Understanding the Modus Operandi of Class II KNOX Transcription Factors in Secondary Cell Wall Biosynthesis
Abstract
1. Introduction
1.1. KNOX Genes and Encoded KNOX Proteins in Plants
1.2. The Expression Patterns of Class II KNOX Genes in Plants Provide Some Clues about Their Functionality in SCW Formation
1.3. Genetic Mutations in Class II KNOX Genes Further Clarify Their Role in SCW Formation
1.4. Targeted Genetic Manipulations in Class II KNOX Genes Confirm Their Role in SCW Formation
1.5. Transcriptional Network of the Class II KNOX Genes Involved in SCW Formation
1.6. Upstream Top- and Mid-Level Master Switches Control the Expression of KNAT7
1.7. Physical Interactions of Class II KNOX TF Proteins with Other Proteins
2. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BLH | BEL-like homeodomain |
| BiFc | Bimolecular fluorescence complementation |
| CesA | Cellulose synthase |
| ChIP | Chromatin immunoprecipitation assays |
| F5H | Ferulate 5-hydroxylase |
| HD | Homeodomain |
| irx | Irregular xylem |
| KNOX | Knotted-like homeobox |
| OFPs | Ovate family proteins |
| SCW | Secondary cell wall |
| S/G | Syringyl to Guaicyl lignin ratio |
| TALE | Three amino acid loop extension |
| TFs | Transcription factors |
| VIGS | Virus-induced gene silencing |
| Y1H | Yeast-one hybrid |
| Y2H | Yeast-two hybrid |
References
- Mohr, A.; Raman, S. Lessons from First Generation Biofuels and Implications for the Sustainability Appraisal of Second Generation Biofuels. Energy Policy 2013, 63, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Vega-Sánchez, M.E.; Ronald, P.C. Genetic and Biotechnological Approaches for Biofuel Crop Improvement. Curr. Opin. Biotechnol. 2010, 21, 218–224. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mansfield, S.D. Solutions for Dissolution—Engineering Cell Walls for Deconstruction. Curr. Opin. Biotechnol. 2009, 20, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.; Hatcher, C.; Mazarei, M.; Haynes, E.; Baxter, H.; Kim, K.; Hamilton, C.; Sykes, R.; Turner, G.; Davis, M.; et al. Development and Field Assessment of Transgenic Hybrid Switchgrass for Improved Biofuel Traits. Euphytica 2020, 216, 25. [Google Scholar] [CrossRef]
- Bryant, N.D.; Pu, Y.; Tschaplinski, T.J.; Tuskan, G.A.; Muchero, W.; Kalluri, U.C.; Yoo, C.G.; Ragauskas, A.J. Transgenic Poplar Designed for Biofuels. Trends Plant Sci. 2020, 25, 881–896. [Google Scholar] [CrossRef]
- de Vries, L.; Guevara-Rozo, S.; Cho, M.; Liu, L.-Y.; Renneckar, S.; Mansfield, S.D. Tailoring Renewable Materials via Plant Biotechnology. Biotechnol. Biofuels 2021, 14, 167. [Google Scholar] [CrossRef]
- Demura, T.; Ye, Z.-H. Regulation of Plant Biomass Production. Curr. Opin. Plant Biol. 2010, 13, 298–303. [Google Scholar] [CrossRef]
