Impacts of Mn, Fe, and Oxidative Stressors on MnSOD Activation by AtMTM1 and AtMTM2 in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Generation of AtMnSOD-Overexpressing Plants and Characterisation of AtMnSOD T-DNA Insertion Mutants
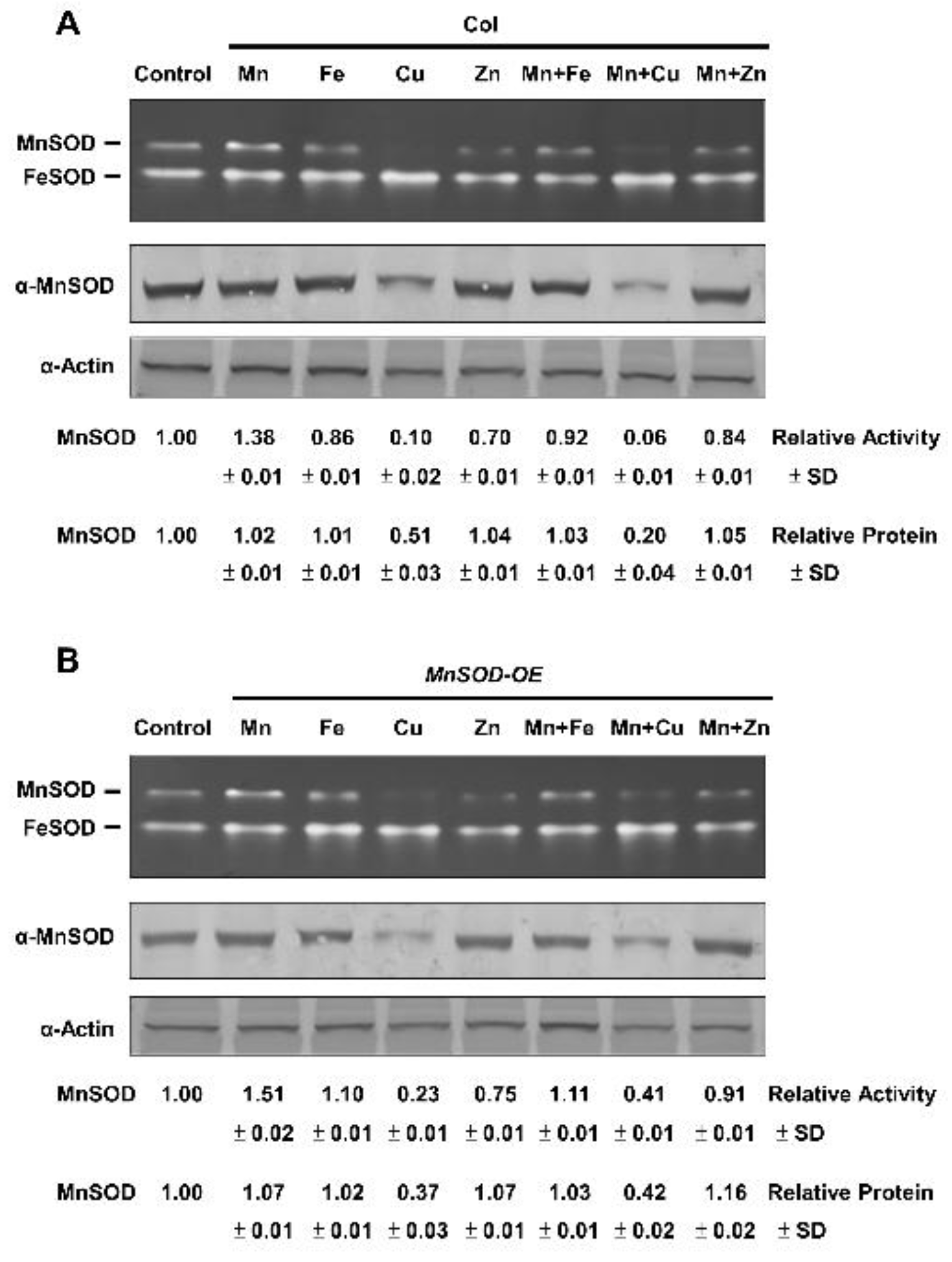
2.2. Effect of Mn and Fe on Transgenic AtMnSOD-Overexpressing Plants
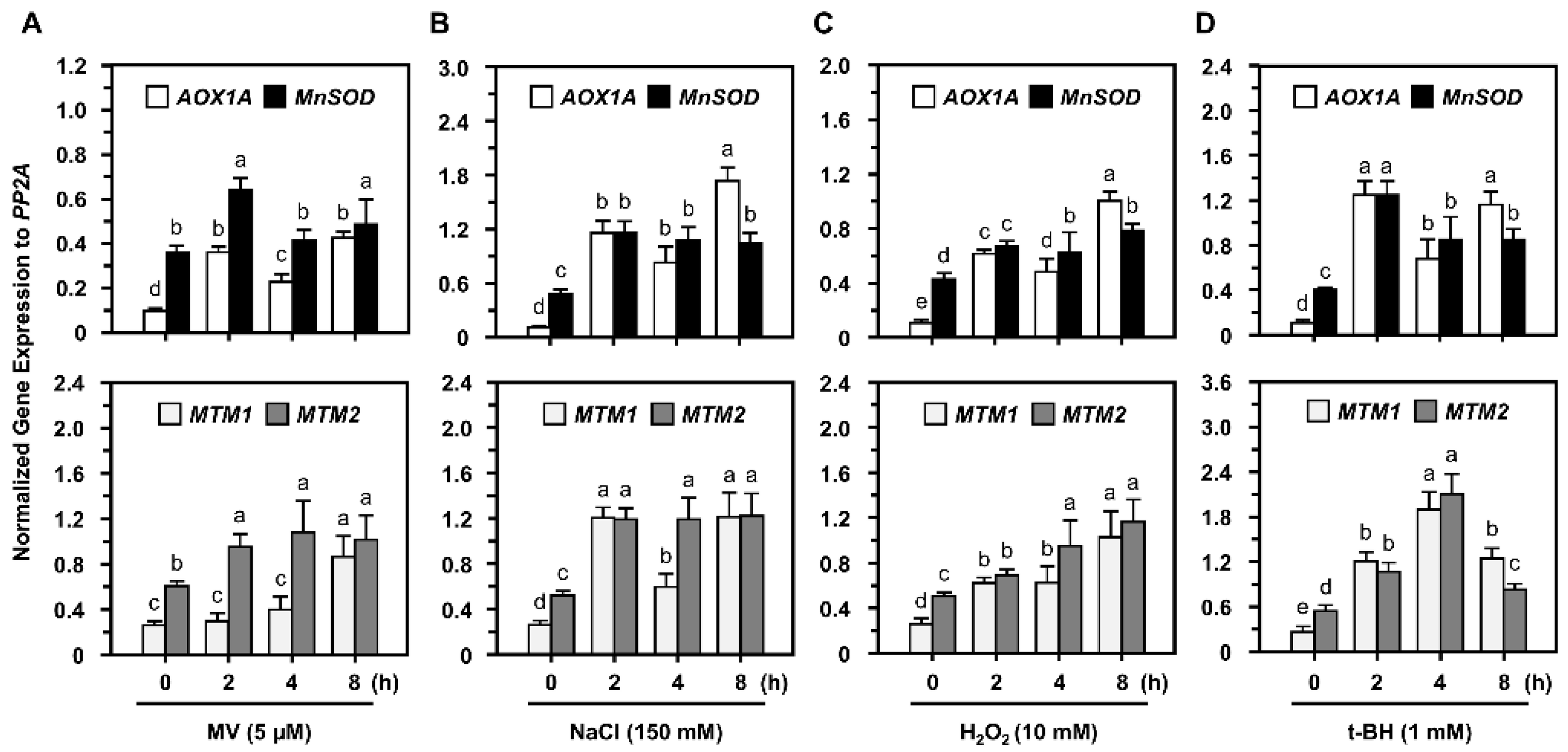
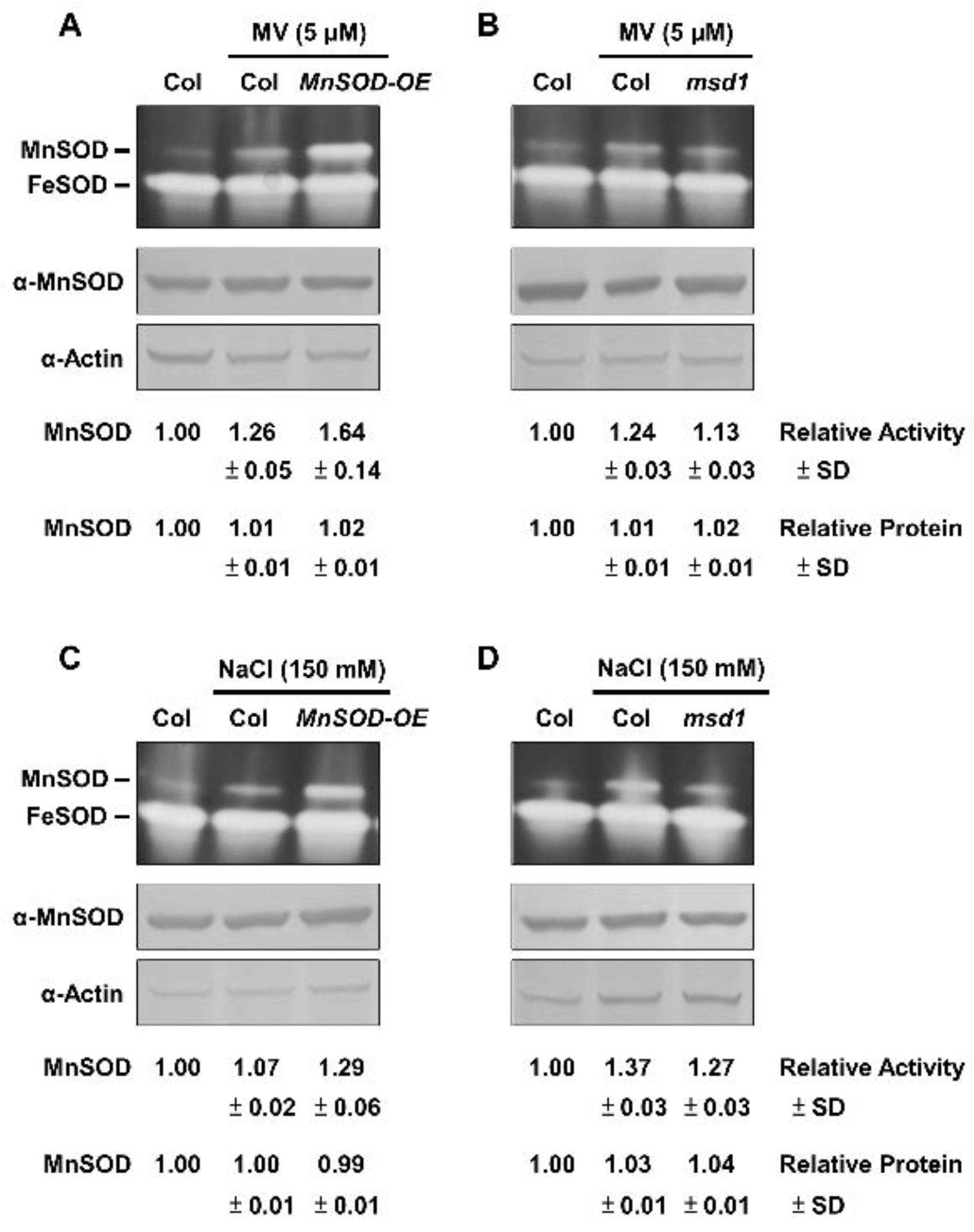
2.3. Post-Translational Regulation of MnSOD through Oxidative Stressors
2.4. Role of MnSOD in the Control of Primary Root Growth during Oxidative Stress
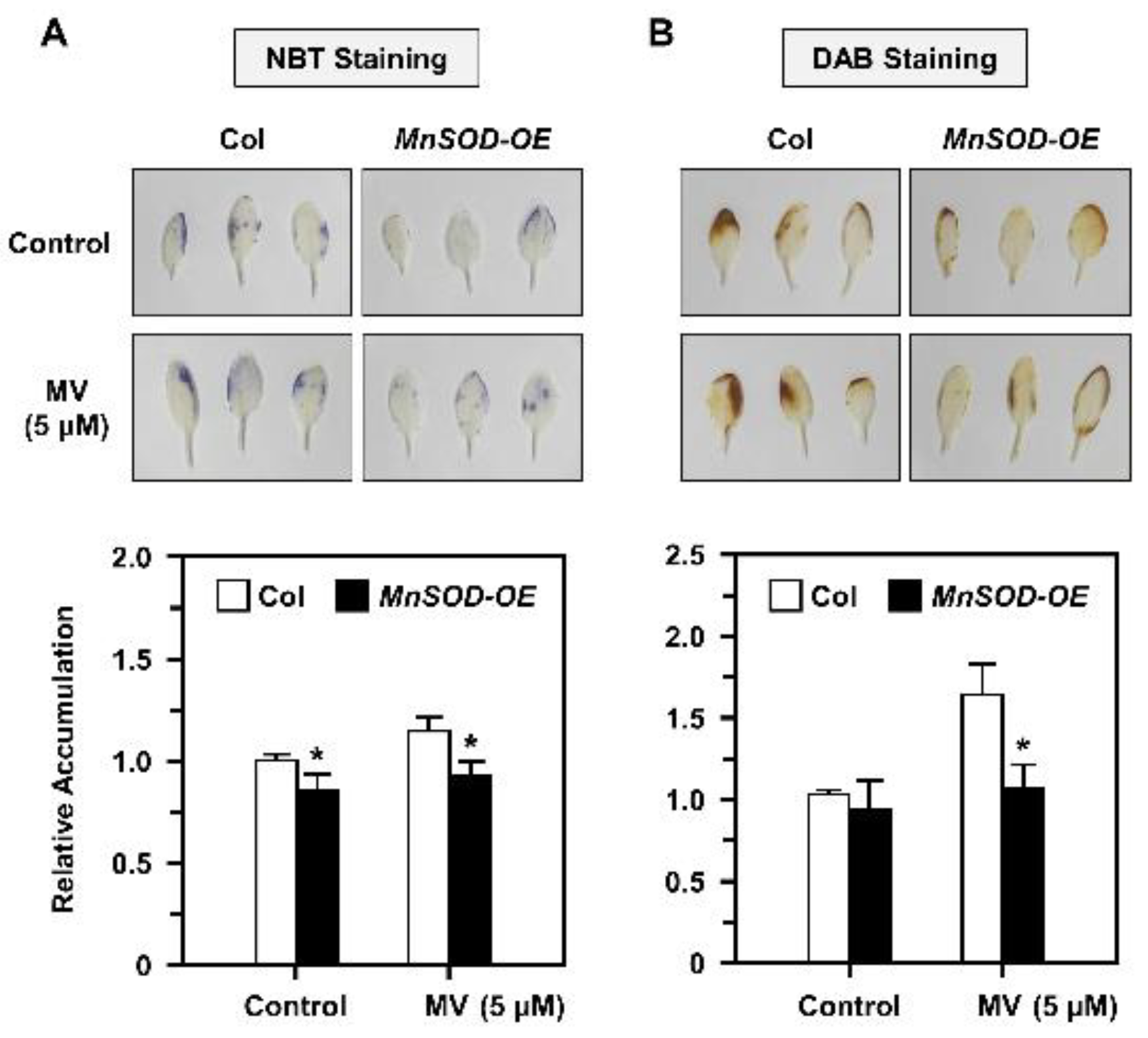
2.5. O2•− and H2O2 Accumulation and Distribution in AtMnSOD-Overexpressing Plants under Stress
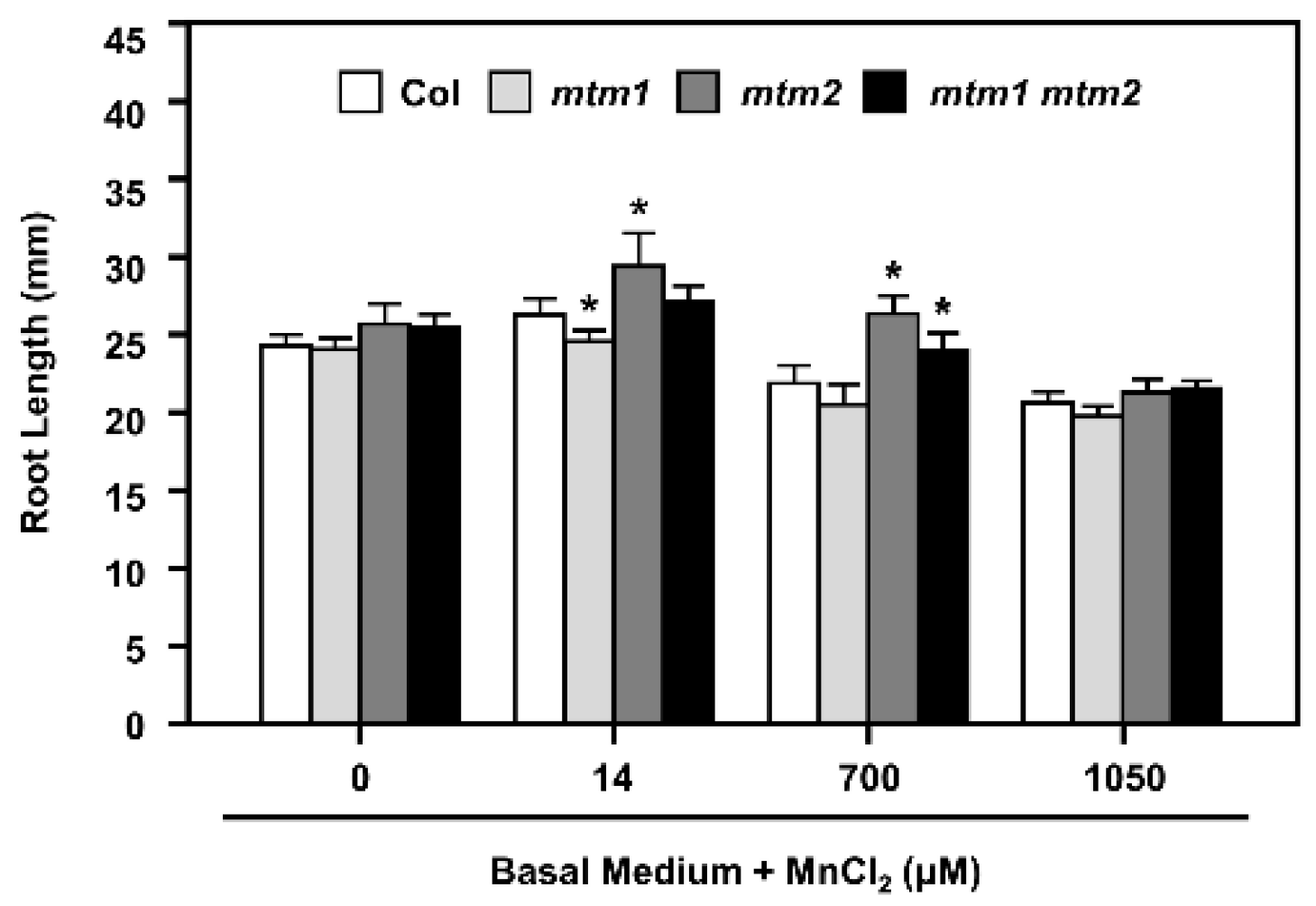
2.6. Root-Length Phenotype of AtMTM1- and AtMTM2-Mutated Seedlings Analysed by Extra Mn Supply
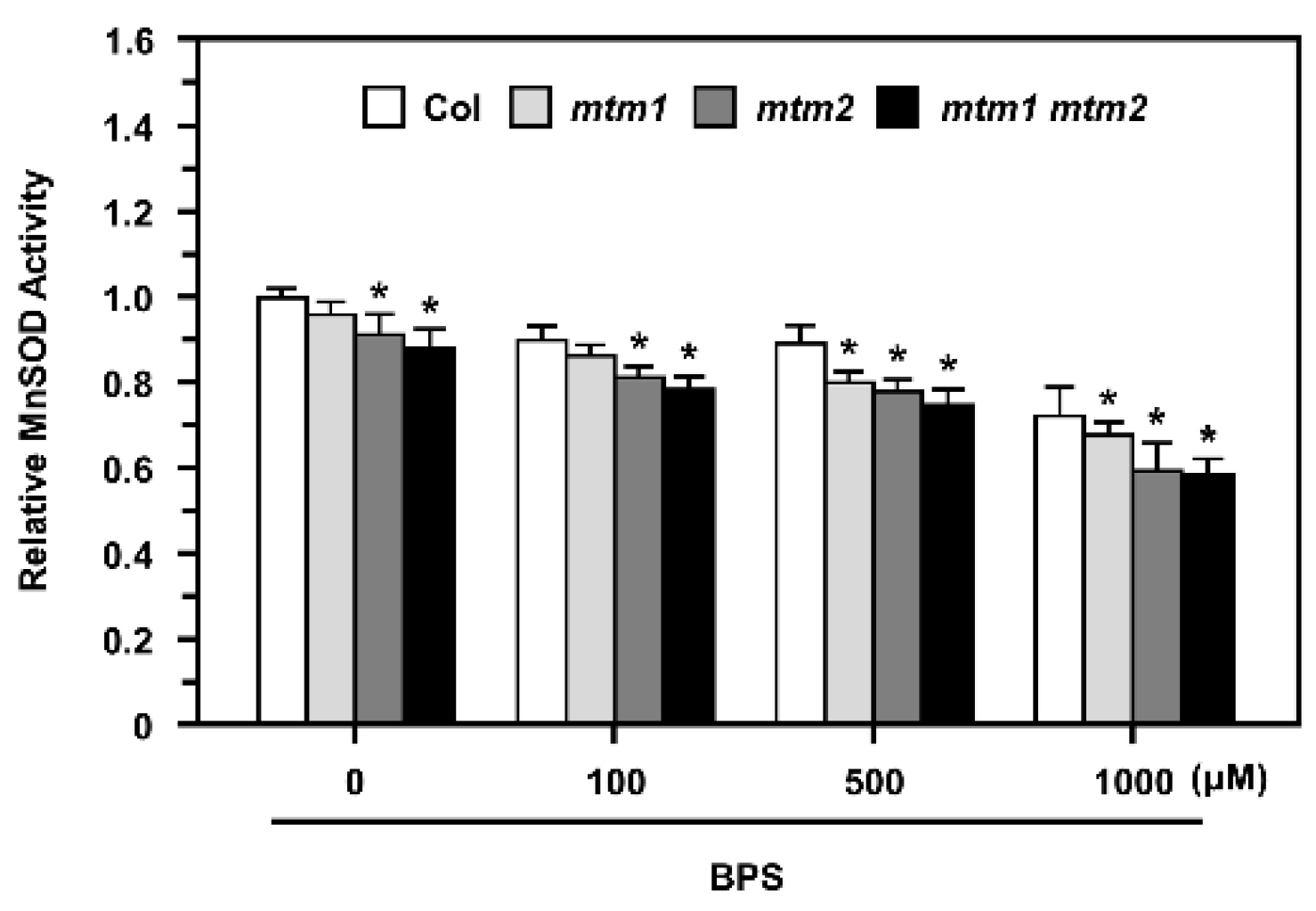
2.7. MnSOD Activity in AtMTM1- and AtMTM2-Mutated Protoplasts after Fe Chelation
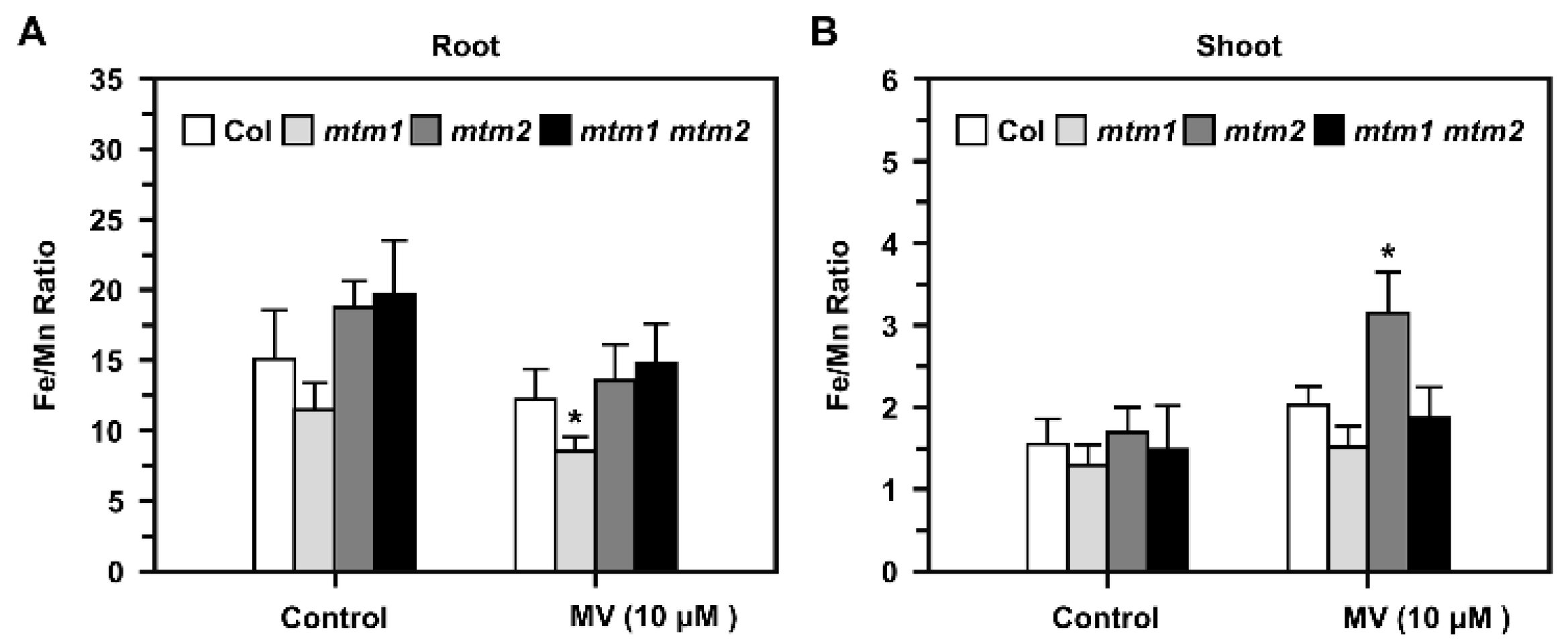
2.8. Fe/Mn Ratio in AtMTM1- and AtMTM2-Mutant Seedlings Treated with Methyl Viologen (MV)
3. Discussion
4. Materials and Methods
4.1. Plants and Growth Conditions
4.2. Generation of AtMnSOD-Overexpressing Plants
4.3. Genotyping, RT-qPCR, In-Gel SOD Activity, and Immunoblotting Assay
4.4. Analysis of O2•− and H2O2 Accumulations by Nitrobule Tetrazolium (NBT) and Diaminobenzidine (DAB) Staining
4.5. Root Length Assay in Response to Oxidative Stressors and Mn Treatment
4.6. Fe chelation in Arabidopsis Protoplasts
4.7. Fe/Mn Ratios in Roots and Shoots in Response to MV
4.8. Statistical Analysis
