc., hinder growth and development of rice plant [1]. Development of climate resilient varieties with multiple stress tolerance is needed for preventing yield losses and increasing the income of rice farmers in an environmentally sustainable manner.
Conventional and marker-assisted backcross breeding has traditionally been used to introduce useful agronomic traits into elite cultivars; however, combining high yield with multiple stress tolerance using these approaches is tedious. The simultaneous occurrences of multiple abiotic and biotic stresses have demanded the development of climate-smart rice by combining quantitative trait loci (QTL) and genes for tolerance or resistance to various stresses in the genetic background of high yielding cultivars to confer a wider range of tolerance or resistance [1]. Enhanced capability of climate-smart cultivars would enable the crop to thrive under adverse environmental conditions. During the last 40 years, molecular marker systems have become well established, enabling precision in selection. Gene/QTLs conferring resistance/tolerance for various biotic/abiotic stresses are well characterized, and the recent advances in molecular marker technology and genomics have played an important role in developing single [2,3] and multiple stress-tolerant rice cultivars [1,4,5,6].
Among the biotic stress, BB caused by Xanthomonas oryzae pv. oryzae with about 22 pathotypes identified from diverse geographies [7] significantly reduces the rice yields. To date, at least 45 BB resistance genes [8,9] have been identified, and 11 of them have been cloned (Xa1, xa5, xa10, xa13, Xa21, Xa23, Xa25 and Xa27 [10,11]) and characterized [12], and 7 genes (Xa4, Xa7, Xa22, Xa30, Xa31, Xa33 and Xa34) have been fine-mapped. Some of them have been introgressed in genetic background of elite cultivars and few of these have been released as cultivars. The obvious difference in the level of resistance among the genes to a number of virulent pathogens encouraged plant breeders to pyramid two or more genes. Xa4 from TKM6 and xa5 from DZ192 [13], xa13 from long grain [14] and Xa21 from O. longistaminata [15] were used as donors in the development of near isogenic lines (NILs)-IRBB60 (Xa4+xa5+xa13+Xa21) and IRBB62 (xa5+xa13+Xa21), with four and three genes introgressed, respectively, in the genetic background IR24 [16]. Further, an improved version of ‘PR106’ an elite cultivar with xa5+xa13+Xa21 introgression was developed from IRBB62 [17] and improved PR106 (SS1113) was used in the transfer of xa5+xa13+Xa21 into the background of Samba Mahsuri, a fine grain medium slender grain type variety quite popular in Southern India, which resulted in the development of NILs ‘RP Bio226’ released as a variety in the name of ‘Improved Samba Mahsuri’ (ISM) with xa5+xa13+Xa21 [1]. The incorporation of Xa33 alone in the background of ‘Akshayadhan’ and Xa38, in combination with xa5, xa13 and Xa21 in the background of Improved Samba Mahsuri, has proved to provide broad-spectrum resistance to BB, and the same varieties have been released as DRR Dhan 58 [18] and DRR Dhan 53 [7,19], respectively.
Blast, caused by the fungus Magnaporthe oryzae Barr, is the most devastating fungal disease in rice causing up to 50% yield losses [20]. It is also referred as rice fever disease and has been reported in approximately 85 rice growing countries across the world [21]. Several (>100) blast-R genes have been identified and 31 genes (Pi37, Pit, Pish, Pi35, Pi64, Pib, pi21, Pi63/Pikahei-1(t), Pid2, Pi9, Pi2, Pizt, Pid3, Pi25, Pi50, Pigm, Pid3-I1, Pi36, Pi5, Pikm, Pb1, Pi54, Pia, Pikp, Pik, Pi1, Pi-Co39, Pike, Pita and Ptr) have been cloned and characterized [22,23,24,25]. Pi1 from West African cv. LAC23 and Pi2 from cv. 5173 [26], Pi9 from O. minuta [27] and Pi54 from Tetep [28], when deployed in elite varietal backgrounds either singly or in combination, are effective against a wide range of blast pathotypes. Enhanced resistance to blast disease was observed in IET 25484 (RP 5960 Patho 7-5-9) with Pi2 and IET 25480 (Pusa 1850-27) with three genes—Pi54, Pi1 and Pita, now released as DRR Dhan 51 and Pusa Samba, respectively, in India [29]. Many of the resistant genes have been incorporated in modern rice cultivars [30]. These studies indicate that the use of multiple blast resistance gene combinations could be effective against blast disease.
Among the insect pests, BPH is the most devastating insect pest of rice with symptoms popularly known as hopper burn. Bph20 and Bph21 from O. minuta in the introgression line, IR 71033-121-15-B, showed BPH resistance [31]. The pyramided or single-gene introgression lines with six dominant BPH resistance genes (Bph3, Bph14, Bph15, Bph18, Bph20 and Bph21) showed enhanced resistance to the recurrent parent line Jin 23B [32].
Water scarcity consequent to climate change is the most serious climatic challenge, particularly to the rainfed area of rice cultivation in Southern and southeastern Asia, affecting >23 million ha of rice area [33]. The reproductive stage is the most sensitive to drought stress with significant yield loss [34]. As many as 14 QTLs for drought tolerance (qDTY1.1, qDTY2.1, qDTY3.1, qDTY6.1, qDTY3.2, qDTY12.1, qDTY2.2, qDTY4.1, qDTY9.1 and qDTY10.1) have been identified [35,36,37,38,39]. qDTY1.1, qDTY2.1, qDTY3.1 and qDTY12.1 have been effectively used in the development of rice cultivars with 10–30 yield advantage over the recurrent parents under drought stress [40,41,42].
Recently, efforts have been directed in sequential introgression of two or more biotic and abiotic traits. In the background of Improved Samba Mahsuri with inherent xa5, xa13 and Xa21, and DRR Dhan 58 (IET 28784), DRR Dhan 60 (IET 28061) and DRR Dhan 62 (IET 28804) have been developed and released as cultivars with introgression of Saltol QTL for salinity tolerance, Pup1 QTL for low soil P tolerance and Pi2 and Pi54 for blast resistance, respectively [6,18]. Additionally, there are successful examples of simultaneous introgression of multiple QTL and genes for biotic and abiotic stress in rice like drought and submergence tolerance in the background of Swarna [41], blast, BB, gall midge and drought tolerance in the background of Swarna [1] and Naveen [4], and blast, BB and drought tolerance in Lalat [5]. In India, boro season is a highly productive rice growing ecology in the Eastern and northeastern regions of the country from November to April. “Boro” means a special type of rice cultivation on residual or stored water in low-lying areas after the harvest of wet season (kharif) rice. Farmers have a limited choice in this ecology as only 35 cultivars have been released for cultivation to date, compared to several hundreds of cultivars in other rice growing ecologies of the country. Hence, the present study is aimed at improvement of ‘Krishna Hamsa’, an elite cultivar suitable for boro areas, by combining resistance against BB, blast, BPH and drought tolerance. Repeated cycles of intercrossing to combine traits from different donors, genotyping with foreground markers to track alleles and stringent phenotyping were deployed at different generations.
2. Results
2.1. Selective InterCrossing to Combine Genes and QTLs
In the genetic background of the elite rice cultivar, ‘Krishna Hamsa’ genes and QTLs conferring tolerance to various biotic and abiotic stresses were combined using selective intercrossing approach and forward breeding aided by marker-assisted selection. In kharif 2013, six simultaneous single cross F1’s were generated with 63 to 126 seeds by crossing ‘Krishna Hamsa’ with each of the six donors. Hybridity of 595 F1 plants was confirmed using gene-specific or tightly linked polymorphic SSRs for target genes and polymorphic markers at peak and flanking regions of the QTL (Supplementary Table S1). The first cycle of intercrosses were attempted during rabi 2014 among three pairs of single cross F1’s: (1) Krishna Hamsa/IRBB60 (xa5+xa13+Xa21)//Krishna Hamsa/IR74371-46-1-1-13 (qDTY12.1), (2) Krishna Hamsa/Tetep (Pi9+Pi54)//Krishna Hamsa/IR96321-1447-561-B-1 (qDTY1.1+qDTY3.1) and (3) Krishna Hamsa/IR71033-121-15-B (Bph20+Bph21)//Krishna Hamsa/IR74371-46-1-1-13 (qDTY2.1) (Figure 1).
