Identification and Functional Analysis of the CgNAC043 Gene Involved in Lignin Synthesis from Citrusgrandis “San Hong”
Abstract
:1. Introduction
2. Results
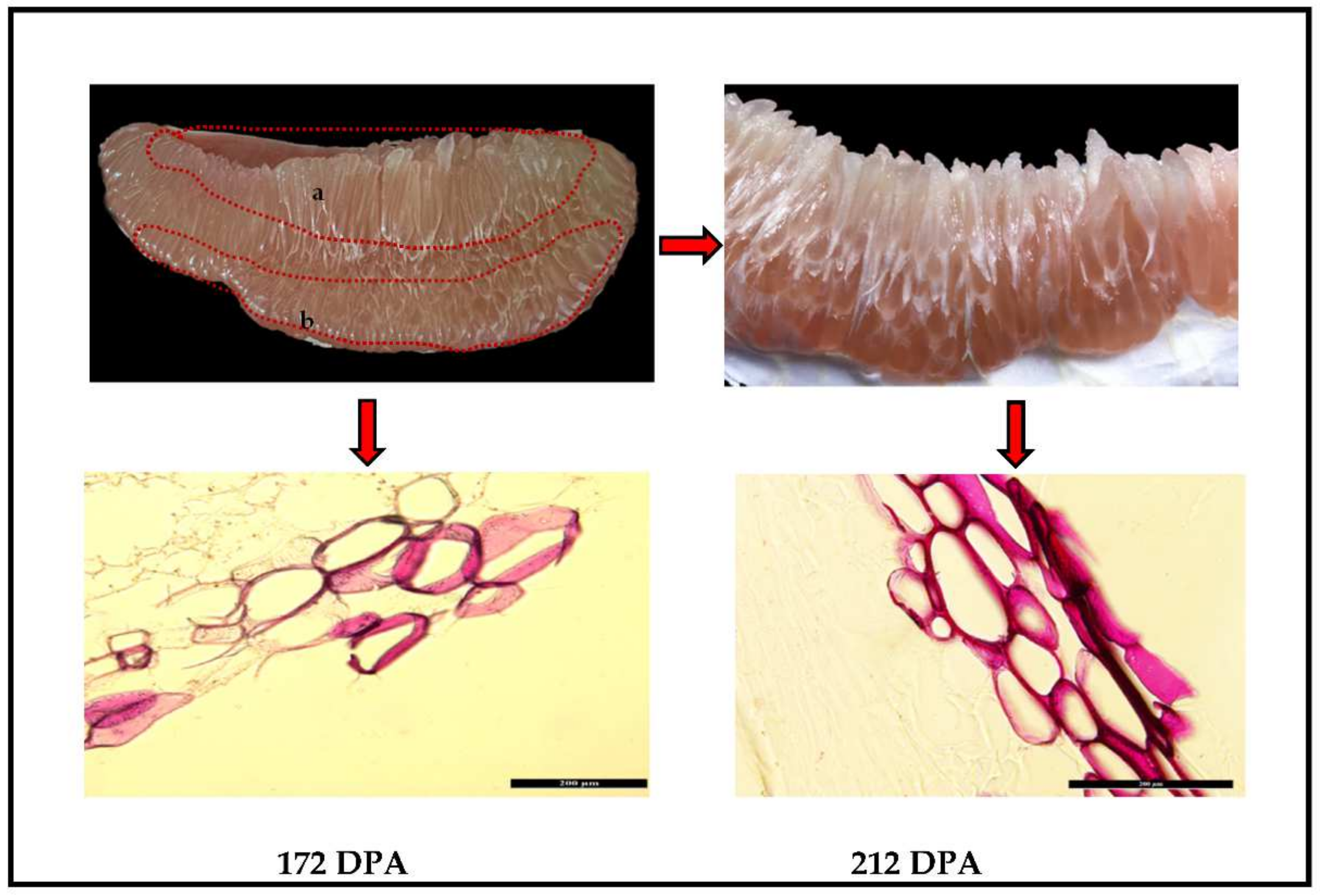
2.1. Microscopic Observation of Lignin Determination of Lignin Juice Sacs
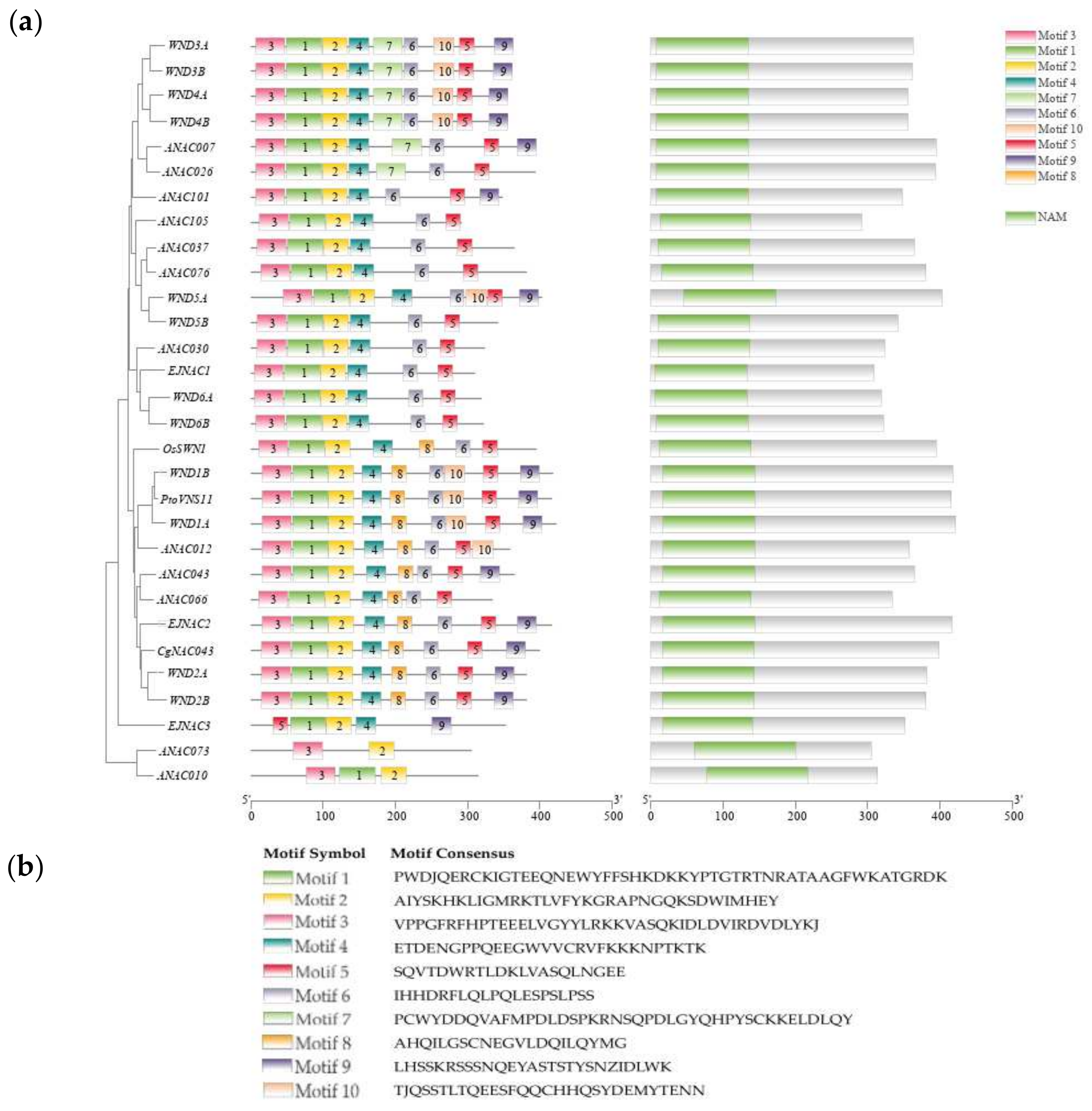
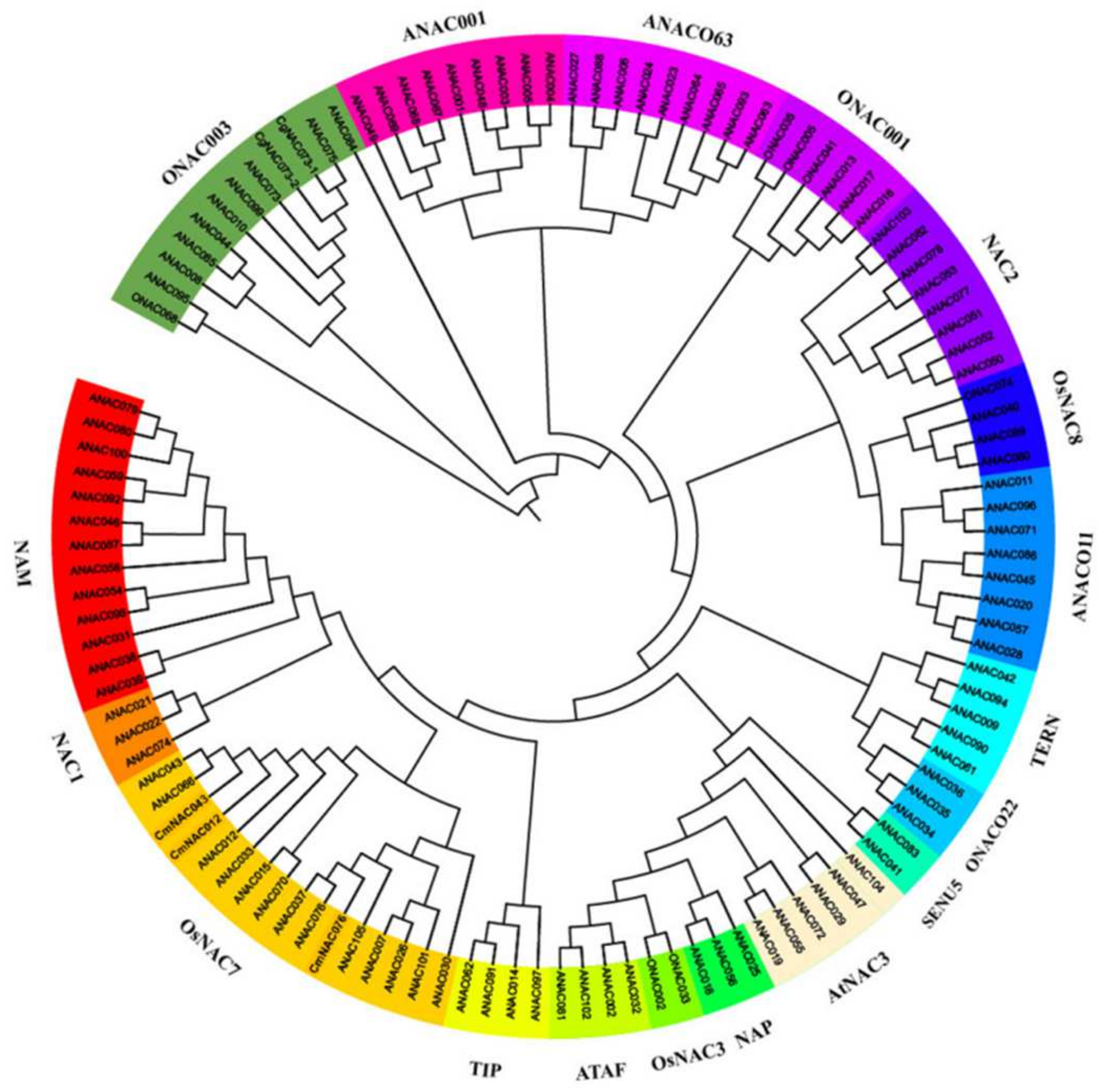
2.2. Identification of CgNAC043 and Sequence Alignment
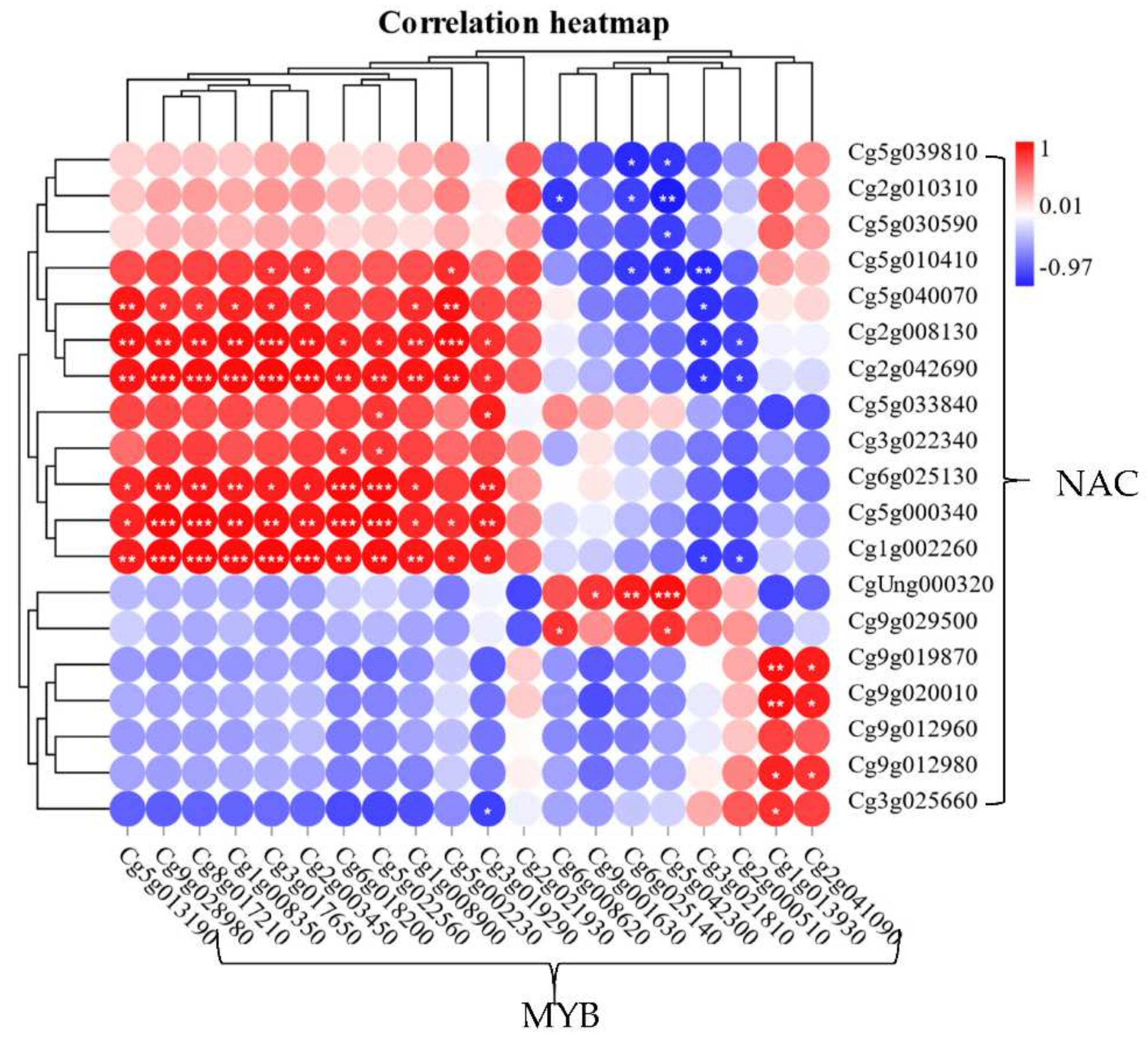
2.3. The Correlation Analysis of CgNACs and CgMYBs in the Process of Lignin Synthesis
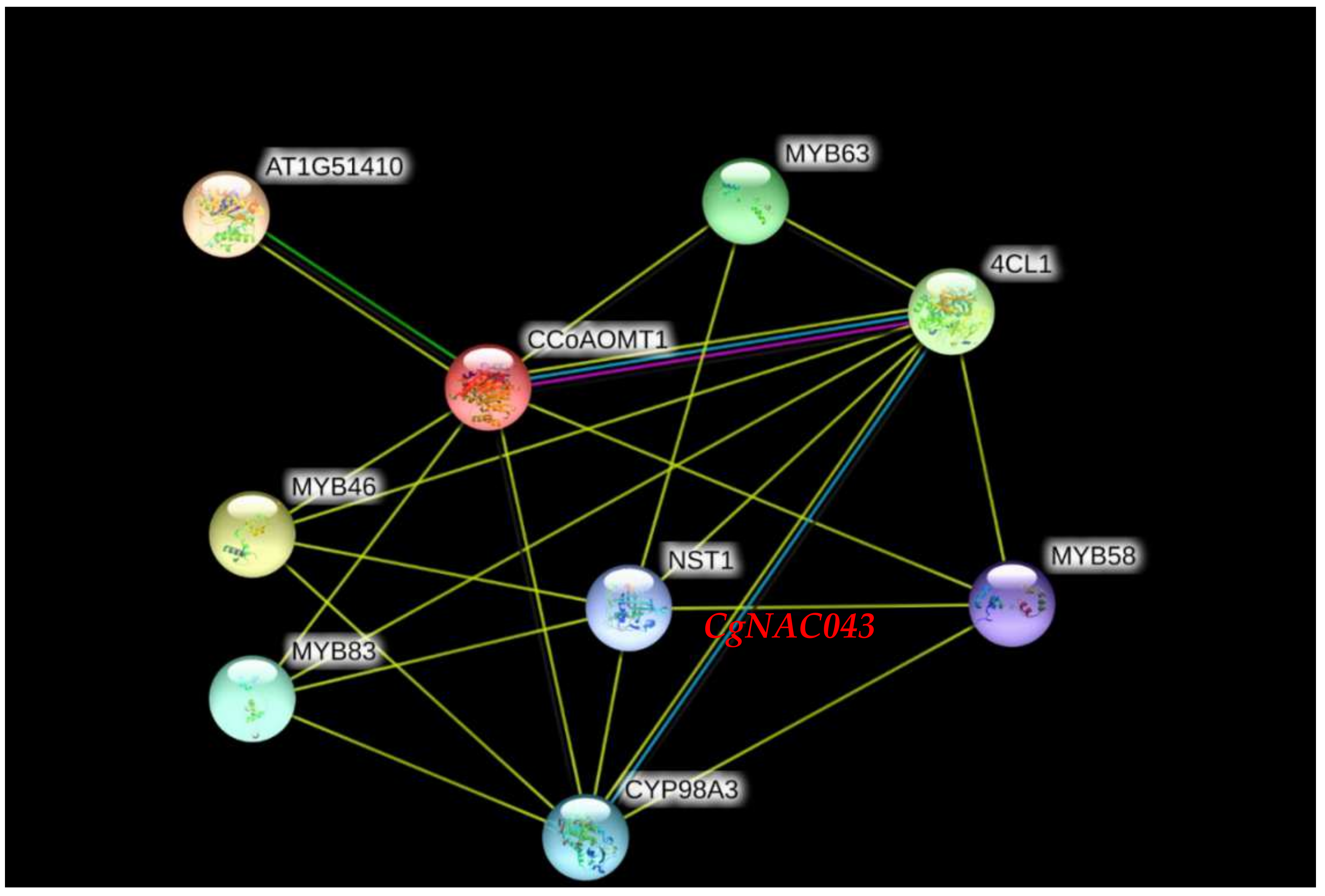
2.4. Protein-Protein Interaction Analysis of CgNAC043
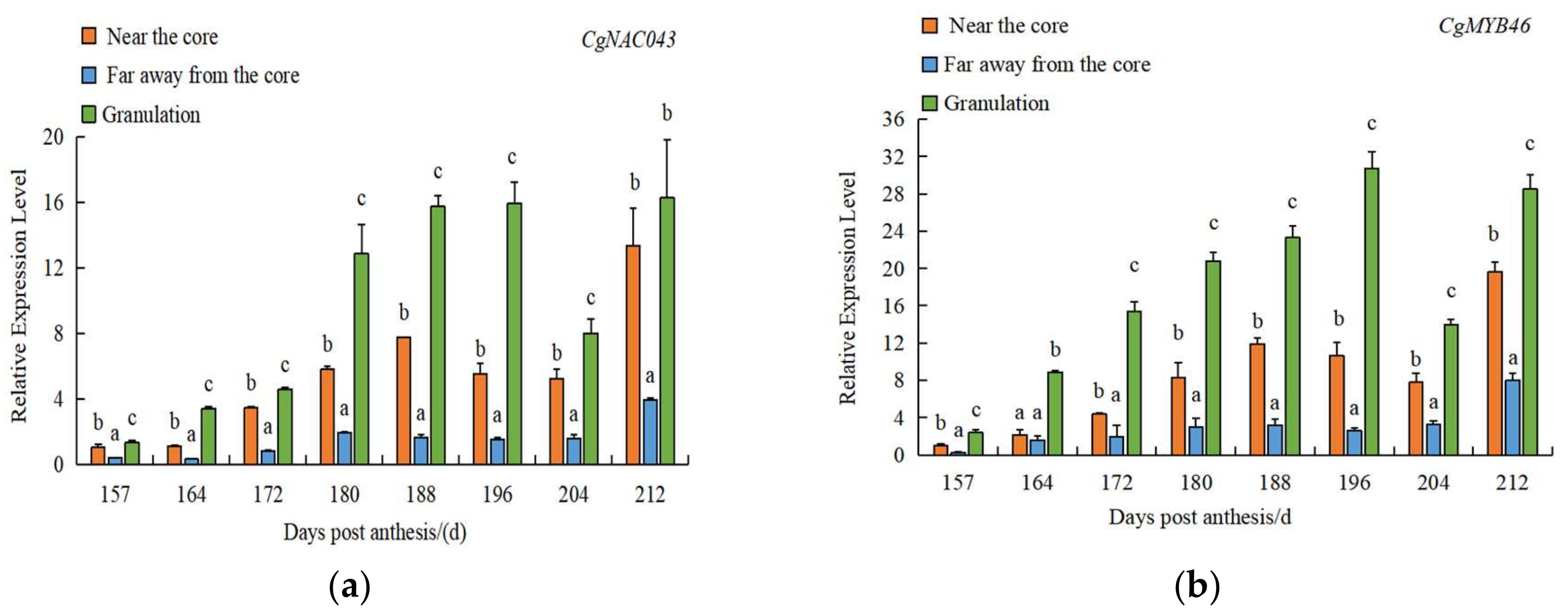
2.5. Expression Patterns of CgNAC043 Gene
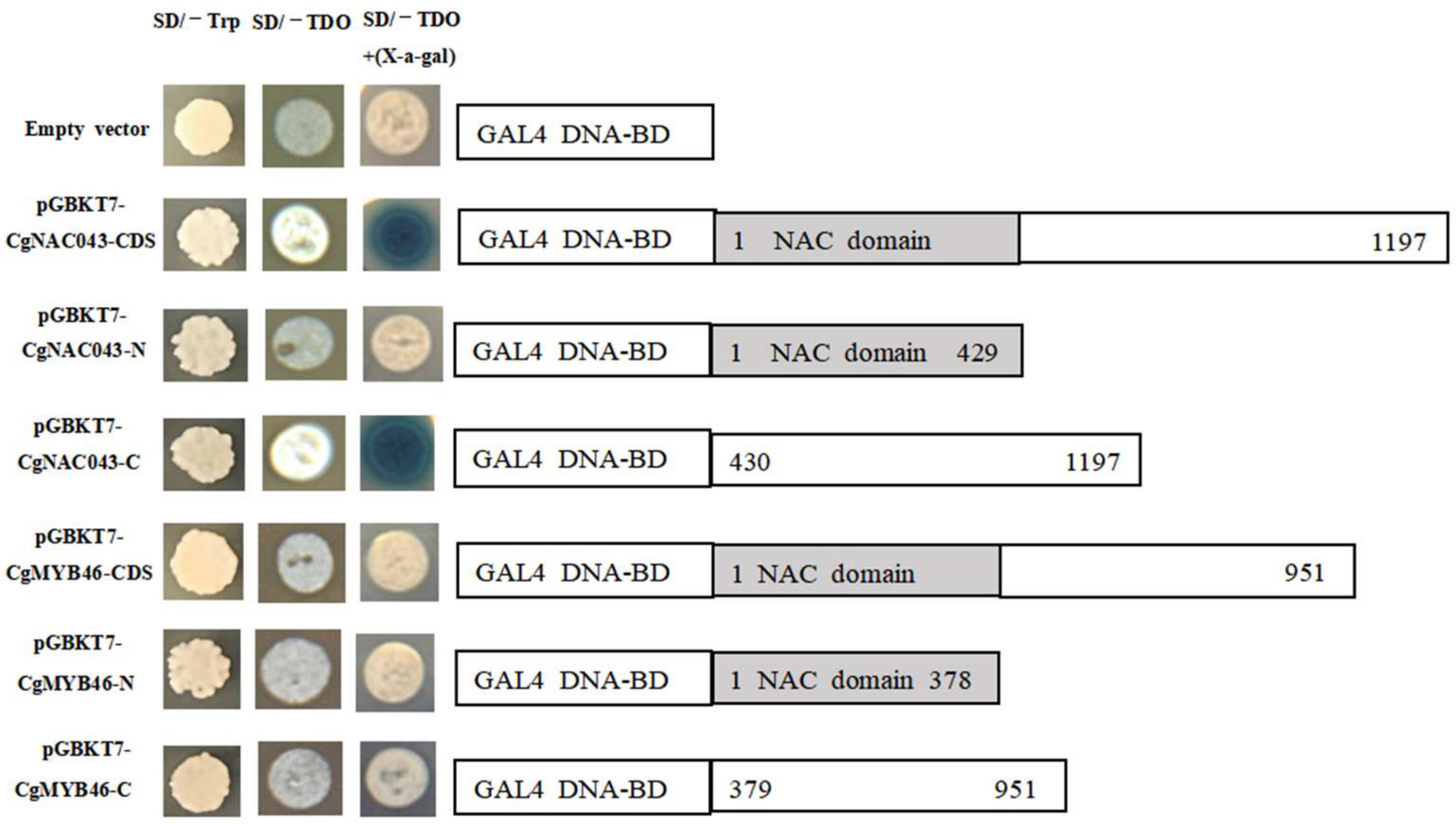
2.6. Transcriptional Activation of CgNAC043 and CgMYB46 in Yeast
2.7. Transient Expression in Mesocarp
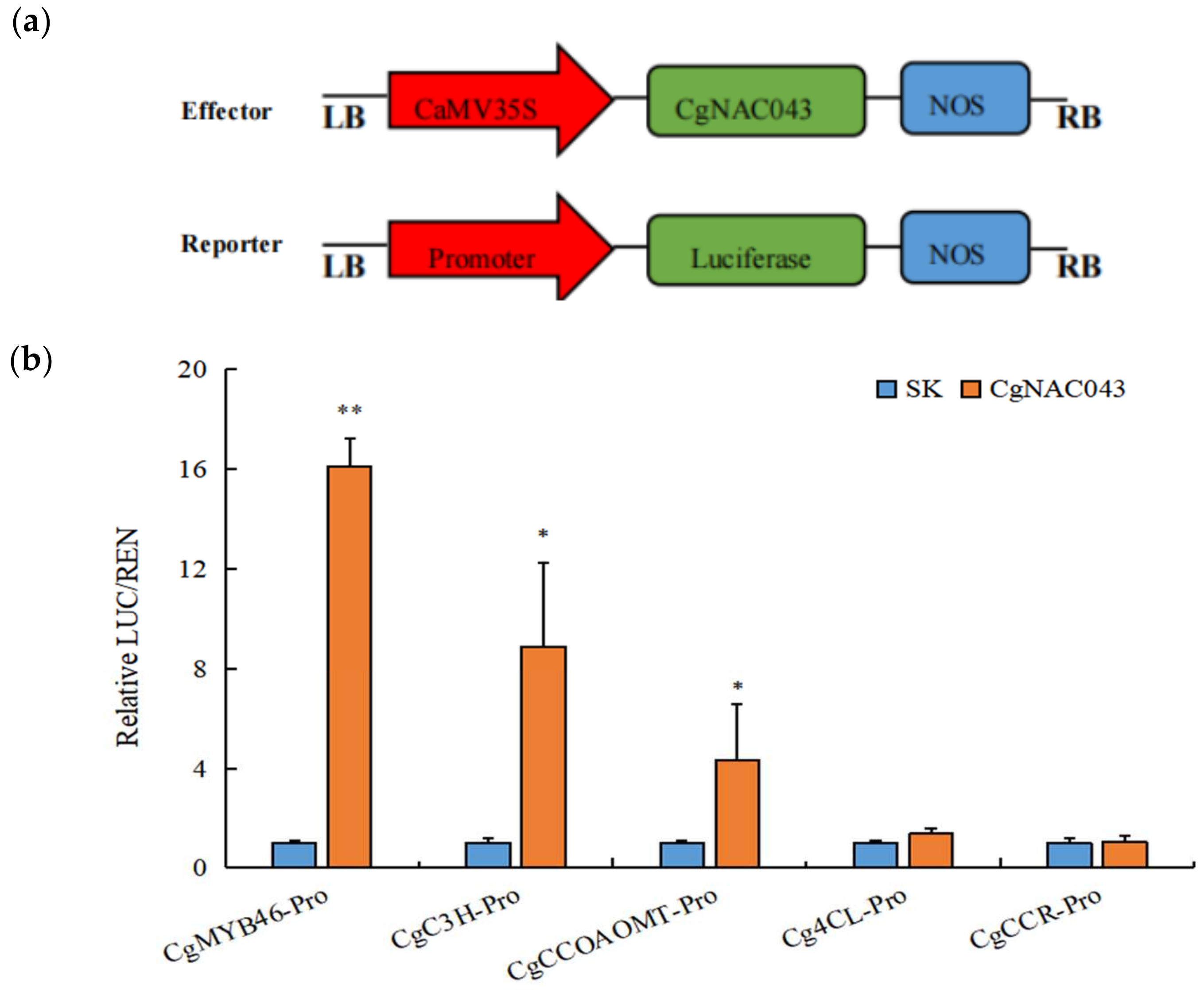
2.8. Trans-Activation by CgNAC043 of Promoters of Lignin Transcription Factors and Biosynthesis Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Observation of Cell Walls of the Fruits Juice Sacs
4.3. Gene Isolation
4.4. Bioinformatics Analysis
4.5. Construction of Interaction Networks of CgNAC043 Protein in Pomelo
4.6. Transcriptional Activity Assay in Yeast
4.7. RNA Extraction and Real-Time PCR Analysis
4.8. Transient Expression in Pomelo Mesocarp
4.9. Dual Luciferase Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shomer, I.; Chalutz, E.; Vasiliver, R.; Lomaniec, E.; Berman, M. Scierification of juice sacs in pummelo (Citrus grandis) fruit. Can. J. Bot. 1989, 67, 625–632. [Google Scholar] [CrossRef]