4.9. PCR Primers and GenBank Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fridovich, I. Superoxide dismutases. Adv. Enzymol. Relat. Areas Mol. Biol. 1986, 58, 61–97. [Google Scholar] [PubMed]
- McCord, J.M.; Fridovich, I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar] [CrossRef]
- Pauling, L. The discovery of the superoxide radical. Trends Biochem. Sci. 1979, 4, N270–N271. [Google Scholar] [CrossRef]
- Bowler, C.; Camp, W.V.; Montagu, M.V.; Inzé, D. Superoxide dismutase in plants. Crit. Rev. Plant Sci. 1994, 13, 199–218. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Monde, R.A.; Last, R.L. Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.F. Superoxide dismutases: Ancient enzymes and new insights. FEBS Lett. 2012, 586, 585–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinalducci, S.; Murgiano, L.; Zolla, L. Redox proteomics: Basic principles and future perspectives for the detection of protein oxidation in plants. J. Exp. Bot. 2008, 59, 3781–3801. [Google Scholar] [CrossRef] [Green Version]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [Green Version]
- Bowler, C.; Montagu, M.; Inzé, D. Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Møller, I.M. Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. Rep. 2001, 52, 561–591. [Google Scholar] [CrossRef] [Green Version]
- Taylor, N.L.; Tan, Y.F.; Jacoby, R.P.; Millar, A.H. Abiotic environmental stress induced changes in the Arabidopsis thaliana chloroplast, mitochondria and peroxisome proteomes. J. Proteom. 2009, 72, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ying, Y.; Chen, J.; Wang, X. Transgenic Arabidopsis overexpressing Mn-SOD enhanced salt-tolerance. Plant Sci. 2004, 167, 671–677. [Google Scholar] [CrossRef]
