Comparative Transcriptome and Co-Expression Network Analyses Reveal the Molecular Mechanism of Calcium-Deficiency-Triggered Tipburn in Chinese Cabbage (Brassica rapa L. ssp. Pekinensis)
Abstract
1. Introduction
2. Results
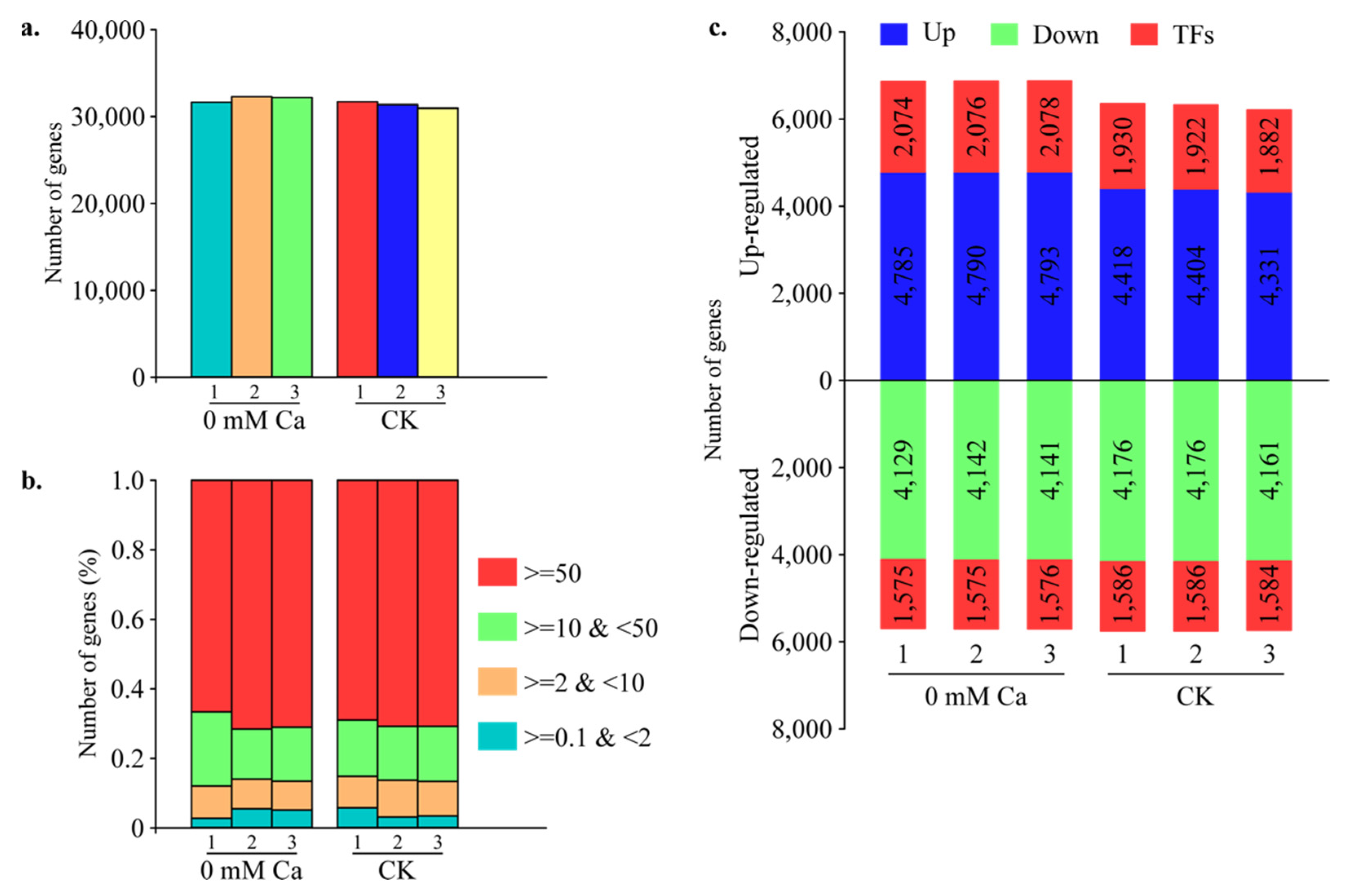
2.1. Global Transcriptome Analysis in Chinese Cabbage
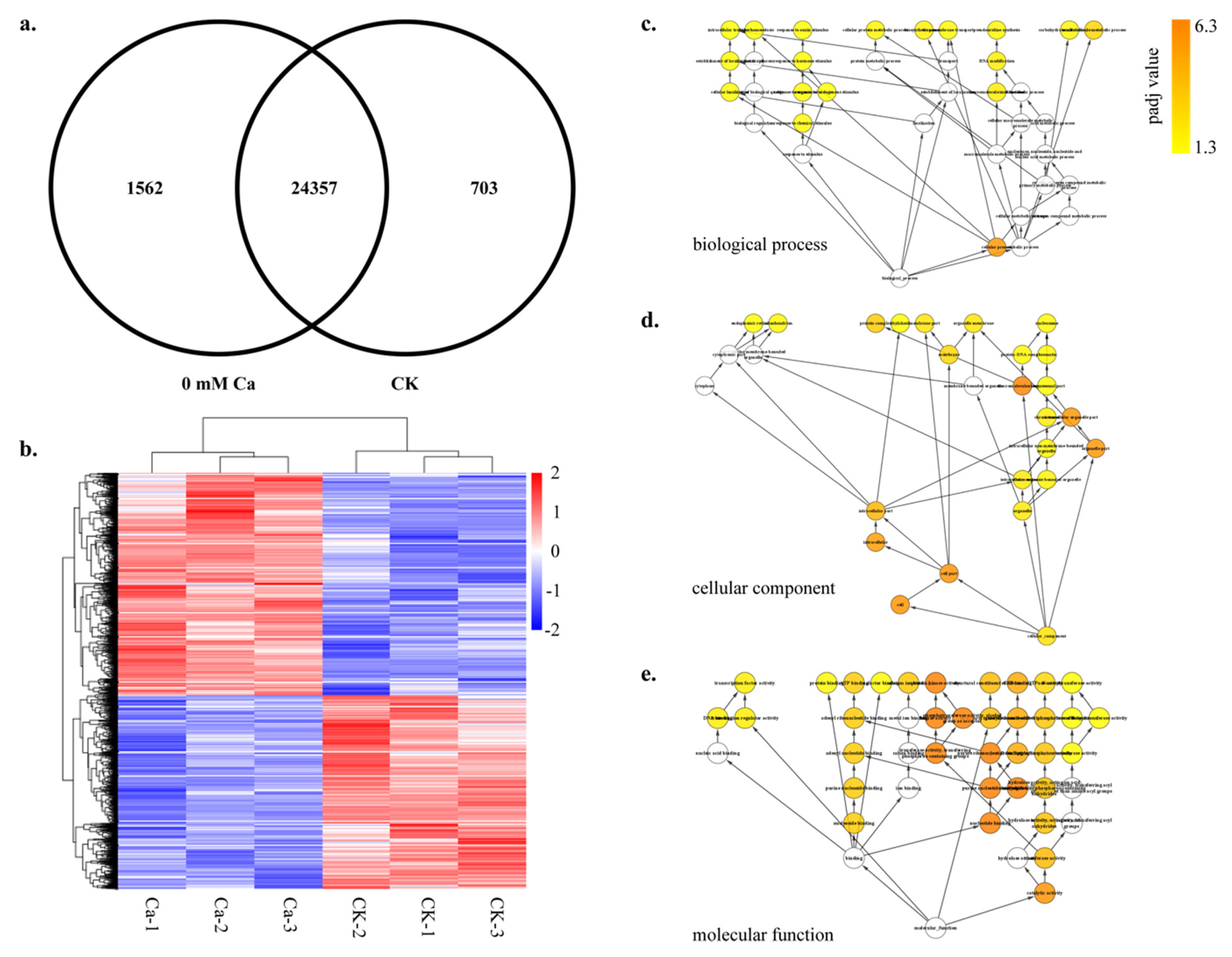
2.2. Global Comparison of Transcriptomes of Calcium Treatment
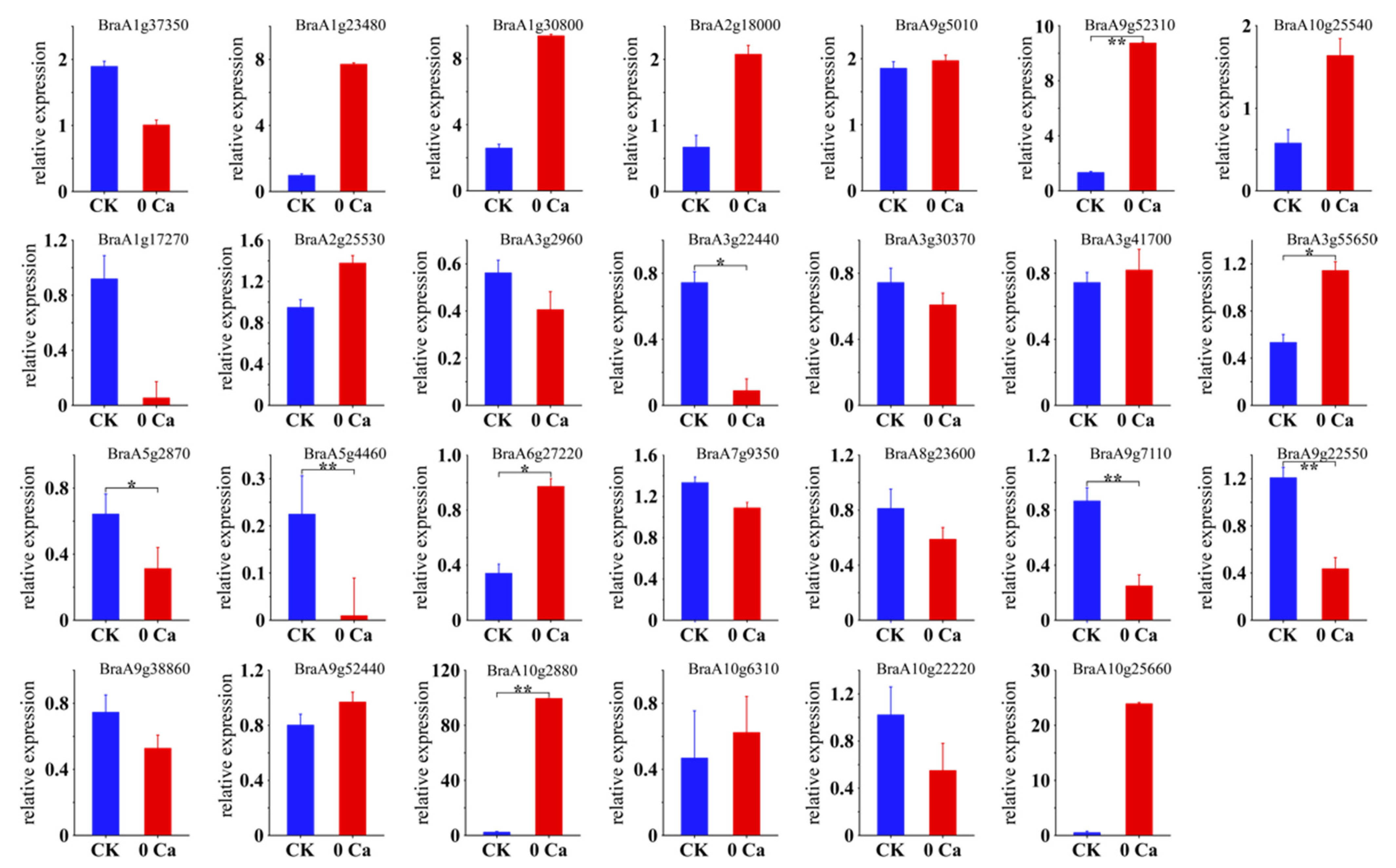
2.3. Differential Gene Expression during Tipburn
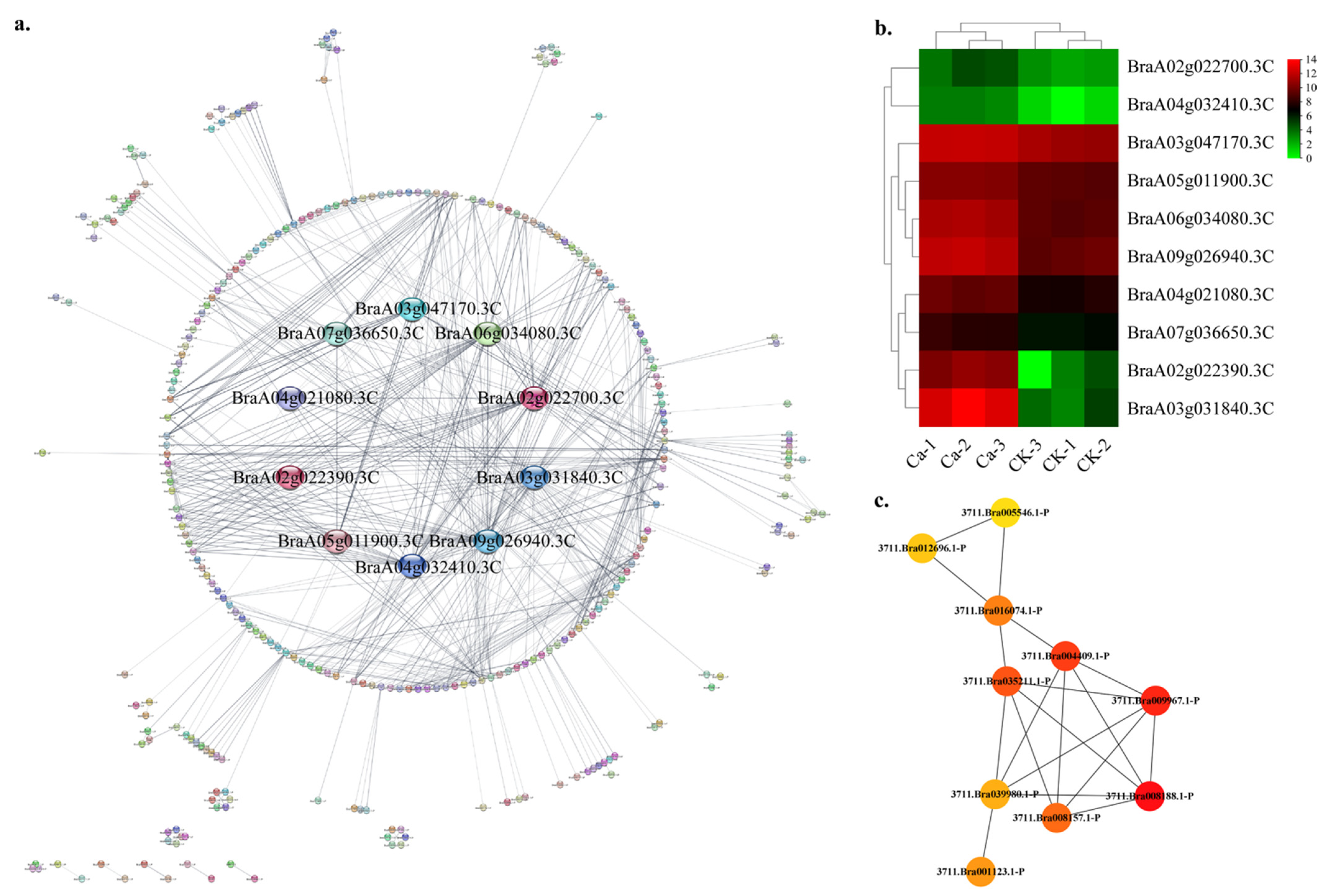
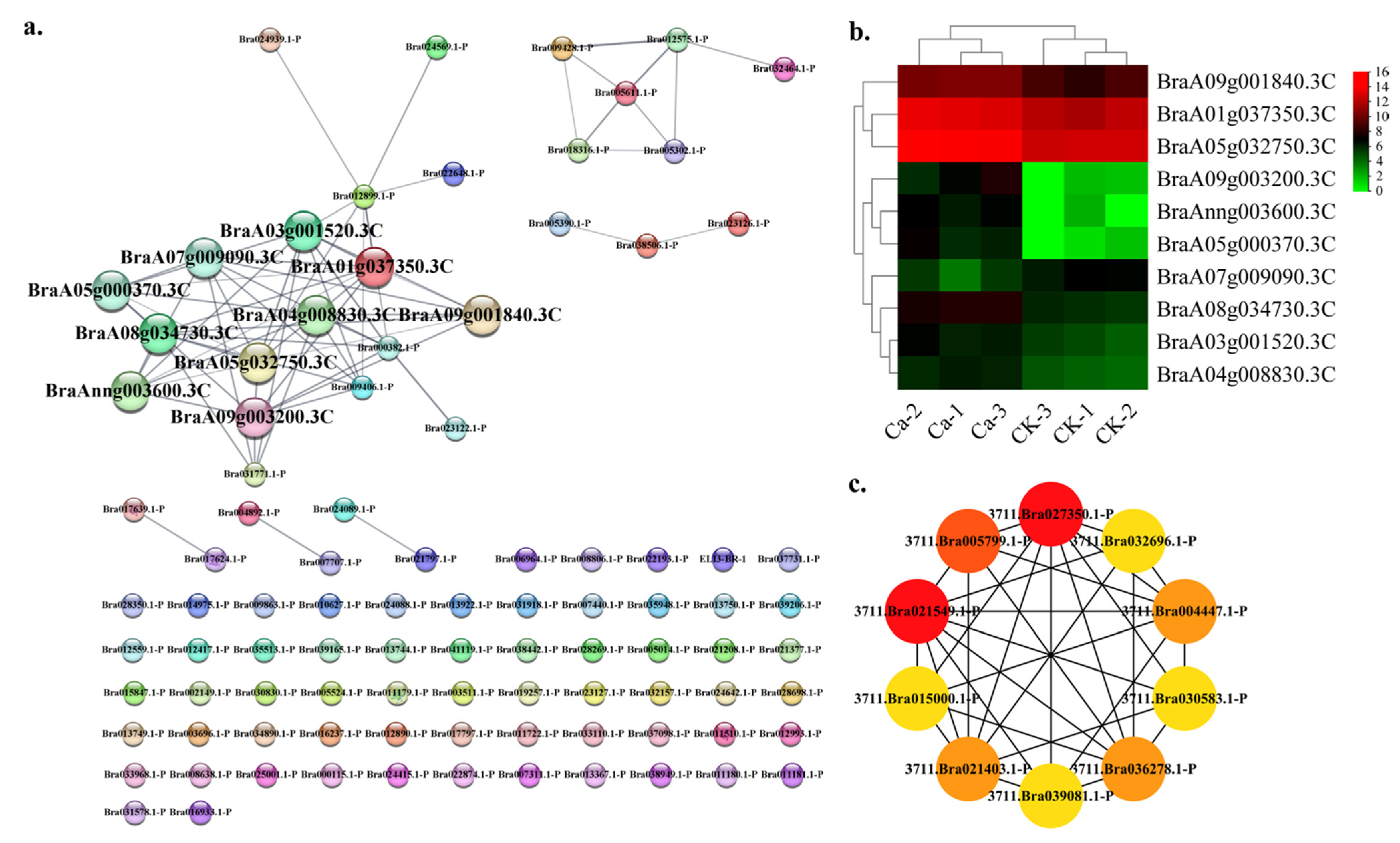
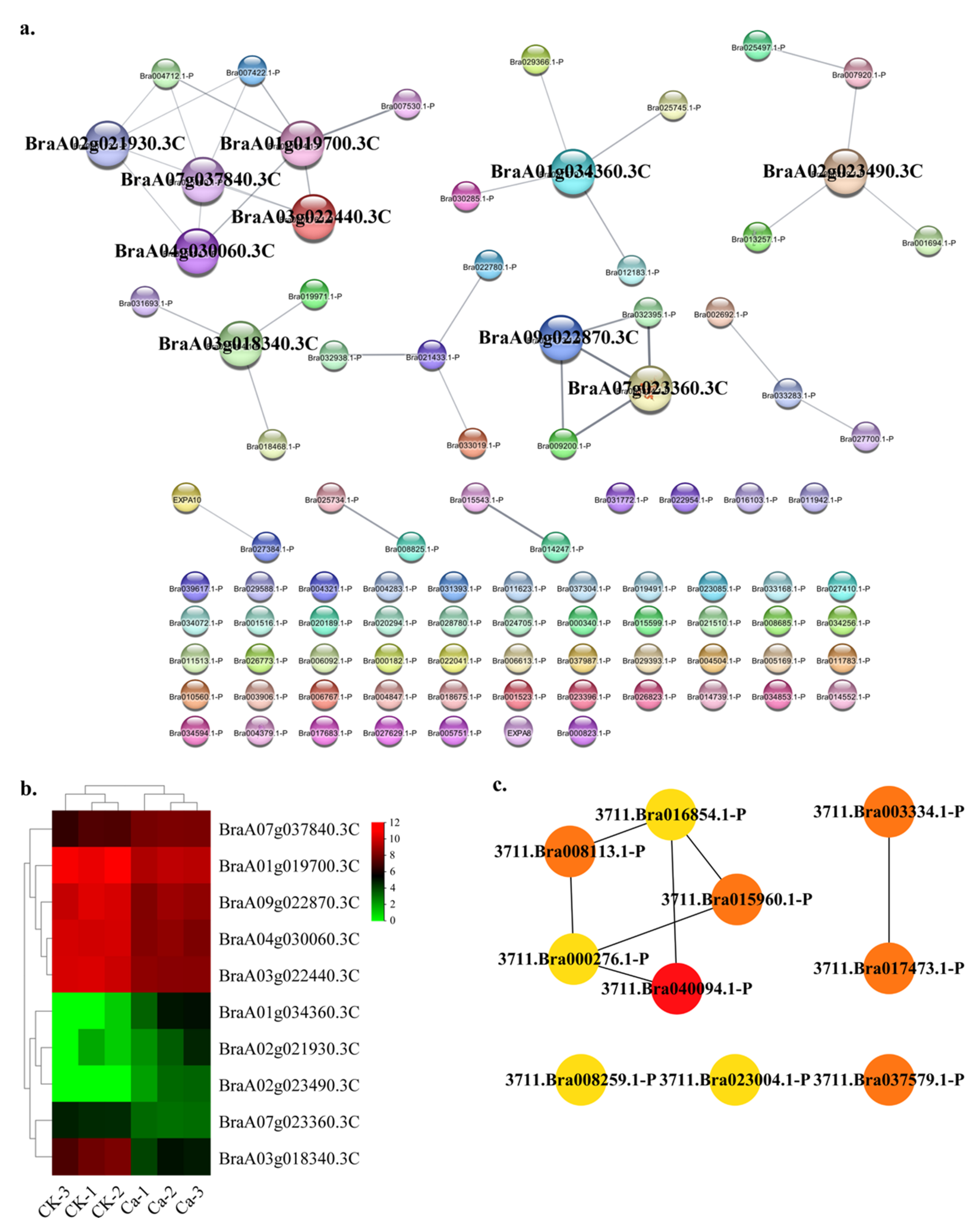
2.4. Protein—Protein Interaction (PPI) Regulatory Networks Related to Chinese Cabbage Tipburn
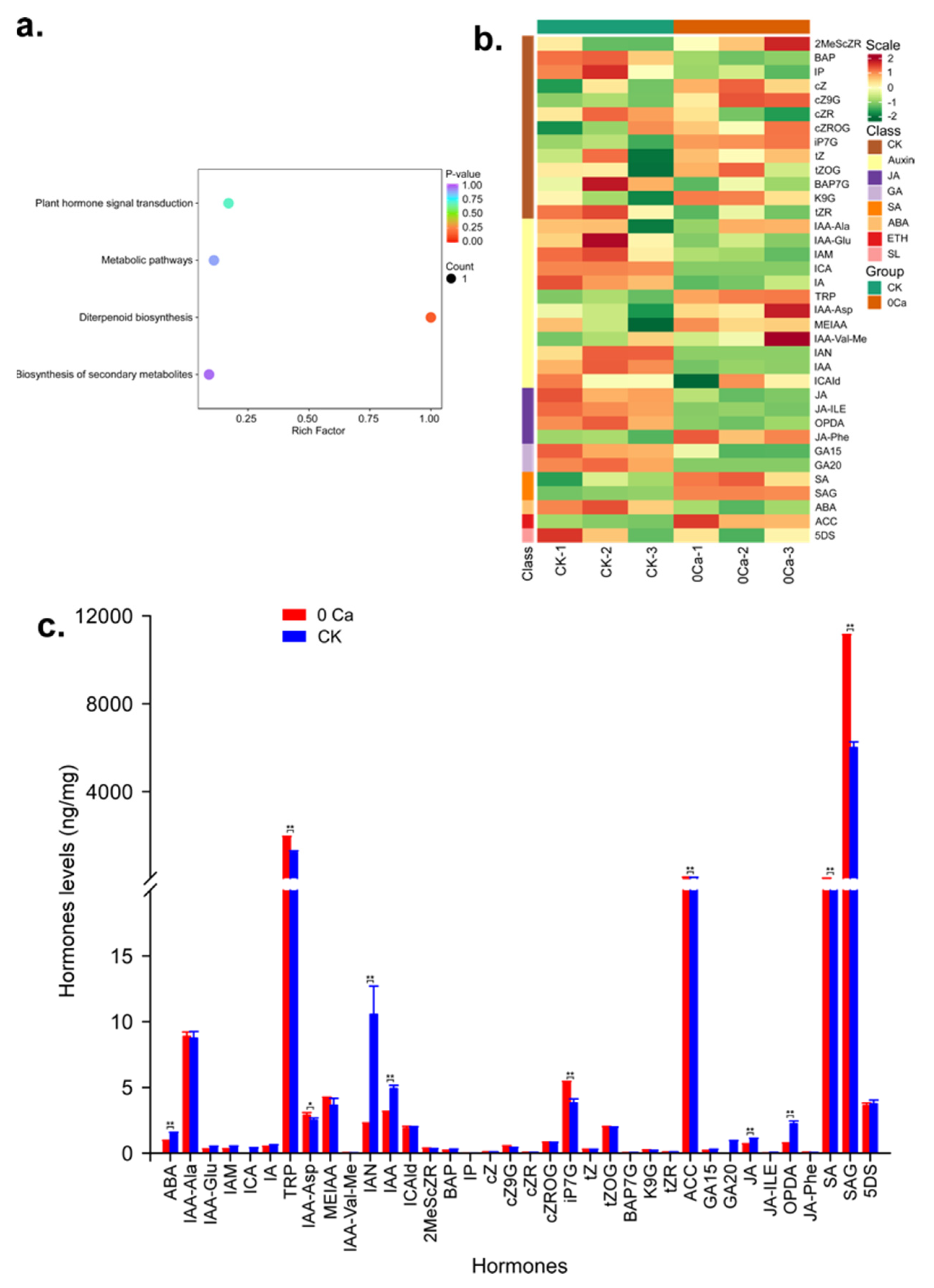
2.5. Metabolome Analysis to Identify the Hormones Related to Chinese Cabbage Tipburn
3. Discussion
4. Material and Methods