- Jia, N.; Liu, J.; Sun, Y.; Tan, P.; Cao, H.; Xie, Y.; Wen, B.; Gu, T.; Liu, J.; Li, M. Citrus sinensis MYB transcription factors CsMYB330 and CsMYB308 regulate fruit juice sac lignification through fine-tuning expression of the Cs4CL1 gene. Plant Sci. 2018, 277, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, M.; Cheng, F.S.; Dai, C.; Sun, Y.F.; Lu, J.; Huang, Y.T.; Li, M.M.; He, Y.; Wang, F.Z.; et al. Identification of microRNAs correlated with citrus granulation based on bioinformatics and molecular biology analysis. Postharvest Biol. Technol. 2016, 118, 59–67. [Google Scholar] [CrossRef]
- Sharma, R.R.; Singh, R.; Saxena, S.K. Characteristics of citrus fruits in relation to granulation. Sci. Hortic. 2006, 111, 91–96. [Google Scholar] [CrossRef]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Pan, T.; Guo, Z.; Pan, D. Specific lignin accumulation in granulated juice sacs of Citrus maxima. Agr. Food Chem. 2014, 62, 12082–12089. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. Nac transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Tasaka, F.M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [Green Version]
- Souer, E.; Van, H.A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Ye, D. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [Green Version]
- Golfier, P.; Volkert, C.; He, F.; Rausch, T.; Wolf, S. Regulation of secondary cell wall biosynthesis by a NAC transcription factor from miscanthus. Plant Direct 2017, 1, e00024. [Google Scholar] [CrossRef] [PubMed]
- Taichi, K.; Naoki, Y.; Yuki, T.; Masaomi, Y.; Shiro, S.; Takefumi, H.; Mai, M.; Soichiro, N.; Daisuke, S.; Masahiro, S. MYB-mediated upregulation of lignin biosynthesis in Oryza sativa towards biomass refinery. Plant Biotechnol. J. 2017, 34, 7–15. [Google Scholar]