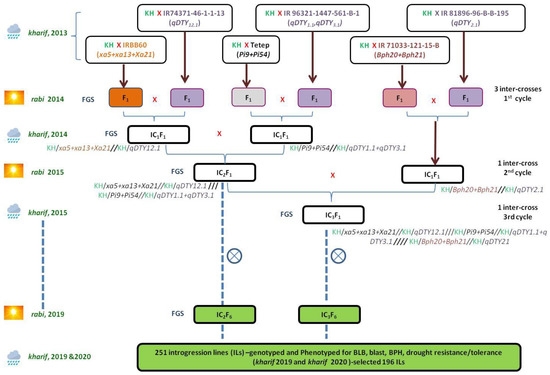
Figure 1.
Introgression scheme involving elite recurrent parent, ‘Krishna Hamsa’, and six donor parents for biotic and abiotic traits. IRBB 60 with xa5+xa13+Xa21 for BB, Tetep with Pi9 for blast and IR 71033-121-15-B with Bph20+Bph21 for BPH genes, as well as IR 96321-1447-561-B-1 with qDTY1.1+qDTY3.1, IR 81896-96-B-B-195 with qDTY2.1 and IR74371-46-1-1-13 with qDTY12.1 for drought-tolerant QTLs. kharif is the wet season with crop growing period from June to November, and rabi is the dry season with crop growing period from December to May.
Foreground selection in 635 IC1F1 plants revealed one to four gene/QTL in 37 different combinations of which three gene/QTL combinations—xa5+xa13+Xa21+qDTY12.1, Pi9+Pi54+qDTY1.1+qDTY3.1 and Bph20+Bph21+qDTY2.1—were selected for crossing. In kharif 2014, second cycle of intercross was attempted between IC1F1 plants with gene/QTL combinations of xa5+xa13+Xa21+qDTY12.1 and Pi9+Pi54+qDTY1.1+qDTY3.1, while five IC1F1 plants with Bph20+Bph21+qDTY2.1 were maintained as stubbles. Among 234 IC2F1 plants, four plants were identified with eight gene/QTL combinations of xa5+xa13+Xa21+Pi9+Pi54+qDTY1.1+qDTY3.1+qDTY12.1, and one to seven gene/QTLs in 25 different combinations in the remaining 230 IC2F1 plants. In the third cycle of intercrossing during rabi 2015, the four IC2F1 plants with eight gene/QTL combinations of xa5+xa13+Xa21+Pi9+Pi54+qDTY1.1+qDTY3.1+qDTY12.1 were simultaneously advanced by selfing and crossed with five IC1F1 plants containing Bph20+Bph21+qDTY2.1, which were maintained as stubbles. Foreground selection in 72 IC3F1 plants identified five plants with all the target alleles xa5+xa13+Xa21+Pi9+Pi54+Bph20+Bph21+qDTY1.1+qDTY2.1+qDTY3.1+qDTY12.1 and one to eight gene/QTL in myriad of combinations.
The four IC2F1 plants with eight gene/QTL combinations of xa5+xa13+Xa21+Pi9+Pi54+qDTY1.1+qDTY3.1+qDTY12.1 and five IC3F1 plants with all the 11 target alleles of xa5+xa13+Xa21+Pi9+Pi54+Bph20+Bph21+qDTY1.1+qDTY2.1 +qDTY3.1+qDTY12.1 were advanced by selfing up to IC2F6 and IC3F6 generations, respectively. Out of 3328 F2 plants from the two sets of populations, 578 single panicle selections were made and genotyped. In F3 generation, 49 plants were selected with five to nine target alleles in homozygous condition and grown. A total of 251 plants in F4 generation of both IC2 and IC3 populations were selected with desirable agronomic traits, genotyped and further advanced as families up to F6 generation (Figure 1).
2.2. Gene/QTL Introgression vis-à-vis Phenotypic Response
All the 251 IC2F6 lines were genotyped with foreground markers and phenotyped for BB, blast and BPH during kharif 2019 and kharif 2020 and for grain yield both under non-stress, as well as reproductive-stage drought stress conditions during kharif 2019. Out of 251 introgression lines (ILs), 196 ILs with positive phenotypic reaction to one or more targeted traits were broadly classified into seven phenotypic classes, as follows: (1) phenotypic class I with 5 ILs resistant/tolerant to BB, blast and drought; (2) phenotypic class II with 10 ILs resistant/tolerant to BB and blast; (3) phenotypic class III with 9 ILs resistant/tolerant to BB and drought; (4) phenotypic class IV with 42 ILs resistant/tolerant to blast and drought; (5) phenotypic class V with 3 ILs resistant to BB; (6) phenotypic class VI with 84 ILs resistant/tolerant to blast; and (7) phenotypic class VII with 43 ILs tolerant to drought stress (Figure 2). None of the ILs were resistant to BPH despite possessing the resistant alleles of Bph20 and Bph21.
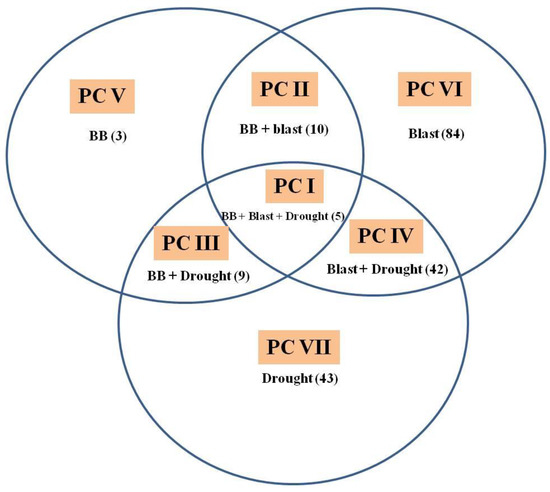
Figure 2.
Venn diagram depicting resistance/tolerance of 196 introgression lines (ILs) in seven major phenotypic classes (PC) of bacterial blight (BB), blast and drought in various combinations. Phenotypic classes are represented as PC-I to PC-VII. Number in parenthesis represents the total ILs in that PC.
The introgression of gene/QTL in various combinations in the 196 ILs is given in Table 1, Table 2 and Table 3. Yield evaluation of all the 196 ILs under non-stress conditions and 97 ILs under reproductive-stage drought stress revealed significant phenotypic differences among the ILs, their recurrent parent—Krishna Hamsa—and four checks (Supplementary Tables S2 and S3). Broad sense heritability (H2) for the grain yield was 86.00 and 70.73 under the control and reproductive-stage drought stress conditions, respectively. The descriptive statistics and critical difference (CD) at 1% and 5% level of significance (p-value) for days to 50% flowering and grain yield under the control and reproductive-stage drought stress are summarized in Supplementary Tables S4 and S5.

Table 1.
Gene/QTL introgressions in the introgression lines (ILs) of phenotypic classes with resistance/tolerance to three (BB + blast + drought) and two (BB + blast; BB + drought; blast + drought) traits.

Table 2.
Gene/QTL introgressions in the ILs of phenotypic class with resistance/tolerance to single trait (BB; blast).

Table 3.
Gene/QTL introgressions in the ILs of phenotypic class-VII with tolerance to drought.
2.2.1. Phenotypic Class-I Resistance/Tolerance to BB, Blast and Drought
Among the five ILs, IL-19246 was resistant to BB with standard evaluation system (SES) score of 1 and blast with an SES score of 1 both during kharif 2019 and kharif 2020. The percentage yield advantage of IL-19246 over the recurrent parent Krishna Hamsa was +180% (Supplementary Table S7) with a yield of 560 g/m2 under non-stress conditions (Supplementary Table S6) (Figure 3). Similarly, IL-19247 was resistant to BB (SES score 1), moderately resistant to blast (SES score 4) and recorded +131% yield advantage over Krishna Hamsa under drought stress conditions and with a yield of 598 g/m2 under non-stress conditions. The remaining three ILs—19174, 19193 and 19196—were moderately resistant to both BB and blast with yield advantage of 82%, 68% and 120% over Krishna Hamsa under reproductive-stage drought stress (Supplementary Table S7) and a yield of 737, 374 and 544 g/m2 under non-stress conditions, respectively (Supplementary Table S6). Interestingly, only one IL-19196 has at least one gene Xa21 for BB and Pi9 for blast and two QTL-qDTY2.1+qDTY3.1 for drought (Supplementary Table S6). Resistant alleles for blast were not observed in ILs 19174 and 19193, while resistant alleles of both BB and blast were absent in ILs 19246 and 19247 (Table 1).