4.1. Plant Material and Sampling
4.2. Illumina Sequencing, Read Mapping and Differential Gene Expression Analyses
4.3. GO KEGG and Pathway Enrichment Analysis
4.4. Real-Time Quantitative PCR Analysis
4.5. Protein—Protein Interaction Analysis
4.6. Hormone Identification and Quantification for Metabolomic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marschner, H. Mineral nutrition of higher plants. J. Ecol. 1995, 76, 1250. [Google Scholar]
- McLaughlin, S.B.; Wimmer, R. Tansley Review No. 104. New Phytol. 2002, 142, 373–417. [Google Scholar] [CrossRef]
- Lee, J.; Park, I.; Lee, Z.-W.; Kim, S.W.; Baek, N.; Park, H.-S.; Park, S.U.; Kwon, S.; Kim, H. Regulation of the major vacuolar Ca2+ transporter genes, by intercellular Ca2+ concentration and abiotic stresses, in tip-burn resistant Brassica oleracea. Mol. Biol. Rep. 2013, 40, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Heuvelink, E.; Kierkels, T. Many physiogene problems due to poor calcium distribution: Balance can be restored at night. Greenh. Int. Mag. Greenh. Grow. 2013, 2, 10–11. [Google Scholar]
- Krebs, J.; Agellon, L.B.; Michalak, M. Ca2+ homeostasis and endoplasmic reticulum (ER) stress: An integrated view of calcium signaling. Biochem. Biophys. Res. Commun. 2015, 460, 114–121. [Google Scholar] [CrossRef]
- Khalaj, K.; Ahmadi, N.; Souri, M.K. Improvement of Postharvest Quality of Asian Pear Fruits by Foliar Application of Boron and Calcium. Horticulturae 2016, 3, 15. [Google Scholar] [CrossRef]
- Souri, M.K.; Hatamian, M. Aminochelates in plant nutrition: A review. J. Plant Nutr. 2018, 42, 67–78. [Google Scholar] [CrossRef]
- Pantoja, O. Recent Advances in the Physiology of Ion Channels in Plants. Annu. Rev. Plant Biol. 2021, 72, 463–495. [Google Scholar] [CrossRef]