- Ko, J.-H.; Kim, W.-C.; Han, K.-H. Ectopic Expression of MYB46 Identifies Transcriptional Regulatory Genes Involved in Secondary Wall Biosynthesis in Arabidopsis. Plant J. 2009, 60, 649–665. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. Complexity of the Transcriptional Network Controlling Secondary Wall Biosynthesis. Plant Sci. 2014, 229, 193–207. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. Secondary Cell Walls: Biosynthesis, Patterned Deposition and Transcriptional Regulation. Plant Cell Physiol. 2015, 56, 195–214. [Google Scholar] [CrossRef]
- Ehlting, J.; Mattheus, N.; Aeschliman, D.S.; Li, E.; Hamberger, B.; Cullis, I.F.; Zhuang, J.; Kaneda, M.; Mansfield, S.D.; Samuels, L.; et al. Global Transcript Profiling of Primary Stems from Arabidopsis Thaliana Identifies Candidate Genes for Missing Links in Lignin Biosynthesis and Transcriptional Regulators of Fiber Differentiation: Global Transcript Profiling of Stems. Plant J. 2005, 42, 618–640. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.-H. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Bhargava, A.; Qiang, W.; Friedmann, M.C.; Forneris, N.; Savidge, R.A.; Johnson, L.A.; Mansfield, S.D.; Ellis, B.E.; Douglas, C.J. The Class II KNOX Gene KNAT7 Negatively Regulates Secondary Wall Formation in Arabidopsis and Is Functionally Conserved in Populus. New Phytol. 2012, 194, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.K.; Nookaraju, A.; Fujino, T.; Pattathil, S.; Joshi, C.P. Virus-Induced Gene Silencing (VIGS)-Mediated Functional Characterization of Two Genes Involved in Lignocellulosic Secondary Cell Wall Formation. Plant Cell Rep. 2016, 35, 2353–2367. [Google Scholar] [CrossRef]
- Wang, S.; Yamaguchi, M.; Grienenberger, E.; Martone, P.T.; Samuels, A.L.; Mansfield, S.D. The Class II KNOX Genes KNAT3 and KNAT7 Work Cooperatively to Influence Deposition of Secondary Cell Walls That Provide Mechanical Support to Arabidopsis Stems. Plant J. 2020, 101, 293–309. [Google Scholar] [CrossRef]
- Qin, W.; Yin, Q.; Chen, J.; Zhao, X.; Yue, F.; He, J.; Yang, L.; Liu, L.; Zeng, Q.; Lu, F.; et al. The Class II KNOX Transcription Factors KNAT3 and KNAT7 Synergistically Regulate Monolignol Biosynthesis in Arabidopsis. J. Exp. Bot. 2020, 71, 5469–5483. [Google Scholar] [CrossRef]
- Ahlawat, Y.K.; Nookaraju, A.; Harman-Ware, A.E.; Doeppke, C.; Biswal, A.K.; Joshi, C.P. Genetic Modification of KNAT7 Transcription Factor Expression Enhances Saccharification and Reduces Recalcitrance of Woody Biomass in Poplars. Front. Plant Sci. 2021, 12, 762067. [Google Scholar] [CrossRef]
- Sakakibara, K.; Ando, S.; Yip, H.K.; Tamada, Y.; Hiwatashi, Y.; Murata, T.; Deguchi, H.; Hasebe, M.; Bowman, J.L. KNOX2 Genes Regulate the Haploid-to-Diploid Morphological Transition in Land Plants. Science 2013, 339, 1067–1070. [Google Scholar] [CrossRef]