Figure 3.
Performance of the IL-19246 of phenotypic class-I with resistance to blast in the universal blast nursery (UBN) during kharif 2019 and kharif 2020 and resistance to BB in glass house and field screening during kharif 2019 and kharif 2020, as well as tolerance to drought during kharif 2019. IL-19246 with (a) blast resistance score of 1 as per standard evaluation system (SES) in UBN during kharif 2020. ‘HR12’ was used as susceptible check (SES score 9). (b) BB resistance (SES score 1) during kharif 2019 in glass house screening and (c) yield of 538 g/m2 under reproductive-stage drought stress conditions with +180% yield advantage over ‘Krishna Hamsa’ during kharif 2019.
2.2.2. Phenotypic Class-II Resistance/Tolerance to BB and Blast
Among the 10 ILs, IL-19378 with xa5+Xa21+Pi54, IL 19031 with xa5+Xa21+Bph20+Bph21, IL 19030 with Pi54+Bph20+Bph21 and two ILs 19019 and 19471 with xa5+Pi9 were resistant to both BB and blast (Table 1) (Figure 4b and Figure 5b,c). IL-19030 has resistant alleles Pi54 for blast but no resistant alleles for BB (Table 1), and IL19031 has resistant alleles for BB but not for blast. Except for xa5 for BB resistance, none of the blast-R genes were introgressed in the remaining five ILs in this group viz., ILs-19007, 19020, 19025, 19406 and 19039, but they were seen with Bph20+Bph21 introgressions. Grain yield varied from 311 in IL 19406 to 1010 g/m2 in IL 19030 under non-stress conditions (Supplementary Table S6).
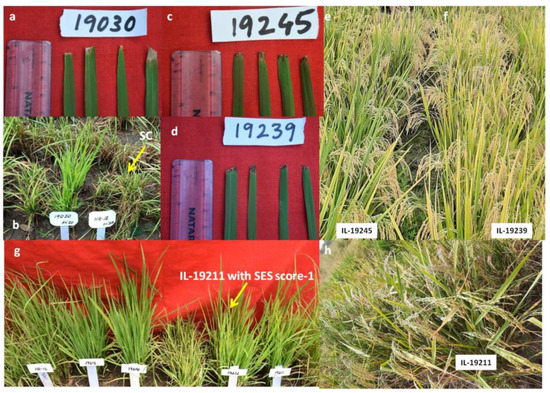
Figure 4.
Performance of the ILs of phenotypic classes-II, -III and -IV with resistance/tolerance to two traits. (a,b): IL-19030 of PC-II with BB resistance (SES score 1) during kharif 2019 in glass house screening and blast resistance (SES score 1) in UBN during kharif 2020. (c–f): ILs 19245 and 19239 of PC-III with BB resistance (SES score 1) during kharif 2019 and yield advantage of +231 and +59.9% over Krishna Hamsa under reproductive-stage drought stress during kharif 2019. (g,h): IL 19211 of PC-IV with blast resistance (SES score 3) and +135% yield advantage over Krishna Hamsa under reproductive-stage drought stress during kharif 2019.

Figure 5.
Performance of the ILs-19,378, 19,379 and 19,031 of phenotypic class-V with resistance to BB in glass house and field screening during kharif 2019 and kharif 2020. ILs can be seen with immune reaction to BB. BB resistance with SES score-1 in (a) IL-19,378 during kharif 2019 glass house screening, (b) IL-19,031 during kharif 2020 field screening, (c) IL-19,379 during kharif 2019 glass house screening and (d) IL-19031 during kharif 2020 glass house screening. TN1 was used as a susceptible check and Improved Samba Mahsuri as a resistant check for BB disease.
2.2.3. Phenotypic Class-III Resistance/Tolerance to BB and Drought
The target resistant alleles of BB were missing in all except IL 19233, 19245 and 19248, and all 10 ILs were resistant to BB (SES score 1) (Figure 4c–f) and recorded +30 to +232% yield advantage over Krishna Hamsa under reproductive-stage drought stress (Supplementary Table S7) and 395 to 693 g/m2 of grain yield under non-stress conditions (Supplementary Table S6). ILs 19241, 19248, 19238 19240, 19232 and 19239 recorded high yield and ILs 19245, 19244 and 19233 recorded comparable yield levels with Krishna Hamsa (Supplementary Table S6).
2.2.4. Phenotypic Class-IV Resistance/Tolerance to Blast and Drought
A total of 42 ILs with one to five introgressions of three genes (xa5, Xa21 and Pi9) and four QTLs (qDTY1.1, qDTY2.1, qDTY3.1 and qDTY12.1) in 25 different combinations were resistant to blast and tolerant to drought (Table 1) (Figure 4g,h). In as many as 19 ILs, resistant alleles of BB were without positive phenotypic reaction, while 17 ILs without resistant allele of blast were with positive phenotype to blast disease (Table 1). In this ILs, percentage yield advantage over Krishna Hamsa under reproductive drought stress varied from +14.88% to +261% (Supplementary Table S7) and yield levels varied from 180 to 1736 g/m2 under non-stress conditions with 19 ILs viz., 19185, 19211, 19249, 19273, 19221, 19191, 19201, 19267, 19263, 19237, 19176, 19195, 19274, 19178, 19264, 19214, 19271, 19250 and 19189, showing a higher yield than Krishna Hamsa under non-stress conditions (Supplementary Table S6).
2.2.5. Phenotypic Class-V Resistance/Tolerance to BB
All the three ILs were resistant to BB with SES score of 1; however, only two ILs—19379 (Figure 5a) and 19460—have resistant alleles of Xa21 for BB along with resistant alleles of Pi54 for blast. None of the target resistant alleles of BB were seen in IL-19046; instead, Bph20 and Bph21 were introgressed in them (Table 2). Except IL-19379, the other two ILs with resistance to BB recorded a high grain yield of 2244 and 631 g/m2 in ILs 19046 and 19460, respectively (Supplementary Table S6).
2.2.6. Phenotypic Class-VI Resistance/Tolerance to Blast
In this class, 84 ILs were resistant to blast with one to five introgressions of 10 gene/QTLs (xa5, xa13, Xa21, Pi9, Pi54, Bph20, Bph21, qDTY2.1, qDTY3.1 and qDTY12.1) combined in 31 different combinations (Table 2) (Figure 6). Out of 83 ILs with resistance to BB, 38 ILs recorded a high yield (>500 g/m2 to 2998 g/m2) and 7 ILs recorded comparable yield levels with Krishna Hamsa under non-stress conditions (Supplementary Table S6).
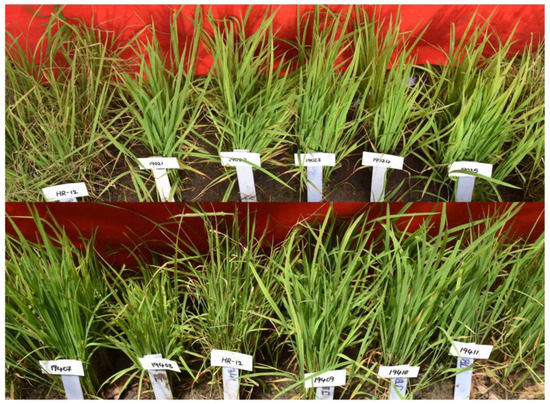
Figure 6.
Performance of the ILs of PC-VI with resistance to blast in universal blast nursery (UBN) during kharif 2019 and kharif 2020. ‘HR-12’ susceptible check (SC) with SES score of 9, and ILs 19021 to 19025 and 19407 to 19411 with blast resistance (SES score—1) during kharif 2019.
2.2.7. Phenotypic Class-VII Tolerance to Drought
The percentage yield advantage in 43 ILs ranged from 23.47% to 276% over the recurrent parent Krishna Hamsa under reproductive-stage drought stress (Supplementary Table S7) and yield levels of 282 to 1282 g/m2 under non-stress conditions (Supplementary Table S6). Introgression of one to five gene/QTLs in 18 different combinations of three genes—xa5, Xa21 and Pi9, and three QTLs—qDTY2.1, qDTY3.1 and qDTY12.1, was observed in these ILs (Table 3). Among the 43 drought-tolerant ILs, 31 ILs recorded high yield and 6 ILs recorded comparable yield with Krishna Hamsa (Supplementary Table S6).