- Xi, D.M.; Liu, W.S.; Yang, G.D.; Wu, C.A.; Zheng, C.C. Seed-specific overexpression of antioxidant genes in Arabidopsis enhances oxidative stress tolerance during germination and early seedling growth. Plant Biotechnol. J. 2010, 8, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.J.; Lehmann, M.; Schwarzländer, M.; Baxter, C.J.; Sienkiewicz-Porzucek, A.; Williams, T.C.R.; Schauer, N.; Fernie, A.R.; Fricker, M.D.; Ratcliffe, R.G.; et al. Decreased in manganese superoxide dismutase leads to reduced root growth and affects tricarboxylic acid cycle flux and mitochondrial redox homeostasis. Plant Physiol. 2008, 147, 101–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Tinaut, M.C.; Leal, A.; Recalde Martínez, L. Iron-manganese interaction and its relation to boron levels in tomato plants. Plant Soil 1980, 55, 377–388. [Google Scholar] [CrossRef]
- Tanaka, A.; Navasero, S.A. Interaction between iron and manganese in the rice plant. Soil Sci. Plant Nutr. 1966, 12, 29–33. [Google Scholar] [CrossRef]
- Twyman, E.S. The iron and manganese requirements of plants. New Phytol. 1951, 50, 210–226. [Google Scholar] [CrossRef]
- Hassan, H.M.; Sun, H.C. Regulatory roles of Fnr, Fur, and Arc in expression of manganese-containing superoxide dismutase in Escherichia coli. Proc. Natl. Acad. Sci. USA 1992, 89, 3217–3221. [Google Scholar] [CrossRef] [Green Version]
- Privalle, C.; Fridovich, I. Transcriptional and maturational effects of manganese dismutase 2 is Saccharomyces serevisiae requires MTM1, a member of the mitochondrial carrier family. Proc. Natl. Acad. Sci. USA 1992, 100, 10353–10357. [Google Scholar]
- Whittaker, J.W. The irony of manganese superoxide dismutase. Biochem. Soc. Trans. 2003, 31, 1318–1321. [Google Scholar] [CrossRef] [Green Version]