- Desplan, C.; Theis, J.; O’Farrell, P.H. The Sequence Specificity of Homeodomain-DNA Interaction. Cell 1988, 54, 1081–1090. [Google Scholar] [CrossRef]
- Burglin, T.R. Analysis of TALE Superclass Homeobox Genes (MEIS, PBC, KNOX, Iroquois, TGIF) Reveals a Novel Domain Conserved between Plants and Animals. Nucleic Acids Res. 1997, 25, 4173–4180. [Google Scholar] [CrossRef]
- Kerstetter, R.; Vollbrecht, E.; Lowe, B.; Veit, B.; Yamaguchi, J.; Hake, S. Sequence Analysis and Expression Patterns Divide the Maize Knotted1-like Homeobox Genes into Two Classes. Plant Cell 1994, 6, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Bharathan, G.; Janssen, B.J.; Kellogg, E.A.; Sinha, N. Phylogenetic Relationships and Evolution of the KNOTTED Class of Plant Homeodomain Proteins. Mol. Biol. Evol. 1999, 16, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Shi, A.; Wu, D.; Weng, Y.; Qin, J.; Ravelombola, W.S.; Shu, X.; Zhou, W. Genome-Wide Identification, Classification and Evolutionary Expansion of KNOX Gene Family in Rice (Oryza Sativa) and Populus (Populustrichocarpa). Am. J. Plant Sci. 2018, 9, 1071–1092. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, N.; Hao, P.; Sun, H.; Wang, C.; Ma, L.; Wang, H.; Zhang, X.; Wei, H.; Yu, S. Genome-Wide Identification and Characterization of TALE Superfamily Genes in Cotton Reveals Their Functions in Regulating Secondary Cell Wall Biosynthesis. BMC Plant Biol. 2019, 19, 432. [Google Scholar] [CrossRef] [PubMed]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The Developmental Gene Knotted-1 Is a Member of a Maize Homeobox Gene Family. Nature 1991, 350, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Hake, S.; Smith, H.M.S.; Holtan, H.; Magnani, E.; Mele, G.; Ramirez, J. The Role of Knox Genes in Plant Development. Annu. Rev. Cell Dev. Biol. 2004, 20, 125–151. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Sato, Y.; Matsuoka, M. Function of KNOX Homeodomain Proteins in Plant Development. Plant Biotechnol. 2001, 18, 85–92. [Google Scholar] [CrossRef]
- Kimura, S.; Koenig, D.; Kang, J.; Yoong, F.Y.; Sinha, N. Natural Variation in Leaf Morphology Results from Mutation of a Novel KNOX Gene. Curr. Biol. 2008, 18, 672–677. [Google Scholar] [CrossRef]
- Di Giacomo, E.; Sestili, F.; Iannelli, M.A.; Testone, G.; Mariotti, D.; Frugis, G. Characterization of KNOX Genes in Medicago Truncatula. Plant Mol. Biol. 2008, 67, 135–150. [Google Scholar] [CrossRef]
- Brown, D.M.; Zeef, L.A.H.; Ellis, J.; Goodacre, R.; Turner, S.R. Identification of Novel Genes in Arabidopsis Involved in Secondary Cell Wall Formation Using Expression Profiling and Reverse Genetics. Plant Cell 2005, 17, 2281–2295. [Google Scholar] [CrossRef]