2.3. Background Selection
Parental polymorphism between pairs of Krishna Hamsa and each of the six donors revealed 26 to 58 polymorphic markers out of the 687 SSRs screened between them with a total of 124 polymorphic markers (Supplementary Table S8). A subset of 27 ILs viz., 19202, 19206, 19211, 19019, 19020, 19021, 19022, 19023, 19024, 19026, 19027, 19181, 19182, 19185, 19198, 19247, 19396, 19030, 19241, 19471, 19002, 19004, 19009, 19016, 19017, 19025 and 19186 with days to 50% flowering in the range of 88 to 100 days, plant height of 81 to 87 cm and a long slender grain type similar to Krishna Hamsa were selected and screened with polymorphic background markers. Background selection (BGS) revealed 73.32% to 96.43% recovery of the RP genome in the subset.
3. Discussion
Several high yielding crop varieties have been developed in the past using conventional breeding approaches but combining high yield with multiple stress-resistant/tolerance using this approach is tedious. Use of molecular markers not only enables trait(s) introgression from multiple donors into a single background, but also ensures retaining desirable agronomic traits of the elite recurrent parent by way of marker-assisted breeding (MAB). There are a few recent reports about effective utilization of marker-assisted breeding for the improvement of multiple stress tolerance in the background of the varieties Lalat [5], Naveen [4], Swarna [1] and ASD16 and ADT45 [43]. In the current study, a simultaneous but selective intercrossing strategy, assisted by gene-specific and tightly linked markers for target genes and peak and flanking markers of the QTLs, was deployed to combine multiple biotic and abiotic stress resistance into the genetic background of the elite rice cultivar, Krishna Hamsa.
We employed both molecular and conventional breeding strategies in the development of multiple stress tolerance. Selective intercrossing of multiparent-derived F1’s was adopted in stacking multiple genes/QTL into a single background assisted by foreground selection (FGS) up to IC3F1 followed by stringent phenotypic selection for the desirable agronomic traits in the later segregating generations. Accumulating maximum gene/QTL introgressions in a common background was the main challenge in the entire process, and several hundreds of selective intercrosses in pairs were attempted from 2013 to 2015 in five consecutive seasons. Intercrossing has its own limitations, as it led to constant re-shuffling of the earlier achieved gene/QTL combinations. However, large populations were generated, which enabled the efficient selection of phenotypically acceptable plant type in the populations.
The widespread prevalence of numerous genetically distinct virulent Xoo strains demanded pyramiding of multiple BB-resistant genes, which can provide broad-spectrum, durable resistance in BB-prone rice growing areas. An additional BB-R gene, Xa33, was introgressed into Improved Samba Mahsuri possessing xa5+xa13+Xa21 to enhance and provide broad-spectrum resistance to BB and has been released as ‘DRR Dhan 53’ for cultivation in BB endemic areas of India [7]. Additionally, recently the ICAR-Indian Institute of Rice Research (ICAR-IIRR) released an improved version of ‘Akshaydhan’ as ‘DRR Dhan 58’ introgressed with a single BB-R gene, Xa33, as a cultivar. Among the several resistant genes identified for blast, Pi2, Pi9 and Pi54 were mostly used in the development of blast-resistant cultivars. Improved versions of ‘Swarna’ with intorgession of Pi2 have been released as ‘DRR Dhan 51’ for cultivation [29]. Similarly, in the present study, a combination of three BB-R genes, two blast-R genes, two BPH-R genes and four drought-tolerant QTLs was targeted, but we observed a positive phenotypic response to both BB and blast in t2 ILs, BB in 9 ILs and blast in 64 ILs despite the absence of corresponding R genes. Inheritance of resistance to BB and/or blast in ILs without corresponding R genes could be due to the unknown genes from other parents of the breeding scheme. Genes other than those targeted could be responsible for the tolerance, as multiple parents were involved in the development of ILs. For instance, IRBB60 was used as donor for BB genes, but it also has moderate resistance to blast [44]. Sometimes resistance pathways of different stress genes may influence each other in phenotypic expression. Expression profiling of stress-inducible genes was carried out in rice varieties and reported variability in gene expression patterns indicating the complex network of pathways for regulation of multiple stresses [45]. Detailed studies on crosstalk between defense pathways are essential [46] when genes are pyramided to engineer plants resistant to multiple stress conditions [47].
In the present study, we observed a negative phenotypic response in 9 ILs that were marker positive to blast-R genes, 9 ILs harboring QTLs for drought tolerance, 44 ILs that were marker positive to BPH-R genes and 67 ILs that were marker positive to BB-R genes. Significant variation was observed in the phenotypic response of the ILs in the background of Swarna despite the presence/absence of corresponding R genes and BPH susceptibility in ILs with Bph3 and Bph17, which could be due to the difference in the genome recovery and background interaction of genes/QTLs of the ILs [1]. Negative interaction is specific to the recurrent parent background ‘Triguna’ and might be due to still unidentified modifier gene/s that affect the expression of resistance in plants, having the gene combinations of either Xa21, xa13 and xa5 or Xa21 and xa5 [48]. Lines with single gene introgression of Bph20, Bph21, Bph3 or Bph18 in the background of rice maintainer line ‘Jin 23B’ had a moderate susceptible level [32]. The real effects of genes cannot be compared using the original source genotypes due to the diverse genetic backgrounds [49]. The negative phenotype in marker positive plants in our study can be the effect of varying expression of a specific gene in different background genomes. Many of these genes or QTLs were identified in different varietal or subspecies backgrounds and will not always show a consistent performance in every background; similarly, the varying pest biotypes, pathogen strains or stress load also can explain the negative phenotypic expression. Interestingly, it was observed in our lines that there is a selective exclusion of BPH resistance genes especially when BB genes are present. However, such interactions are not observed with Pi genes, and this requires further validation in different backgrounds to confirm the results. Multiple genes introgression in single background may result in the selective combination of compatible genes or selective exclusion of some combinations due to recombination events or chromosome-related factors [50].
We targeted four QTLs—qDTY1.1, qDTY2.1, qDTY3.1 and qDTY12.1—for reproductive-stage drought tolerance. In previous studies, the introgression of these four QTLs in different combination along with Sub1 for submergence tolerance resulted in development and release of drought- and submergence-tolerant versions of mega varieties like Swarna as CR dhan 801 (qDTY1.1 +qDTY2.1 +qDTY3.1 +Sub1) and CR Dhan 802 (qDTY2.1+qDTY3.1+Sub1) and Samba Mahsuri as DRR Dhan 50 (qDTY2.1+qDTY3.1+Sub1) as new cultivars in various rice growing countries of Southern Asia [41,51,52,53]. In the present study, we identified five ILs viz., IL 19,196 with Xa21+Pi9+qDTY3.1, ILs 19,174 and 19,193 with xa5+Xa21+qDTY3.1 and ILs 19,246 and 19,247 with qDTY2.1 introgressions and yield advantage of 131 to 346 g/m2 under reproductive-stage drought stress over Krishna Hamsa, accounting to a percentage yield advantage of 68%–180% along with BB and blast resistance. Additionally, qDTY2.1 in combination with BB resistance resulted in a yield advantage under reproductive-stage drought stress in nine ILs in the range of 59–446 g/m2 (30–282%). Four QTLs—qDTY1.1, qDTY2.1, qDTY3.1 and qDTY12.1—in different combinations along with blast resistance in 42 ILs resulted in the yield advantage of 29–502 g/m2 (14–261%) over Krishna Hamsa under reproductive-stage drought stress.
Generally, crosses involving landraces or wild species of distant gene pools are difficult due to delayed flowering or no flowering in rabi season. Consideration of elite donor background helped in attempting several hundreds of intercrosses in both the seasons (i.e., twice a year). Using the same recurrent parent in multiple crosses, selective intercrossing with intensive genotypic/phenotypic selection and morphological similarity among recurrent parent and donors has led to the selection of ILs with close proximity to Krishna Hamsa despite using multiple donors and not following the typical marker-assisted backcross breeding strategy. In the present study, a subset of 27 ILs were similar to Krishna Hamsa in plant type features and the data are supported retrospectively by BGS. Stacking of multiple QTLs and genes in one step with the use of a simultaneous crossing program coupled with marker-assisted forward breeding is an effective breeding approach using the elite donors used in the crossing program [5].