- Whittaker, J.W. Metal uptake by manganese superoxide dismutase. Biochem. Biophys. Acta 2010, 1804, 298–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, D.H.; Tuminello, J.F.; Emptage, M.H. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 1993, 268, 22369–22376. [Google Scholar] [CrossRef]
- Aguirre, J.D.; Culotta, V.C. Battles with iron: Manganese in oxidative stress protection. J. Biol. Chem. 2012, 287, 13541–13548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maringanti, S.; Imlay, J.A. An intracellular iron chelator pleiotropically suppresses enzymatic and growth defects of superoxide dismutase-deficient Escherichia coli. J. Bacteriol. 1999, 181, 3792–3802. [Google Scholar] [CrossRef] [Green Version]
- Luk, E.; Carroll, M.; Baker, M.; Culotta, V.C. Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a membrane of the mitochondrial carrier family. Proc. Natl. Acad. Sci. USA 2003, 100, 10353–10357. [Google Scholar] [CrossRef] [Green Version]
- Luk, E.; Culotta, V.C. Manganese superoxide dismutase in Saccharomyces cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transporter, Smf2p. J. Biol. Chem. 2001, 276, 47556–47562. [Google Scholar] [CrossRef] [Green Version]
- Luk, E.; Yang, M.; Jensen, L.T.; Bourbonnais, Y.; Culotta, V.C. Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 22715–22720. [Google Scholar] [CrossRef] [Green Version]
- Portnoy, M.E.; Liu, X.F.; Culotta, V.C. Saccharomyces cerevisiae expresses three functionally distinct homologues of the Nramp family of metal transporters. Mol. Cell. Biol. 2000, 20, 7893–7902. [Google Scholar] [CrossRef]