- Demidchik, V.; Bowen, H.C.; Maathuis, F.J.; Shabala, S.N.; Tester, M.A.; White, P.J.; Davies, J.M. Arabidopsis thaliana root non-selective cation channels mediate calcium uptake and are involved in growth. Plant J. 2002, 32, 799–808. [Google Scholar] [CrossRef]
- Miedema, H.; Demidchik, V.; Véry, A.; Bothwell, J.H.F.; Brownlee, C.; Davies, J.M. Two voltage-dependent calcium channels co-exist in the apical plasma membrane of Arabidopsis thaliana root hairs. New Phytol. 2008, 179, 378–385. [Google Scholar] [CrossRef]
- Thor, K. Calcium—Nutrient and Messenger. Front. Plant Sci. 2019, 10, 440. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kang, Y.; Ma, C.; Miao, R.; Wu, C.; Long, Y.; Ge, T.; Wu, Z.; Hou, X.; Zhang, J.; et al. CNGC2 Is a Ca2+ Influx Channel That Prevents Accumulation of Apoplastic Ca2+ in the Leaf. Plant Physiol. 2017, 173, 1342–1354. [Google Scholar] [CrossRef] [PubMed]
- Von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zheng, H.; Chen, Q.; Chen, Y.; Wang, S.; Lu, L.; Zhang, S. The Lectin Chaperone Calnexin Is Involved in the Endoplasmic Reticulum Stress Response by Regulating Ca2+ Homeostasis in Aspergillus nidulans. Appl. Environ. Microbiol. 2017, 83, e00673-17. [Google Scholar] [CrossRef]
- La Verde, V.; Dominici, P.; Astegno, A. Towards Understanding Plant Calcium Signaling through Calmodulin-Like Proteins: A Biochemical and Structural Perspective. Int. J. Mol. Sci. 2018, 19, 1331. [Google Scholar] [CrossRef]
- Kami, C.; Lorrain, S.; Hornitschek, P.; Fankhauser, C. Light-Regulated Plant Growth and Development. Curr. Top. Dev. Biol. 2010, 91, 29–66. [Google Scholar]
- Zemanová, V.; Pavlík, M.; Pavlíková, D.; Tlustoš, P. The significance of methionine, histidine and tryptophan in plant responses and adaptation to cadmium stress. Plant Soil Environ. 2014, 60, 426–432. [Google Scholar] [CrossRef]