Resistance to two or more biotic and abiotic traits was achieved earlier by sequential introgressions. In the background of Improved Samba Mahsuri with inherent xa5, xa13 and Xa21, DRR Dhan 58, DRR Dhan 60 and DRR Dhan 62 were developed and released as cultivars with introgression of Saltol QTL for salinity tolerance, Pup1 QTL for low soil P tolerance and Pi2 and Pi54 for blast resistance, respectively [6,18]. The present study successfully illustrates simultaneous introgression of BB and blast resistance and reproductive-stage drought tolerance, adopting selective intercrossing assisted by foreground selection and stringent phenotyping. Similar to our findings, there are successful examples of simultaneous introgression of multiple QTL and genes for biotic and abiotic stress in rice like blast, BB, gall midge and drought tolerance in the background of Swarna [1] and Naveen [4], as well as blast, BB and drought tolerance in Lalat [5].
In the present study, as mentioned earlier, we observed a negative phenotypic response in a total of 85 ILs marker positive to resistant/tolerant gene/QTLs (67 BB+9 blast + 9 drought). However, we also witnessed a positive phenotypic response to BB and/or blast in as many as 75 ILs despite the absence of the target-resistant allele for the corresponding genes. Marker-assisted selection enables rapid introgression of targeted gene/QTLs and recovery of recurrent parent genome but does not always ensure a positive phenotypic response. Marker-assisted selection-derived genotypes resulting from the breeding strategy involving multiple parents are not always a true reflection of the targeted phenotype due to complex genic interactions and recombination events. Genome shuffling and complex recombination events due to repeated cycles of intercrossing result in an altered phenotype. Our findings suggest the need of stringent phenotyping for the targeted traits and not relying only on FGS, even more so when multiple parents are involved. Stringent phenotyping of all the lines is imperative irrespective of the targeted gene introgression when multiple donors are involved in the breeding scheme. It is also equally important to validate the marker–trait associations for each marker before venturing into such studies. The genotypes other than the donors of targeted genes may harbor unknown resistance genes, which could be the possible cause of inheritance of resistance in the ILs.
The present study demonstrates that simultaneous introgression to multiple biotic and abiotic stress resistance/tolerance can be achieved following marker-assisted selective intercrossing among multiple single cross F1’s and their intercrossed F1’s, followed by forward breeding coupled with stringent phenotyping for the target traits. Maintenance of many plants and progenies in the early segregating generations and phenotypic selection for desirable agronomic traits enabled us to get all combinations of a positive phenotype for target traits with superior plant types. Six promising ILs—IL 19,246 with resistance to BB, blast and tolerance to drought stress, IL 19,241 with resistance to BB and drought, ILs 19,471 and 19,378 with resistance to BB and blast, IL 19,177 with resistance to blast and tolerance to drought, and IL 19001 with resistance to blast—are currently under evaluation in All India Coordinated Improvement Project (AICRIP) trials. Furthermore, several other promising ILs in various phenotypic classes from the present study will be nominated to AICRIP and once available for cultivation will benefit the farmers of boro areas. The 196 ILs from the present study falling into seven major phenotypic classes serve as a repertoire of genetic resource to choose from the myriad of gene/QTL combinations to combine resistance to more multiple abiotic and biotic stresses. The use of these ILs in future breeding programs owing to their elite cultivar background and preferred plant type traits will enable in avoiding background noise and undesirable linkage drag.
4. Materials and Methods
4.1. Plant Material and Introgression Scheme
The present investigation was taken up at the ICAR-Indian Institute of Rice Research (ICAR-IIRR), Rajendranagar, Hyderabad, India. In the introgression scheme, the elite recurrent parent chosen was the variety, ‘Krishna Hamsa’, which was released in 1998 for cultivation in Andhra Pradesh, Tripura, West Bengal and Bihar states of India. It has a mid-early duration (115 to 120 days) and is suitable for cultivation both in kharif (i.e., wet season) and rabi (i.e., dry season) and is tolerant to low temperatures during the vegetative phase; hence, it is suitable for the highly productive Boro season in the Eastern and northeastern states of India. It also has very good grain quality and fetches good price in the local markets.
Initially, Krishna Hamsa was simultaneously crossed with six different donors in kharif, 2013. (1) IRBB60 with xa5, xa13 and Xa21 was used as a source of BB resistance. (2) Tetep possessing Pi9 and Pi54 was used as a source of blast resistance. (3) IR 71033-121-15-B possessing Bph20 and Bph21 was used as a donor for introgression of BPH resistance. (4) IR 96,321-1447-561-B-1 with qDTY1.1 and qDTY3.1, (5) IR 81,896-96-B-B-195 with qDTY2.1 and (6) IR 74,371-46-1-1-13 with qDTY12.1 were used as sources for incorporating drought tolerance in the background of Krishna Hamsa. In rabi 2014, intercrosses were attempted in three pairs of single cross F1’s. Two more cycles of selective intercrossing were attempted in the subsequent seasons by selecting sets of intercrossed F1’s with different gene/QTL combinations up to kharif 2015 to generate IC2F1 and IC3F1 progenies. The IC2F1 and IC3F1 progenies with maximum number of targeted gene/QTLs in various combinations were advanced further by selfing. At every generation of crossing and selfing, individual plants were genotyped using tightly linked foreground markers for the target alleles for selecting plants possessing maximum gene/QTL combinations. Stringent phenotypic selection for desirable agronomic traits was performed from IC2F2 to IC2F6 and IC3F2 to IC3F6. In IC2F6 and IC3F6 generations, lines were both genotyped and phenotyped for target traits. The introgression scheme is graphically represented in Figure 1.
4.2. Genotyping
Genomic DNA was extracted by the cetyl trimethyl ammonium bromide (CTAB) method [54] from fresh leaf samples collected for all the donor and recipient parents and progenies in each generation at 2 weeks after transplanting to check for the presence of targeted gene/QTLs. For all four qDTY varieties (qDTY1.1, qDTY2.1, qDTY3.1 and qDTY12.1) and eight genes (xa5, xa13, Xa21, Pi2, Pi9, Pi54, Bph20 and Bph21), polymerase chain reaction (PCR)-based genotyping was performed. Both peak and flanking markers were used for QTL, while gene-specific and linked markers were used for genes. Here, xa5, xa13, Xa21 and Pi9 are functional markers, whereas Bph20 and Bph21 are gene-linked markers. QTL and genes were surveyed between recurrent parent and donors, and the markers with clearly distinguishable polymorphism between them were selected for foreground selection (FGS) (Supplementary Table S1). Additionally, a set of 687 SSRs spread across the rice genome were surveyed among the parents to identify polymorphic markers for use in background selection (BGS).
PCR reaction was performed in a total volume of 10 μL containing 50 ng of DNA template, 1 μL 10XPCR buffer, 2.5 picoM of each forward and reverse primer, 75 μM of each dNTP and 0.5U of Taq DNA polymerase (Geneilabs, India). The PCR amplification cycle was performed based on standardized annealing temperatures specific to each marker representing the gene/QTL. Products were resolved in a 3.0% agarose gel stained with EtBr and the gel images were captured using Gel documentation unit).
4.3. Phenotyping
4.3.1. Bacterial Leaf Blight Screening
The ILs were evaluated against BB resistance during kharif 2019 and 2020 both under field conditions and glass house conditions using the artificial clip inoculation method along with Krishna Hamsa, Improved Samba Mahsuri (resistant check) and TN1 (susceptible check). A highly virulent, local isolate of BB pathogen, Xanthomonas oryzae pv. oryzae (Xoo) IX-020 maintained at ICAR-IIRR, was used for screening the ILs under field conditions at two locations, the ICAR-IIRR farm, Rajendranagar, Hyderabad, Telangana State, India and the International Crop Research Institute for Semi-Arid Tropics (ICRISAT), Ramachandrapuram, Hyderabad, Telangana State, India in kharif 2019 and the ICAR-IIRR farm, Rajendranagar in 2020, while glass house screening was done at the IIRR farm, Rajendranagar. Under field conditions, leaf tips of three plants in each IL were cut with scissors dipped in a BB suspension of 109 cfu/mL at 40 days after transplanting (DAT), coinciding with the maximum tillering stage. Similarly, under glass house conditions, 45–50-day-old seedlings were inoculated. Inoculated plants were scored at 20 days after inoculation (DAI) on a 0–9 SES scale according to the Standard Evaluation System, IRRI [55], based on lesion length measured from the cut end of the leaf. A plant was classified as resistant if the average lesion length was shorter than 3 cm, moderately resistant if the lesion was >3–6 cm, moderately susceptible if the lesion was >6–9 cm and susceptible if the lesion was longer than >9 cm [7].