- He, J.-B.; Zhao, X.-H.; Du, P.-Z.; Zeng, W.; Beahan, C.T.; Wang, Y.-Q.; Li, H.-L.; Bacic, A.; Wu, A.-M. KNAT7 Positively Regulates Xylan Biosynthesis by Directly Activating IRX9 Expression in Arabidopsis: KNAT7 Positively Regulates Xylan Biosynthesis. J. Integr. Plant Biol. 2018, 60, 514–528. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.; Wei, H.; Milne, J.; Page, G.P.; Somerville, C.R. Identification of Genes Required for Cellulose Synthesis by Regression Analysis of Public Microarray Data Sets. Proc. Natl. Acad. Sci. USA 2005, 102, 8633–8638. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.-Y.; Huang, G.-Q.; Sun, X.; Qin, L.-X.; Li, Y.; Zhou, L.; Li, X.-B. Cotton KNL1, Encoding a Class II KNOX Transcription Factor, Is Involved in Regulation of Fibre Development. J. Exp. Bot. 2014, 65, 4133–4147. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Yuan, Y.; Spiekerman, J.J.; Guley, J.T.; Egbosiuba, J.C.; Ye, Z.-H. Functional Characterization of NAC and MYB Transcription Factors Involved in Regulation of Biomass Production in Switchgrass (Panicum Virgatum). PLoS ONE 2015, 10, e0134611. [Google Scholar] [CrossRef]
- Wang, S.; Yang, H.; Mei, J.; Liu, X.; Wen, Z.; Zhang, L.; Xu, Z.; Zhang, B.; Zhou, Y. Rice Homeobox Protein KNAT7 Integrates the Pathways Regulating Cell Expansion and Wall Stiffness. Plant Physiol. 2019, 181, 669–682. [Google Scholar] [CrossRef]
- Yoo, C.G.; Dumitrache, A.; Muchero, W.; Natzke, J.; Akinosho, H.; Li, M.; Sykes, R.W.; Brown, S.D.; Davison, B.; Tuskan, G.A.; et al. Significance of Lignin S/G Ratio in Biomass Recalcitrance of Populus Trichocarpa Variants for Bioethanol Production. ACS Sustain. Chem. Eng. 2018, 6, 2162–2168. [Google Scholar] [CrossRef]
- Pimrote, K. Transcriptional Regulatory Network Controlling Secondary Cell Wall Biosynthesis and Biomass Production in Vascular Plants. Afr. J. Biotechnol. 2012, 11, 13928–13937. [Google Scholar] [CrossRef]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-Based Transcriptional Regulation of Secondary Cell Wall Biosynthesis in Land Plants. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Rao, X.; Dixon, R.A. Current Models for Transcriptional Regulation of Secondary Cell Wall Biosynthesis in Grasses. Front. Plant Sci. 2018, 9, 399. [Google Scholar] [CrossRef]
- Yu, Y. OsKNAT7 Bridges Secondary Cell Wall Formation and Cell Growth Regulation. Plant Physiol. 2019, 181, 385–386. [Google Scholar] [CrossRef]
- Joshi, C.P.; Bhandari, S.; Ranjan, P.; Kalluri, U.C.; Liang, X.; Fujino, T.; Samuga, A. Genomics of Cellulose Biosynthesis in Poplars. New Phytol. 2004, 164, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin Biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Teng, Q.; Zhong, R.; Ye, Z.-H. Molecular Dissection of Xylan Biosynthesis during Wood Formation in Poplar. Mol. Plant 2011, 4, 730–747. [Google Scholar] [CrossRef] [PubMed]