- Galeriani, T.M.; Neves, G.O.; Ferreira, J.H.S.; Oliveira, R.N.; Oliveira, S.L.; Calonego, J.C.; Crusciol, C.A.C. Calcium and Boron Fertilization Improves Soybean Photosynthetic Efficiency and Grain Yield. Plants 2022, 11, 2937. [Google Scholar] [CrossRef]
- Rangnekar, P.V. Effect of calcium deficiency on the carbon metabolism in photosynthesis and respiration of tomato leaf. Plant Soil 1975, 42, 565–583. [Google Scholar] [CrossRef]
- Hochmal, A.K.; Schulze, S.; Trompelt, K.; Hippler, M. Calcium-dependent regulation of photosynthesis. Biochim. et Biophys. Acta Bioenerg. 2015, 1847, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Stael, S.; Wurzinger, B.; Mair, A.; Mehlmer, N.; Vothknecht, U.C.; Teige, M. Plant organellar calcium signalling: An emerging field. J. Exp. Bot. 2012, 63, 1525–1542. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, W.; Karapetyan, S.; Mwimba, M.; Marqués, J.; Buchler, N.E.; Dong, X. Redox rhythm reinforces the circadian clock to gate immune response. Nature 2015, 523, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Jahan, S.; Ogawa, K.; Nakamura, Y.; Shimoishi, Y.; Mori, I.C.; Murata, Y. Deficient Glutathione in Guard Cells Facilitates Abscisic Acid-Induced Stomatal Closure but Does Not Affect Light-Induced Stomatal Opening. Biosci. Biotechnol. Biochem. 2008, 72, 2795–2798. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Ding, L.; Gu, B.; Zhang, H.; Guan, P.; Zhang, J. A stress associated protein from Chinese wild Vitis amurensis, VaSAP15, enhances the cold tolerance of transgenic grapes. Sci. Hortic. 2021, 285, 110147. [Google Scholar] [CrossRef]