- Takada, S.; Hibara, K.I.; Ishida, T.; Tasaka, M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 2001, 128, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- Vroemen, C.W.; Mordhorst, A.P.; Albrecht, C.; Kwaaitaal, M.A.; de Vries, S.C. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 2003, 15, 1563–1577. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Ohtani, M.; Mitsuda, N.; Kubo, M.; Ohme-Takagi, M.; Fukuda, H.; Demura, T. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 2010, 22, 1249. [Google Scholar] [CrossRef] [Green Version]
- Xie, Q.; Guo, H.S.; Dallman, G.; Fang, S.; Weissman, A.M.; Chua, N.H. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 2002, 419, 167–170. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, S.G.; Park, J.E.; Park, H.Y.; Lim, M.H.; Chua, N.H.; Park, C.M. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 2006, 18, 3132–3144. [Google Scholar] [CrossRef] [Green Version]
- Jensen, M.K.; Hagedorn, P.H.; De Torres-Zabala, M.; Grant, M.R.; Lyngkjaer, M.F. Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2010, 56, 867–880. [Google Scholar] [CrossRef]
- Wei, S.; Kuang, J.F.; Chen, L.; Xie, H.; Peng, H.H.; Xiao, Y.Y.; Li, X.P.; Chen, W.X.; He, Q.G.; Chen, J.Y. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 2012, 63, 5171–5187. [Google Scholar]