- Polko, J.K.; Kieber, J.J. The Regulation of Cellulose Biosynthesis in Plants. Plant Cell 2019, 31, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Behr, M.; Guerriero, G.; Grima-Pettenati, J.; Baucher, M. A Molecular Blueprint of Lignin Repression. Trends Plant Sci. 2019, 24, 1052–1064. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Y.; Pei, S.; Lu, M.; Kong, Y.; Zhou, G.; Hu, R. KNAT7 Regulates Xylan Biosynthesis in Arabidopsis Seed-Coat Mucilage. J. Exp. Bot. 2020, 71, 4125–4139. [Google Scholar] [CrossRef]
- Zhong, R.; Demura, T.; Ye, Z.-H. SND1, a NAC Domain Transcription Factor, Is a Key Regulator of Secondary Wall Synthesis in Fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. The MYB46 Transcription Factor Is a Direct Target of SND1 and Regulates Secondary Wall Biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.-H. Global Analysis of Direct Targets of Secondary Wall NAC Master Switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. MYB46 and MYB83 Bind to the SMRE Sites and Directly Activate a Suite of Transcription Factors and Secondary Wall Biosynthetic Genes. Plant Cell Physiol. 2012, 53, 368–380. [Google Scholar] [CrossRef]
- Kim, W.-C.; Kim, J.-Y.; Ko, J.-H.; Kang, H.; Han, K.-H. Identification of Direct Targets of Transcription Factor MYB46 Provides Insights into the Transcriptional Regulation of Secondary Wall Biosynthesis. Plant Mol. Biol. 2014, 85, 589–599. [Google Scholar] [CrossRef]
- Ko, J.-H.; Jeon, H.-W.; Kim, W.-C.; Kim, J.-Y.; Han, K.-H. The MYB46/MYB83-Mediated Transcriptional Regulatory Programme Is a Gatekeeper of Secondary Wall Biosynthesis. Ann. Bot. 2014, 114, 1099–1107. [Google Scholar] [CrossRef]
- Romano, J.M.; Dubos, C.; Prouse, M.B.; Wilkins, O.; Hong, H.; Poole, M.; Kang, K.; Li, E.; Douglas, C.J.; Western, T.L.; et al. At MYB61, an R2R3-MYB Transcription Factor, Functions as a Pleiotropic Regulator via a Small Gene Network. New Phytol. 2012, 195, 774–786. [Google Scholar] [CrossRef]
- Bellaoui, M.; Pidkowich, M.S.; Samach, A.; Kushalappa, K.; Kohalmi, S.E.; Modrusan, Z.; Crosby, W.L.; Haughn, G.W. The Arabidopsis BELL1 and KNOX TALE Homeodomain Proteins Interact through a Domain Conserved between Plants and Animals. Plant Cell 2001, 13, 2455–2470. [Google Scholar] [CrossRef]
- Hackbusch, J.; Richter, K.; Muller, J.; Salamini, F.; Uhrig, J.F. A Central Role of Arabidopsis Thaliana Ovate Family Proteins in Networking and Subcellular Localization of 3-Aa Loop Extension Homeodomain Proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4908–4912. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Guo, J.; Zeng, Q.; Ellis, B.E.; Chen, J.-G. Arabidopsis Ovate Family Proteins, a Novel Transcriptional Repressor Family, Control Multiple Aspects of Plant Growth and Development. PLoS ONE 2011, 6, e23896. [Google Scholar] [CrossRef]