- Yang, M.; Cobine, P.A.; Molik, S.; Naranuntarat, A.; Lill, R.; Winge, D.R.; Culotta, V.C. The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 2006, 25, 1775–1783. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.H.; Lin, S.F.; Huang, Y.C.; Huang, C.H.; Kuo, W.Y.; Jinn, T.L. Significance of AtMTM1 and AtMTM2 for mitochondrial MnSOD activation in Arabidopsis. Front. Plant Sci. 2021, 12, 690064. [Google Scholar] [CrossRef]
- Su, Z.; Chai, M.F.; Lu, P.L.; An, R.; Chen, J.; Wang, X.C. AtMTM1, a novel mitochondrial protein, may be involved in activation of the manganese-containing superoxide dismutase in Arabidopsis. Planta 2007, 226, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Alejandro, S.; Höller, S.; Meier, B.; Peiter, E. Manganese in plants: From acquisition to subcellular allocation. Front. Plant Sci. 2020, 26, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, A.; Connolly, E.L. Mitochondrial iron transport and homeostasis in plants. Front. Plant Sci. 2013, 4, 348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M.V.; Fiol, D.F.; Sundaresan, V.; Zabaleta, E.J.; Pagnussat, G.C. oiwa, a female gametophytic mutant impaired in a mitochondrial manganese-superoxide dismutase, reveals crucial roles for reactive oxygen species during embryo sac development and fertilization in Arabidopsis. Plant Cell 2013, 25, 1573–1591. [Google Scholar] [CrossRef] [Green Version]
- Martin, M.V.; Distéfano, A.M.; Zabaleta, E.J.; Pagnussat, G.C. New insights into the functional roles of reactive oxygen species during embryo sac development and fertilization in Arabidopsis thaliana. Plant Signal Behav. 2013, 8, e25714. [Google Scholar] [CrossRef] [Green Version]
- Millar, A.H.; Heazlewood, J.L. Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol. 2003, 131, 443–453. [Google Scholar] [CrossRef] [Green Version]