4.3.2. Blast Screening
Blast screening was performed in universal blast nursery (UBN) facility at ICAR-IIRR, Rajendranagar during kharif 2019 and kharif 2020. In raised nursery beds with a row spacing of 10 cm, one row of the susceptible check (HR-12) was planted between every four entries and also along the borders to facilitate the build-up of inoculum for uniform and rapid spread of the disease. The inoculums with the concentration of 1 × 105 spore/mL were sprayed onto young seedlings at four leaf stages using fine sprayer, and high relative humidity was maintained for disease development. Tetep was used as resistant check, and plants were scored for blast disease reaction at 20 DAI on a 0–9 SES scale IRRI [53]. SES scores of 0–3 were considered resistant (R), 4–5 as moderately resistant (MR) and 6–9 as susceptible.
4.3.3. Brown Plant Hopper Screening
The seedling box technique was used for screening of ILs against BPH during kharif 2018 at ICAR-IIRR, Rajendranagar. BPH biotype 4 from IIRR Rajendranagar was used to screen BPH under controlled glass house conditions. Newly hatched nymphs or adults were utilized for screening ILs during kharif 2018 along with recurrent parents and TN1 and PTB33 as susceptible (S) and resistant (R) checks, respectively, following the standard protocol [56]. Seedlings were sown in a tray in three replications with two border rows of the S check and one row of R check in the center. Seedlings of 3–4 leaf stage at 10 days after sowing were infested with 6–8 instars nymphs per seedling, and the evaluation of damage was based on SES scores on a scale of 0–9 [57].
4.3.4. Yield Evaluation under Non-Stress and Drought Stress
The ILs were evaluated for their agronomic performance both under control and drought stress situations during kharif 2019 at the IIRR farm, ICRISAT. For the non-stress experiment, the ILs along with their recurrent parents were sown in raised nursery beds on 19 June 2019. Each line was sown in a 1 m row length with spacing of 10 cm between the rows. Seedlings at the age of 31 days were transplanted to the main field on 13 July 2019. Since the drought trial in 2019 was conducted during kharif, sowing and transplanting were delayed by 20 days to prevent the crop experiencing rainy days during reproductive stage. The standard agronomic package of practices was followed while growing the rice plants. Irrigation was supplied continuously in a non-stress experiment until maturity. In drought stress trial, reproductive-stage drought stress was imposed by withholding the irrigation at 30 DAT and draining out the remaining water in the field. Perforated PVC pipes of about 1 m length were inserted diagonally in the drought trial plot to monitor the below ground soil moisture content. The decline in water table depth was measured on a daily basis with a meter scale inserted into the PVC pipes. Lifesaving irrigation was provided for 24 h when the water table level reached 100 cm below the soil surface and most lines were wilted and exhibited severe leaf drying. Then, a second cycle of the stress was initiated that continued until maturity.
In both the trials, the experiment was laid out in an augmented randomized complete block design with repetition of Krishna Hamsa, MTU1010, IR 64 and DRR Dhan 44 as checks. Each entry was planted in two rows with a row length of 3.45 m following the spacing of 20 cm between the rows and 15 cm between the hills. The experimental material was evaluated for days to 50% flowering and grain yield in g/m2.
4.4. Statistical Analysis
Yield data from non-stress and drought stress trials were subjected to statistical analysis using R Studio (Version 3.5.2, R Core Team, Vienna, Austria) using R-scripts. Analysis of variance (ANOVA) and critical difference (CD) at the 1% and 5% level of significance (p-value) were used for assessing the variance and testing of significant differences among the ILs, respectively, and to understand the variation and descriptive statistics, the broad sense heritability (H2) and F test were calculated. CD, representing confidence intervals of all levels for a parameter of interest, was calculated using CD = SE (d) × t, where S.E (d) = √EMS/r and t is the critical value for a specified level of significance and error degrees of freedom. Heritability in broad sense was computed by using the following:
where,
h2 = Heritability (broad sense);
σ 2g = Genotypic variance;
σ 2p = Phenotypic variance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11050622/s1, Figure S1. Foreground selection in introgression lines in IC4F6 generation for targeted genes xa13, Xa21, xa5, Pi54 and Bph20 in the background of Krishna Hamsa. DP-Donor parent, RP-Recurrent parent; Table S1: Selection of polymorphic markers between recurrent parents and donors for foreground selection; Table S2: Analysis of variance for days to 50% flowering and grain yield (g/m2) traits under control conditions (kharif 2019); Table S3: Analysis of variance for days to 50% flowering and grain yield (g/m2) traits under reproductive-stage drought stress conditions (kharif 2019); Table S4: Descriptive statistics and heritability for days to 50% flowering and grain yield (g/m2) traits under control conditions (kharif 2019); Table S5: Descriptive statistics and heritability for days to 50% flowering and grain yield (g/m2) traits under reproductive-stage drought stress conditions (kharif 2019); Table S6: Phenotypic evaluation for yield under control conditions (kharif 2019) and mean SES scores of BB (kharif 2018 (field screening), kharif 2019 (glass house), kharif 2020 (both field and glass house)) and blast (UBN—kharif 2018, kharif 2019 and kharif 2020); Table S7: Phenotypic evaluation for yield under reproductive-stage drought stress (kharif 2019); Table S8: Polymorphic markers between pairs of parents for use in background selection.
Author Contributions
Conceptualization, J.B., V.K.S., A.K. and T.R.; Data curation, J.B.; Formal analysis, J.B. and R.P.; Funding acquisition, J.B., V.K.S., A.K., T.R. and R.M.S.; Investigation, J.B., G.L., L.R.K.J., M.S.P., G.S.L., V.J.L., S.R.I. and U.M.S.; Methodology, J.B., M.S.P., G.S.L., V.J.L., U.M.S., V.K.S., A.K. and T.R.; Project administration, J.B., U.M.S., V.K.S., A.K. and T.R.; Resources, J.B., M.S.P., G.S.L., A.K.J., U.M.S., V.K.S., A.K., T.R., L.V.S.R. and R.M.S.; Software, R.P.; Supervision, J.B., V.K.S., A.K. and T.R.; Validation, J.B., S.R.I. and Y.V.P.V.; Visualization, J.B.; Writing—original draft, J.B., G.L., S.R.I., R.P. and Y.V.P.V.; Writing—review and editing, J.B., D.B., A.K.J. and R.M.S. Writing—review and editing, J.B., M.S.P., D.B. and A.K.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was conducted as part of the project on “Marker-assisted introgression of different traits to develop new generation rice varieties” at ICAR-IIRR funded by Department of Biotechnology (DBT), grant number: BT/AB/01/ IRRI-India/2012 dt. 01.07.2013”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Nagarjuna E., Ashok J., Laxmi Bhavani P., Swathi G. and Vishnuvardhan Reddy are greatly acknowledged for their technical support in field and lab.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dixit, S.; Singh, U.M.; Singh, A.K.; Alam, S.; Venkateshwarlu, C.; Nachimuthu, V.V.; Yadav, S.; Abbai, R.; Selvaraj, R.; Devi, M.N.; et al. Marker Assisted Forward Breeding to Combine Multiple Biotic-Abiotic Stress Resistance/Tolerance in Rice. Rice 2020, 13, 29. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, R.M.; Vishnupriya, M.R.; Biradar, S.K.; Laha, G.S.; Reddy, G.A.; Rani, N.S.; Sarma, N.P.; Sonti, R.V. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 2008, 160, 411–422. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S.; Anandan, A. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 2015, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ramayya, P.J.; Vinukonda, V.P.; Singh, U.M.; Alam, S.; Venkateshwarlu, C.; Vipparla, A.K.; Dixit, S.; Yadav, S.; Abbai, R.; Badri, J.; et al. Marker-assisted forward and backcross breeding for improvement of elite Indian rice variety Naveen for multiple biotic and abiotic stress tolerance. PLoS ONE 2021, 16, e0256721. [Google Scholar] [CrossRef]