- Li, E.; Wang, S.; Liu, Y.; Chen, J.-G.; Douglas, C.J. OVATE FAMILY PROTEIN4 (OFP4) Interaction with KNAT7 Regulates Secondary Cell Wall Formation in Arabidopsis Thaliana: KNAT7-OFP Complex Regulates Secondary Wall Formation. Plant J. 2011, 67, 328–341. [Google Scholar] [CrossRef]
- Liu, Y.; Douglas, C.J. A Role for OVATE FAMILY PROTEIN1 (OFP1) and OFP4 in a BLH6-KNAT7 Multi-Protein Complex Regulating Secondary Cell Wall Formation in Arabidopsis Thaliana. Plant Signal. Behav. 2015, 10, e1033126. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Ellis, B. Overview of OVATE FAMILY PROTEINS, A Novel Class of Plant-Specific Growth Regulators. Front. Plant Sci. 2016, 7, 417. [Google Scholar] [CrossRef]
- Liu, Y.; You, S.; Taylor-Teeples, M.; Li, W.L.; Schuetz, M.; Brady, S.M.; Douglas, C.J. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 Interact and Regulate Secondary Cell Wall Formation via Repression of REVOLUTA. Plant Cell 2015, 26, 4843–4861. [Google Scholar] [CrossRef]
- Bhargava, A.; Mansfield, S.D.; Hall, H.C.; Douglas, C.J.; Ellis, B.E. MYB75 Functions in Regulation of Secondary Cell Wall Formation in the Arabidopsis Inflorescence Stem. Plant Physiol. 2010, 154, 1428–1438. [Google Scholar] [CrossRef]
- Liu, Y. Investigation of a KNAT7-BLH-OFP Transcription Factor Complex Involved in Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis Thaliana. Master’s Thesis, University of British Columbia, Vancouver, BC, Canada, 2010. [Google Scholar] [CrossRef]
- Osakabe, K.; Tsao, C.C.; Li, L.; Popko, J.L.; Umezawa, T.; Carraway, D.T.; Smeltzer, R.H.; Joshi, C.P.; Chiang, V.L. Coniferyl Aldehyde 5-Hydroxylation and Methylation Direct Syringyl Lignin Biosynthesis in Angiosperms. Proc. Natl. Acad. Sci. USA 1999, 96, 8955–8960. [Google Scholar] [CrossRef]
- Wang, Q.; Dai, X.; Pang, H.; Cheng, Y.; Huang, X.; Li, H.; Yan, X.; Lu, F.; Wei, H.; Sederoff, R.R.; et al. BEL1-like Homeodomain Protein BLH6a Is a Negative Regulator of CAld5H2 in Sinapyl Alcohol Monolignol Biosynthesis in Poplar. Front. Plant Sci. 2021, 12, 695223. [Google Scholar] [CrossRef]
- Bhargava, A.; Ahad, A.; Wang, S.; Mansfield, S.D.; Haughn, G.W.; Douglas, C.J.; Ellis, B.E. The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis. Planta 2013, 237, 1199–1211. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Liu, C.; Ge, C.; Xu, F.; Luo, M. Ectopic expression of GhIQD14 (cotton IQ67 domain-containing protein 14) causes twisted organ and modulates secondary wall formation in Arabidopsis. Plant Physiol. Biochem. 2021, 163, 276–284. [Google Scholar] [CrossRef]