- Møller, I.M.; Rasmusson, A.G.; Van Aken, O. Plant mitochondria—Past, present and future. Plant J. 2021, 108, 912–959. [Google Scholar] [CrossRef]
- Palmieri, F.; Pierri, C.L.; Grassi, A.D.; Nunes-Nesi, A.; Fernie, A.R. Evolution, structure and function of mitochondrial carriers: A review with new sights. Plant J. 2011, 66, 161–181. [Google Scholar] [CrossRef]
- Picault, N.; Hodges, M.; Palmieri, L.; Palmieri, F. The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci. 2004, 9, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Van Aken, O.; Zhang, B.; Carrie, C.; Uggalla, V.; Paynter, E.; Giraud, E.; Whelan, J. Defining the mitochondrial stress response in Arabidopsis thaliana. Mol. Plant 2009, 2, 1310–1324. [Google Scholar] [CrossRef]
- Pinkham, J.L.; Wang, Z.; Alsina, J. Heme regulates SOD2 transcription by activation and repression in Saccharomyces cerevisiae. Curr. Genet. 1997, 1997, 281–291. [Google Scholar] [CrossRef]
- Naranuntarat, A.; Jensen, L.T.; Panicni, S.; Penner-Hahn, J.E.; Culotta, V.C. The interaction of mitochondrial iron with manganese superoxide dismutase. J. Biol. Chem. 2009, 284, 22633–22640. [Google Scholar] [CrossRef] [Green Version]
- Busi, M.V.; Maliandi, M.V.; Valdez, H.; Clemente, M.; Zabaleta, E.J.; Araya, A.; Gomez-Casati, D.F. Deficiency of Arabidopsis thaliana frataxin alters activity of mitochondrial Fe-S proteins and induces oxidative stress. Plant J. 2006, 48, 873–882. [Google Scholar] [CrossRef]