- Singh, U.M.; Dixit, S.; Alam, S.; Yadav, S.; Prasanth, V.V.; Singh, A.K.; Venkateshwarlu, C.; Abbai, R.; Vipparla, A.K.; Badri, J.; et al. Marker-assisted forward breeding to develop a drought-, bacterial-leaf-blight-, and blast-resistant rice cultivar. Plant Genome 2021, e20170. [Google Scholar] [CrossRef]
- Swamy, H.K.M.; Anila, M.; Kale, R.R.; Rekha, G.; Bhadana, V.P.; Anantha, M.S.; Brajendra, P.; Balachiranjeevi, C.H.; Hajira, S.K.; Prasanna, B.L.; et al. Marker assisted improvement of low soil phosphorus tolerance in the bacterial blight resistant, fine-grain type rice variety, Improved Samba Mahsuri. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Yugander, A.; Sundaram, R.M.; Singh, K.; Ladhalakshmi, D.; Rao, L.V.S.; Madhav, M.S.; Badri, J.; Prasad, M.S.; Laha, G.S. Incorporation of the novel bacterial blight resistance gene Xa38 into the genetic background of elite rice variety Improved Samba Mahsuri. PLoS ONE 2018, 13, e0198260. [Google Scholar] [CrossRef]
- Kim, S.-M. Identification of novel recessive gene xa44(t) conferring resistance to bacterial blight races in rice by QTL linkage analysis using an SNP chip. Theor. Appl. Genet. 2018, 131, 2733–2743. [Google Scholar] [CrossRef] [Green Version]
- Neelam, K.; Mahajan, R.; Gupta, V.; Bhatia, D.; Gill, B.K.; Komal, R.; Lore, J.S.; Mangat, G.S.; Singh, K. High-resolution genetic mapping of a novel bacterial blight resistance gene xa-45(t) identified from Oryza glaberrima and transferred to Oryza sativa. Theor. Appl. Genet. 2019, 133, 689–705. [Google Scholar] [CrossRef]
- Suh, J.-P.; Jeung, J.-U.; Noh, T.-H.; Cho, Y.-C.; Park, S.-H.; Park, H.-S.; Shin, M.-S.; Kim, C.-K.; Jena, K.K. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 2013, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sundaram, R.M.; Madhav, M.S.; Balachandran, S.M.; Neeraja, C.N.; Mangrauthia, S.K.; Padmavathi, G. Marker-assisted selection for biotic stress resistance in rice. Dir. Rice Res. Tech. Bull. 2014, 79. [Google Scholar]
- Wang, S.; Liu, W.; Lu, D.; Lu, Z.; Wang, X.; Xue, J.; He, X. Distribution of Bacterial Blight Resistance Genes in the Main Cultivars and Application of Xa23 in Rice Breeding. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, S.; Yoshimura, A.; Nelson, R.; Mew, T.W.; Iwata, N. Tagging Genes for Bacterial Blight Resistance in Rice by RFLP and RAPD Markers. New Horizons Nitrogen Fixation 1993, 15, 250–253. [Google Scholar] [CrossRef]
- Zhang, G.; Angeles, E.R.; Abenes, M.L.P.; Huang, N.; Khush, G.S. RAPD and RFLP mapping of the bacterial blight resistance gene xa-13 in rice. Theor. Appl. Genet. 1996, 93, 65–70. [Google Scholar] [CrossRef]
- Ronald, P.C.; Albano, B.; Tabien, R.; Abenes, L.; Wu, K.-S.; McCouch, S.; Tanksley, S.D. Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol. Genet. Genomics 1992, 236, 113–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, N.; Angeles, E.R.; Domingo, J.; Magpantay, G.; Singh, S.; Zhang, G.; Kumaravadivel, N.; Bennett, J.; Khush, G.S. Pyramiding of bacterial blight resistance genes in rice: Marker-assisted selection using RFLP and PCR. Theor. Appl. Genet. 1997, 95, 313–320. [Google Scholar] [CrossRef]
- Singh, S.; Sidhu, J.S.; Huang, N.; Vikal, Y.; Li, Z.; Brar, D.S.; Dhaliwal, H.S.; Khush, G.S. Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theor. Appl. Genet. 2001, 102, 1011–1015. [Google Scholar] [CrossRef]
- ICAR-Indian Institute of Rice Research. Progress Report, 2020, Varietal Improvement; All India Coordinated Rice Improvement Project; Rajendranagar, Hyderabad-30, TS; ICAR-Indian Institute of Rice Research: New Delhi, India, 2021; Volume 1, p. 542.
- ICAR-Indian Institute of Rice Research. Progress Report, 2019, Varietal Improvement; India Coordinated Rice Improvement Project, ICAR-Indian Institute of Rice Research, Rajendranagar, Hyderabad-30, TS; ICAR-Indian Institute of Rice Research: New Delhi, India, 2020; Volume 1, p. 683.
- Babujee, L.; Gnanamanickam, S.S. Molecular tools for characterization of rice blast pathogen (Magnaporthe grisea) population and molecular marker-assisted breeding for disease resistance. Curr. Sci. 2000, 78, 248–257. [Google Scholar]
- Thulasinathan, T.; Kambale, R.; Ayyenar, B.; Manonmani, S.; Muthurajan, R. Evaluation of blast resistance genes Pi9 and Pi54 in rice against local isolates of Tamil Nadu. Electron. J. Plant Breed. 2020, 11, 1153–1158. [Google Scholar]
- Alam, S.; Imam, J.; Nitin, M.; Prasad, C.; Variar, M. Molecular Screening of Blast Resistance Gene Pi2 in Indian Rice Landraces (Oryza sativa L.) and its Verification by Virulence Analysis. Proc. Natl. Acad. Sci. India Sect. B Boil. Sci. 2017, 87, 67–72. [Google Scholar] [CrossRef]
- Rajashekara, H.; Ellur, R.K.; Khanna, A.P.U.R.V.A.; Nagarajan, M.; Gopalakrishnan, S.; Singh, A.K.; Sharma, P.; Sharma, T.R.; Singh, U.D. Inheritance of blast resistance and its allelic relationship with five major R genes in a rice landrace ‘Vanasurya’. Indian Phytopathol. 2014, 67, 365–369. [Google Scholar]
- Sharma, T.R.; Rai, A.K.; Gupta, S.K.; Vijayan, J.; Devanna, B.N.; Ray, S. Rice Blast Management Through Host-Plant Resistance: Retrospect and Prospects. Agric. Res. 2012, 1, 37–52. [Google Scholar] [CrossRef] [Green Version]
- Ning, X.; Yunyu, W.; Aihong, L. Strategy for Use of Rice Blast Resistance Genes in Rice Molecular Breeding. Rice Sci. 2020, 27, 263–277. [Google Scholar] [CrossRef]
- Mackill, D.J.; Bonman, J.M. Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 1992, 82, 746–749. [Google Scholar] [CrossRef]
- Amante-Bordeos, A.; Sitch, L.A.; Nelson, R.; Dalmacio, R.D.; Oliva, N.P.; Aswidinnoor, H.; Leung, H. Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice, Oryza sativa. Theor. Appl. Genet. 1992, 84, 345–354. [Google Scholar] [CrossRef]
- Sharma, T.R.; Shanker, P.; Singh, B.K.; Jana, T.K.; Madhav, M.S.; Gaikwad, K.; Plaha, P.; Rathour, R. Molecular Mapping of Rice Blast Resistance Gene Pi-kh in the Rice Variety Tetep. J. Plant Biochem. Biotechnol. 2005, 14, 127–133. [Google Scholar] [CrossRef]
- ICAR-Indian Institute of Rice Research. Progress Report, 2016, Varietal Improvement; All India Coordinated Rice Improvement Project ICAR-Indian Institute of Rice Research; Rajendranagar, Hyderabad-500 030, T.S; ICAR-Indian Institute of Rice Research: New Delhi, India, 2017; Volume 1.