- Huang, D.; Wang, S.; Zhang, B.; Shang-Guan, K.; Shi, Y.; Zhang, D.; Liu, X.; Wu, K.; Xu, Z.; Fu, X.; et al. A Gibberellin-Mediated DELLA-NAC Signaling Cascade Regulates Cellulose Synthesis in Rice. Plant Cell 2015, 27, 1681–1696. [Google Scholar] [CrossRef]
- Schmitz, A.J.; Begcy, K.; Sarath, G.; Walia, H. Rice Ovate Family Protein 2 (OFP2) Alters Hormonal Homeostasis and Vasculature Development. Plant Sci. 2015, 241, 177–188. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3- MYB Transcription Factor MYB 6 Promotes Anthocyanin and Proanthocyanidin Biosynthesis but Inhibits Secondary Cell Wall Formation in Populus Tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef]
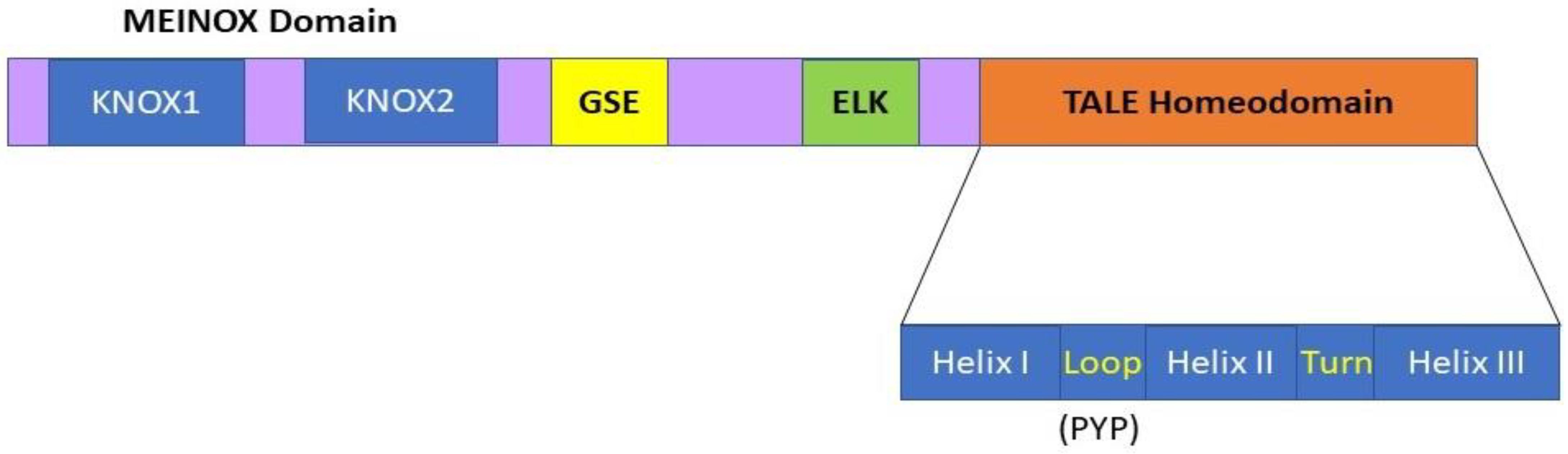
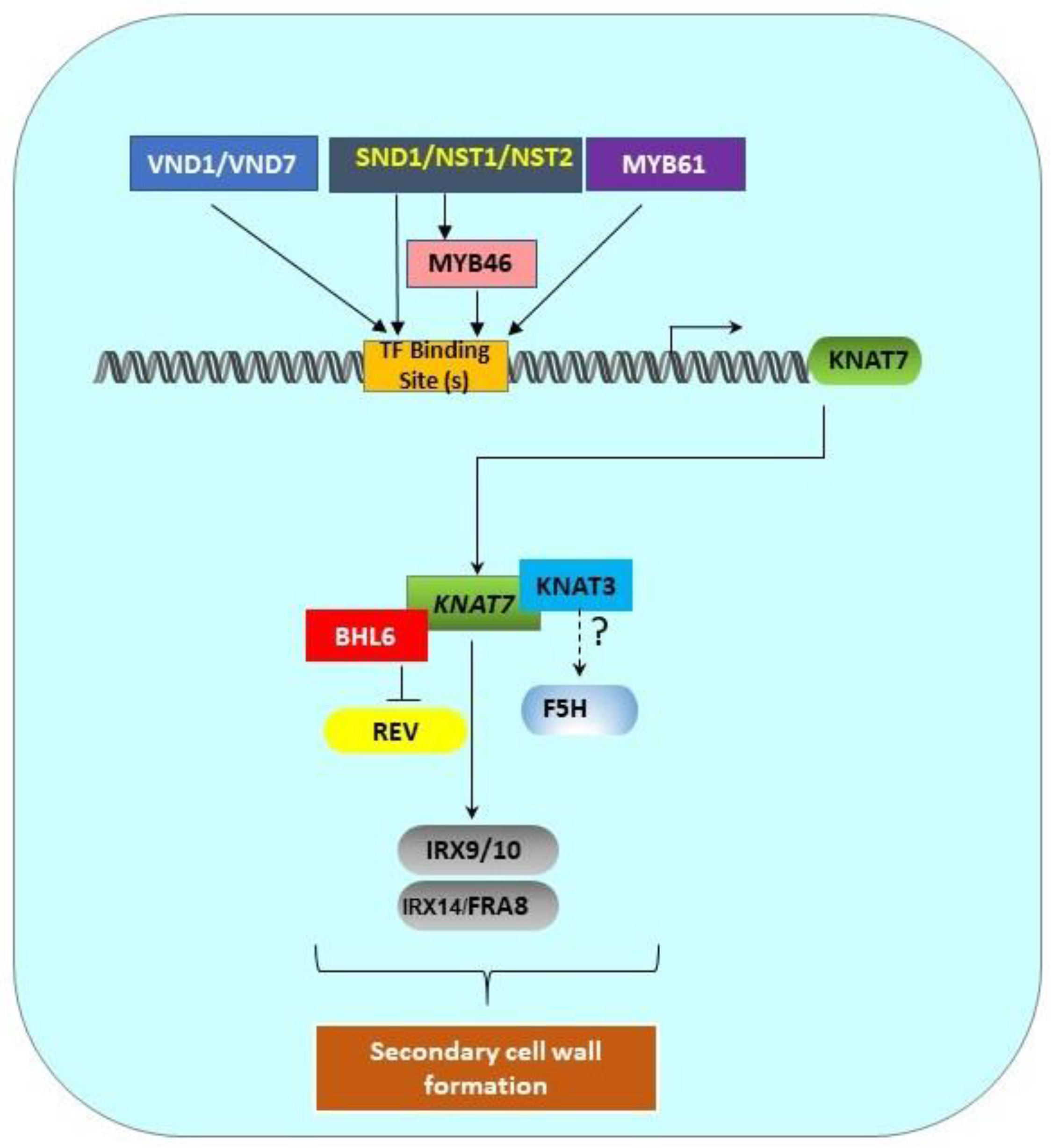
| Target Gene | Mutation | Type of Mutation | Anatomy of Mutants | References |
|---|---|---|---|---|
| AtKNAT7 | irx11 | T-DNA insertion | Irregular xylem with collapsed vessels. | [30] |
| AtKNAT7 | - | Dominant repression | Reduced cell wall thickness of both xylem vessels and fibers; reduced composition of several monosaccharides from the cell walls. | [12] |
| AtKNAT7 | irx11 | Loss-of-function mutation | Thinner vessels walls resulted in a collapse of xylem vessels that showed the irx phenotype and thicker interfascicular fibers compared to controls; increase in lignin content. | [13] |
| AtKNAT3, AtKNAT4, AtKNAT5 | Single mutants | T-DNA insertion | No irx phenotype. | [15] |
| KNAT3/KNAT7 | Double mutant | T-DNA insertion | Enhanced irregular xylem (irx) phenotype characterized by weak inflorescence stem; reduced interfascicular fiber wall thickness and modified cell wall composition. | [15] |
| KNAT3/KNAT7 | Double mutant | Chimeric repression | Thinner interfascicular fiber cell walls compared to single mutants and wild type (WT); reduced cellulose and xylan and reduced S/G lignin ratio. | [16] |
| OsKNAT7 | CRISPR/CAS9 | T-DNA insertion | Thicker fiber cell walls; larger grain size due to cell expansion in spikelet bracts. | [35] |
| GhKNL1 | - | Dominant repression | Abnormal shorter fiber length. | [33] |
| Gene Used | Target Plant | Gene Modification Method | Impact on Transgenic Plants | References |
|---|---|---|---|---|
| AtKNAT7 | Arabidopsis | Overexpression | Thin interfascicular fiber walls, but no change in vessel wall thickness. | [13] |
| Cotton GhKNL1 | Arabidopsis | Overexpression | Thinner interfascicular fibers and slightly thinner vessel walls, but no change in xylary fibers. | [33] |
| Cotton GhKNAT7 | Arabidopsis | Overexpression | Reduced deposition of lignocellulose in interfascicular fibers, but no change in the SCWs of xylem fibers and vessels. | [24] |
| NbKNAT7 | Tobacco | Downregulation by VIGS and RNAi | Increased xylem proliferation with thin-walled fiber cells, increased polysaccharide extractability, and higher saccharification rate. | [14] |