- Shive, J.W. Significant roles of trace elements in the nutrition of plants. Plant Physiol. 1941, 16, 435–445. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.C.; Lee, W.C.; Guo, W.Y.; Pan, S.M.; Chen, L.J.; Li, H.M.; Jinn, T.L. A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant. Physiol. 2005, 139, 425–436. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acid. Res. 1991, 19, 1349. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Remans, T.; Smeets, K.; Opdenakker, K.; Mathijsen, D.; Vangronsveld, J.; Cuypers, A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 2008, 227, 1343–1349. [Google Scholar] [CrossRef] [Green Version]
- Kuo, W.Y.; Huang, C.H.; Shih, C.; Jinn, T.L. Cellular extract preparation for superoxide dismutase (SOD) activity assay. Bio Protoc. 2013, 3, e811. [Google Scholar] [CrossRef]
- Rodríguez-Celma, J.; Pan, I.C.; Li, W.; Lan, P.; Buckhout, T.J.; Schmidt, W. The transcriptional response of Arabidopsis leaves to Fe deficiency. Front. Plant Sci. 2013, 4, 276–285. [Google Scholar] [CrossRef] [Green Version]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Li, C.; Chang, P.P.; Ghebremariam, K.M.; Qin, L.; Liang, Y. Overexpression of tomato SpMPK3 gene in Arabidopsis rnhances the osmotic tolerance. Biochem. Biophys. Res. Commun. 2014, 443, 357–362. [Google Scholar] [CrossRef]
- Song, X.; Fang, J.; Han, X.; He, X.; Liu, M.; Hu, J.; Zhau, R. Overexpression of quinine reductase from Salix matsudana Koidz enhances salt tolerance in transgenic Arabidopsis thaliana. Gene 2016, 576, 520–527. [Google Scholar] [CrossRef]
- Zhai, Z.; Gayomba, S.R.; Jung, H.I.; Vimalakumari, N.K.; Piñeros, M.; Craft, E.; Rutzke, M.A.; Danku, J.; Lahner, B.; Punshon, T.; et al. OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 2014, 26, 2249–2264. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [Green Version]