- Ríos, P.; Argyris, J.; Vegas, J.; Leida, C.; Kenigswald, M.; Tzuri, G.; Troadec, C.; Bendahmane, A.; Katzir, N.; Picó, B.; et al. Ethqv6.3 is involved in melon climacteric fruit ripening and is encoded by a NAC domain transcription factor. Plant J. 2017, 91, 671–683. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, K.; Tran, L.; Dong, V.N.; Fujita, M.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor osnac6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2010, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Wang, Y.; Li, B.; Chang, J.L.; Chen, M.J.; Li, K.X.; Yang, G.X.; He, G.Y. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Zhang, A.; Ye, Y.; Xu, B.; Chen, J.; He, X.; Wang, C.; Zhou, S.; Zhang, X.; Peng, Y. Genome-wide survey of switchgrass NACs family provides new insights into motif and structure arrangements and reveals stress-related and tissue-specific NACs. Sci. Rep. 2017, 7, 3056. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Seo, P.J.; Lee, H.J.; Park, C.M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012, 70, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Bibi, N.; Gan, S.; Li, F.; Yuan, S.; Ni, M.; Wang, M.; Shen, H.; Wang, X. A novel nap member GhNAP is involved in leaf senescence in Gossypium hirsutum. J. Exp. Bot. 2015, 66, 4669–4682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, C.; Lu, S.; Bo, L.; Zhang, B.; Feng, M. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [Green Version]
- Nobutaka, M.; Akira, I.; Hiroyuki, Y.; Masato, Y.; Motoaki, S.; Kazuo, S.; Masaru, O.T. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar]