| AtKNAT7 | Arabidopsis | Dominant repression | Reduced expression of SCW genes that resulted in thinner fiber cell walls with altered cell wall composition. | [12] |
| PtKNAT7 | Poplar | Overexpression | Enhanced expression of SCW genes, CesA8, IRX9, PAL, and CCR. | [17] |
| PtKNAT7 | Poplar | Downregulation by antisense | Reduced expression of SCW genes, reduced lignin content, altered lignin composition (S/G ratio), and increased saccharification. | [17] |
| Species | Class II KNOX Proteins | Interacting Proteins | Biological Function | Reference |
|---|---|---|---|---|
| Arabidopsis | AtKNAT7 | AtMYB75 | SCW formation. | [61,65] |
| AtMYB5 | SCW formation. | [65] | ||
| AtOFP1/4 | KNAT7 transcriptional repression enhanced during SCW formation. | [57] | ||
| AtBLHs | SCW formation | [55,60,62] | ||
| AtKNAT3 | Regulates S-lignin formation. | [55] | ||
| AtKNAT3 | NST1/2 | Possibly regulates F5H gene expression to promote syringyl lignin synthesis. | [16] | |
| AtBLH1 | SCW formation. | [55] | ||
| AtKNAT7 | Possibly regulates S-lignin formation. | [16] | ||
| Cotton | GhKNAT7 | GhMYB75 | SCW biosynthesis. | [24] |
| GhBLH1/5/6 | SCW biosynthesis. | [24] | ||
| GhBEL1 | SCW biosynthesis. | [24] | ||
| GHOFP1/5/4 | SCW biosynthesis. | [24] | ||
| GhIQD14 | SCW biosynthesis. | [66] | ||
| Poplar | PtKNAT7 | PtMYB6 | Promotes anthocyanin synthesis and represses SCW development. | [69] |
| PtMYB75 | SCW formation. | [69] | ||
| PtMYB115 | SCW formation. | [69] | ||
| Rice | OsKNAT7 | OsGRF4 | Negatively regulates cellulose biosynthesis and cell expansion. | [35] |
| OsOFP2 | Vasculature development. | [68] | ||
| OsNAC29/31 | Suppresses the activation of MYB61 expression during SCW formation. | [35] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nookaraju, A.; Pandey, S.K.; Ahlawat, Y.K.; Joshi, C.P. Understanding the Modus Operandi of Class II KNOX Transcription Factors in Secondary Cell Wall Biosynthesis. Plants 2022, 11, 493. https://doi.org/10.3390/plants11040493
Nookaraju A, Pandey SK, Ahlawat YK, Joshi CP. Understanding the Modus Operandi of Class II KNOX Transcription Factors in Secondary Cell Wall Biosynthesis. Plants. 2022; 11(4):493. https://doi.org/10.3390/plants11040493
Chicago/Turabian StyleNookaraju, Akula, Shashank K. Pandey, Yogesh K. Ahlawat, and Chandrashekhar P. Joshi. 2022. "Understanding the Modus Operandi of Class II KNOX Transcription Factors in Secondary Cell Wall Biosynthesis" Plants 11, no. 4: 493. https://doi.org/10.3390/plants11040493
APA StyleNookaraju, A., Pandey, S. K., Ahlawat, Y. K., & Joshi, C. P. (2022). Understanding the Modus Operandi of Class II KNOX Transcription Factors in Secondary Cell Wall Biosynthesis. Plants, 11(4), 493. https://doi.org/10.3390/plants11040493