- Farid, M.; Ali, S.; Shakoor, M.B.; Bharwana, S.A.; Rizvi, H.; Ehsan, S.; Tauqeer, H.M.; Iftikhar, U.; Hannan, F. EDTA assisted phytoremediation of Cadmium, Lead and Zinc. Intl. J. Agron. Plant. Prod. 2013, 4, 2833–2846. [Google Scholar]
- Kociałkowski, W.Z.; Diatta, J.B.; Grzebisz, W. Evaluation of chelating agents as heavy metals extractants in agricultural soils under threat of contamination. Pol. J. Environ. Stud. 1999, 8, 149–154. [Google Scholar]
- Lin, Y.F.; Liang, H.M.; Yang, S.Y.; Boch, A.; Clemens, S.; Chen, C.C.; Wu, J.F.; Huang, J.L.; Yeh, K.C. Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 2009, 182, 392–404. [Google Scholar] [CrossRef]
- Shanmugam, V.; Lo, J.C.; Wu, C.L.; Wang, S.L.; Lai, C.C.; Connolly, E.L.; Huang, J.L.; Yeh, K.C. Differential expression and regulation of iron-regulated metal transporters in Arabidopsis halleri and Arabidopsis thaliana-the role in zinc tolerance. New Phytol. 2011, 190, 125–137. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.-H.; Jinn, T.-L. Impacts of Mn, Fe, and Oxidative Stressors on MnSOD Activation by AtMTM1 and AtMTM2 in Arabidopsis. Plants 2022, 11, 619. https://doi.org/10.3390/plants11050619
Hu S-H, Jinn T-L. Impacts of Mn, Fe, and Oxidative Stressors on MnSOD Activation by AtMTM1 and AtMTM2 in Arabidopsis. Plants. 2022; 11(5):619. https://doi.org/10.3390/plants11050619
Chicago/Turabian StyleHu, Shu-Hsuan, and Tsung-Luo Jinn. 2022. "Impacts of Mn, Fe, and Oxidative Stressors on MnSOD Activation by AtMTM1 and AtMTM2 in Arabidopsis" Plants 11, no. 5: 619. https://doi.org/10.3390/plants11050619
APA StyleHu, S.-H., & Jinn, T.-L. (2022). Impacts of Mn, Fe, and Oxidative Stressors on MnSOD Activation by AtMTM1 and AtMTM2 in Arabidopsis. Plants, 11(5), 619. https://doi.org/10.3390/plants11050619