- Mitsuda, N.; Ohme-Takagi, M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 2008, 56, 768–778. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Zhou, J.; Mccarthy, R.L.; Ye, Z.H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Lin, Y.C.; Sun, Y.H.; Song, J.; Hao, C.; Zhang, X.H.; Sederoff, R.R.; Chiang, V.L. Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl Acad. Sci. USA 2012, 109, 14699–14704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, M.; Bellizzi, M.; Wan, C.; Cui, Z.; Wang, G.L. The NAC transcription factor OsSWN1 regulates secondary cell wall development in Oryza sativa. J. Plant Biol. 2015, 58, 44–51. [Google Scholar] [CrossRef]
- Jie, Z.; Geng, Q.H.; Dan, Z.; Jing, Q.Y.; Yang, L.; Shan, H.; Xue, B.L. The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 2018, 217, 625–640. [Google Scholar]
- Xu, Q.; Wang, W.; Zeng, J.; Zhang, J.; Grierson, D.; Li, X.; Yin, X.; Chen, K. A NAC transcription factor, EjNAC1, affects lignification of loquat fruit by regulating lignin. Postharvest Biol. Technol. 2015, 102, 25–31. [Google Scholar] [CrossRef]
- Hu, P.; Zhang, K.; Yang, C. BpNAC012 positively regulates abiotic stress responses and secondary wall biosynthesis. Plant Physiol. 2019, 179, 700–717. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, R.L.; Zhong, R.; Ye, Z.-H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009, 50, 1950–1964. [Google Scholar] [CrossRef] [Green Version]
- Ko, J.; Kim, W.C.; Han, K.H. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 2009, 60, 649–665. [Google Scholar] [CrossRef]
- Jia, N.; Liu, J.; Tan, P.H.; Sun, Y.F.; Lv, Y.M.; Liu, J.M.; Sun, J.; Huang, Y.T.; Lu, J.; Jin, N.; et al. Citrus sinensis MYB transcription factor CsMYB85 induce fruit juice sac lignification through interaction with other CsMYB transcription factors. Front. Plant Sci. 2019, 10, 213. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Liu, X.; Zhang, H.; He, Z.; Yang, H.; Chen, J.; Feng, J.; Yang, W.; Jiang, Y.; Yao, J.L. The IAA- and ABA-responsive transcription factor CgMYB58 upregulates lignin biosynthesis and triggers juice sac granulation in pummelo. Hortic. Res. 2020, 7, 139. [Google Scholar] [CrossRef]
- Xu, Q.; Yin, X.R.; Zeng, J.K.; Ge, H.; Song, M.; Xu, C.J.; Li, X.; Ferguson, I.B.; Chen, K.S. Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 2014, 65, 4349–4359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Lee, C.; Ye, Z.H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.Q.; Lee, C.H.; McCarthy, R.L.; Reeves, C.K.; Jones, E.G.; Ye, Z.-H. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 2011, 52, 1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, D.; Zheng, G.; Chen, G.; She, W.; Guo, Z.; Shi, M.; Huiying, A.L. Analysis of the reasons caused granulation of juice sacs in Guanximiyou pomelo variety. J. Fruit Sci. 1999, 16, 202–209. [Google Scholar]
- Zhong, R.Q.; Lee, C.H.; Ye, Z.-H. Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 2010, 152, 1044–1055. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef]
- Hussey, S.G.; Mizrachi, E.; Spokevicius, A.V.; Bossinger, G.; Berger, D.K.; Myburg, A.A. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 2011, 11, 173. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.-H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Lee, C.; Ye, Z.-H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010, 15, 625–632. [Google Scholar] [CrossRef]