- Eashkani, S.; Rafii, M.Y.; Eshabanimofrad, M.; Miah, G.; Esahebi, M.; Eazizi, P.; Tanweer, F.A.; Eakhtar, M.S.; Enasehi, A. Molecular Breeding Strategy and Challenges Towards Improvement of Blast Disease Resistance in Rice Crop. Front. Plant Sci. 2015, 6, 886. [Google Scholar] [CrossRef] [Green Version]
- Rahman, L.; Jiang, W.; Chu, S.H.; Qiao, Y.; Ham, T.-H.; Woo, M.-O.; Lee, J.; Khanam, M.S.; Chin, J.-H.; Jeung, J.-U.; et al. High-resolution mapping of two rice brown planthopper resistance genes, Bph20(t) and Bph21(t), originating from Oryza minuta. Theor. Appl. Genet. 2009, 119, 1237–1246. [Google Scholar] [CrossRef]
- Jiang, H.; Hu, J.; Li, Z.; Liu, J.; Gao, G.; Zhang, Q.; Xiao, J.; He, Y. Evaluation and breeding application of six brown planthopper resistance genes in rice maintainer line Jin 23B. Rice 2018, 11, 1–11. [Google Scholar] [CrossRef]
- Pandey, S.; Bhandari, H.; Sharan, R.; Naik, D.; Taunk, S.K.; Sastri, A.S.R.A.S. Economic Costs of Drought and Rainfed Rice Farmers’ Coping Mechanisms in Eastern India; Final Project Report; IRRI: Laguna, Philippines, 2005. [Google Scholar]
- Barnabás, B.; Jäger, K.; Fehér, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008, 31, 11–38. [Google Scholar] [CrossRef]
- Mishra, K.K.; Vikram, P.; Yadaw, R.B.; Swamy, B.M.; Dixit, S.; Cruz, M.T.S.; Maturan, P.; Marker, S.; Kumar, A. qDTY12.1: A locus with a consistent effect on grain yield under drought in rice. BMC Genet. 2013, 14, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swamy, B.P.M.; Ahmed, H.U.; Henry, A.; Mauleon, R.; Dixit, S.; Vikram, P.; Tilatto, R.; Verulkar, S.B.; Perraju, P.; Mandal, N.P.; et al. Genetic, Physiological, and Gene Expression Analyses Reveal That Multiple QTL Enhance Yield of Rice Mega-Variety IR64 under Drought. PLoS ONE 2013, 8, e62795. [Google Scholar] [CrossRef]
- Venuprasad, R.; Dalid, C.O.; Del Valle, M.; Zhao, D.; Espiritu, M.; Cruz, M.T.S.; Amante, M.; Kumar, A.; Atlin, G.N. Identification and characterization of large-effect quantitative trait loci for grain yield under lowland drought stress in rice using bulk-segregant analysis. Theor. Appl. Genet. 2009, 120, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Venuprasad, R.; Bool, M.E.; Quiatchon, L.; Cruz, M.T.S.; Amante, M.; Atlin, G.N. A large-effect QTL for rice grain yield under upland drought stress on chromosome 1. Mol. Breed. 2011, 30, 535–547. [Google Scholar] [CrossRef]
- Yadaw, R.B.; Dixit, S.; Raman, A.; Mishra, K.K.; Vikram, P.; Swamy, B.M.; Cruz, M.T.S.; Maturan, P.T.; Pandey, M.; Kumar, A. A QTL for high grain yield under lowland drought in the background of popular rice variety Sabitri from Nepal. Field Crop. Res. 2013, 144, 281–287. [Google Scholar] [CrossRef]
- Shamsudin, N.A.A.; Swamy, B.P.M.; Ratnam, W.; Cruz, M.T.S.; Sandhu, N.; Raman, A.K.; Kumar, A. Pyramiding of drought yield QTLs into a high quality Malaysian rice cultivar MRQ74 improves yield under reproductive stage drought. Rice 2016, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sandhu, N.; Dixit, S.; Swamy, B.P.M.; Raman, A.; Kumar, S.; Singh, S.P.; Yadaw, R.B.; Singh, O.N.; Reddy, J.N.; Anandan, A.; et al. Marker Assisted Breeding to Develop Multiple Stress Tolerant Varieties for Flood and Drought Prone Areas. Rice 2019, 12, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Teh, S.L.; Ramirez, J.F.; Clark, M.; Gadoury, D.M.; Sun, Q.; Cadle-Davidson, L.; Luby, J.J. Genetic dissection of powdery mildew resistance in interspecific half-sib grapevine families using SNP-based maps. Mol. Breed. 2017, 37, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ramalingam, J.; Savitha, P.; Alagarasan, G.; Saraswathi, R.; Chandrababu, R. Functional Marker Assisted Improvement of Stable Cytoplasmic Male Sterile Lines of Rice for Bacterial Blight Resistance. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Oladosu, Y.; Kolapo, K.; Musa, I.; Halidu, J.; Muhammad, I.; Ahmed, M. Marker-Assisted Introgression of Multiple Resistance Genes Confers Broad Spectrum Resistance against Bacterial Leaf Blight and Blast Diseases in PUTRA-1 Rice Variety. Agronomy 2019, 10, 42. [Google Scholar] [CrossRef] [Green Version]
- Basu, S.; Roychoudhury, A. Expression Profiling of Abiotic Stress-Inducible Genes in response to Multiple Stresses in Rice (Oryza sativaL.) Varieties with Contrasting Level of Stress Tolerance. BioMed Res. Int. 2014, 2014, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Moumeni, A.; Attia, K.; Venuprasad, R.; Serraj, R.; Kumar, A.; Leung, H.; Islam, A.K.M.R.; et al. Comparative transcriptome analysis of AP2/EREBP gene family under normal and hormone treatments, and under two drought stresses in NILs setup by Aday Selection and IR64. Mol. Genet. Genom. 2011, 287, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saidi, A.; Hajibarat, Z. Characterization of cis-elements in hormonal stress-responsive genes in Oryza sativa. Asia Pac. J. Mol. Biol. Biotechnol. 2019, 27, 95–102. [Google Scholar] [CrossRef]
- Sundaram, R.M.; Vishnupriya, M.R.; Laha, G.S.; Rani, N.S.; Rao, P.S.; Balachandran, S.M.; Reddy, G.A.; Sarma, N.P.; Sonti, R.V. Introduction of bacterial blight resistance into Triguna, a high yielding, mid-early duration rice variety. Biotechnol. J. 2009, 4, 400–407. [Google Scholar] [CrossRef]
- Xiao, C.; Hu, J.; Ao, Y.-T.; Cheng, M.-X.; Gao, G.-J.; Zhang, Q.-L.; He, G.-C.; He, Y.Q. Development and evaluation of near-isogenic lines for brown planthopper resistance in rice cv. 9311. Sci. Rep. 2016, 6, 38159. [Google Scholar] [CrossRef] [Green Version]
- Haque, A.; Rafii, M.Y.; Yusoff, M.M.; Ali, N.S.; Yusuff, O.; Datta, D.R.; Anisuzzaman, M.; Ikbal, M.F. Recent Advances in Rice Varietal Development for Durable Resistance to Biotic and Abiotic Stresses through Marker-Assisted Gene Pyramiding. Sustainability 2021, 13, 10806. [Google Scholar] [CrossRef]
- Kumar, A.; Dixit, S.; Ram, T.; Yadaw, R.B.; Mishra, K.K.; Mandal, N.P. Breeding high-yielding drought-tolerant rice: Genetic variations and conventional and molecular approaches. J. Exp. Bot. 2014, 65, 6265–6278. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Sandhu, N.; Dixit, S.; Yadav, S.; Swamy, B.P.M.; Shamsudin, N.A.A. Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice 2018, 11, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Bhandari, A.; Jayaswal, P.; Yadav, N.; Singh, R.; Singh, Y.; Singh, B.; Singh, N.; Singh, S.; Sevanthi, A.; Rai, V.; et al. Genomics-assisted backcross breeding for infusing climate resilience in high-yielding green revolution varieties of rice. Indian J. Genet. Plant Breed. 2019, 79, 160–170. [Google Scholar] [CrossRef]
- Doyle, J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. [Google Scholar] [CrossRef]
- Standard Evaluation System (SES) for Rice, 5th ed.; IRRI: Los Baños, Philippines, 2013; Volume 5.
- Kalode, M.B.; Viswanathan, P.K.; Seshu, D.V. Standard test to characterize host plant resistance to brown planthopper in rice. Indian J. Plant Prot. 1975, 3, 204–206. [Google Scholar]
- Standard Evaluation System (SES) for Rice; IRRI: Los Baños, Philippines, 2002; p. 56.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).