- Xu, Y. Cloning and Expression of Lignin Genes in Citrus maxima (Burm.) Merr[D]; Fujian Agriculture and Forestry University: Fuzhou, China, 2014. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, T.; Fang, F.; Ge, H.; Shi, Y.N.; Luo, Z.R.; Yao, Y.C.; Donald, G.; Yin, X.R.; Chen, K.S.; Zhang, J.S. Two novel anoxia-induced ethylene response factors that interact with promoters of deastringency-related genes from persimmon. PLoS ONE 2014, 9, e97043. [Google Scholar] [CrossRef] [PubMed]


| SNBE Element Position (Forward/Reverse) | Base Distribution | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CgMYB46-SNBE1 -1253+ | T | G | A | T | T | T | C | T | T | T | T | A | A | A | C | G | A | A | A |
| CgMYB46-SNBE2 -479+ | T | T | A | T | G | T | G | A | A | G | T | G | G | A | A | G | C | A | A |
| CgMYB46-SNBE3 -372+ | T | A | C | C | T | T | G | T | A | A | A | T | G | A | A | G | A | A | A |
| CgMYB46-SNBE4 -372- | T | T | T | C | T | T | C | A | T | T | T | A | C | A | A | G | G | T | A |
| CgMYB46-SNBE5 -454- | T | T | G | T | T | T | A | G | T | A | A | C | C | A | A | G | C | T | A |
| CgMYB46-SNBE6 -566- | A | C | A | C | G | T | G | T | A | T | G | T | C | A | A | G | A | T | A |
| CgC3H-SNBE1 -1412+ | T | T | A | C | C | T | A | A | C | A | T | C | T | A | C | G | C | T | T |
| CgC3H-SNBE2 -1378+ | T | A | G | C | T | T | A | A | G | A | A | A | G | A | A | G | G | C | A |
| CgC3H-SNBE3 -142+ | T | A | A | T | T | T | C | T | T | A | A | C | A | A | C | G | T | A | A |
| CgC3H-SNBE4 -34+ | A | A | G | T | T | T | C | A | A | G | A | A | A | A | A | G | G | A | A |
| CgC3H-SNBE -1378- | T | G | C | C | T | T | C | T | T | T | C | T | T | A | A | G | C | T | A |
| CgCCoAOMT1-SNBE1 -974+ | A | T | A | T | G | T | G | A | G | C | G | T | G | A | A | G | A | C | T |
| CgCCoAOMT1-SNBE2 -651+ | A | C | T | C | T | T | A | T | T | T | G | T | C | A | A | G | A | A | A |
| CgCCoAOMT1-SNBE3 -1734- | A | T | A | T | C | T | A | A | T | C | A | T | A | A | C | G | T | T | T |
| SNBE | T | N | N | C | T | T | N | N | N | N | N | N | N | A | A | G | N | A | A |
| consensus | A | T | C | C | C | T | |||||||||||||
| G | T | ||||||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, N.; She, W.; Guo, Z.; Pan, H.; Yu, Y.; Ye, J.; Pan, D.; Pan, T. Identification and Functional Analysis of the CgNAC043 Gene Involved in Lignin Synthesis from Citrusgrandis “San Hong”. Plants 2022, 11, 403. https://doi.org/10.3390/plants11030403
Li X, Wang N, She W, Guo Z, Pan H, Yu Y, Ye J, Pan D, Pan T. Identification and Functional Analysis of the CgNAC043 Gene Involved in Lignin Synthesis from Citrusgrandis “San Hong”. Plants. 2022; 11(3):403. https://doi.org/10.3390/plants11030403
Chicago/Turabian StyleLi, Xiaoting, Naiyu Wang, Wenqin She, Zhixiong Guo, Heli Pan, Yuan Yu, Jianwen Ye, Dongming Pan, and Tengfei Pan. 2022. "Identification and Functional Analysis of the CgNAC043 Gene Involved in Lignin Synthesis from Citrusgrandis “San Hong”" Plants 11, no. 3: 403. https://doi.org/10.3390/plants11030403
APA StyleLi, X., Wang, N., She, W., Guo, Z., Pan, H., Yu, Y., Ye, J., Pan, D., & Pan, T. (2022). Identification and Functional Analysis of the CgNAC043 Gene Involved in Lignin Synthesis from Citrusgrandis “San Hong”. Plants, 11(3), 403. https://doi.org/10.3390/plants11030403