- Hückelhoven, R. Cell Wall–Associated Mechanisms of Disease Resistance and Susceptibility. Annu. Rev. Phytopathol. 2007, 45, 101–127. [Google Scholar] [CrossRef]
- Abbas, A.A. Purification and Charcterization of Pectinesterase from Potato. Al-Nahrain J. Sci. 2016, 19, 134–139. [Google Scholar] [CrossRef]
- Ruggieri, V.; Sacco, A.; Calafiore, R.; Frusciante, L.; Barone, A. Dissecting a QTL into Candidate Genes Highlighted the Key Role of Pectinesterases in Regulating the Ascorbic Acid Content in Tomato Fruit. Plant Genome 2015, 8, 38. [Google Scholar] [CrossRef]
- Chen, R.; Jiang, J.; Hu, Z.; Ye, W.; Yuan, Q.; Li, M.; Wen, J.; Deng, Y. Coordinated Transcriptional Regulation of Cytochrome P450 3As by Nuclear Transcription Factor Y and Specificity Protein 1. Mol. Pharmacol. 2019, 95, 507–518. [Google Scholar] [CrossRef]
- Liu, Y.; Guan, X.; Liu, S.; Yang, M.; Ren, J.; Guo, M.; Huang, Z.; Zhang, Y. Genome-Wide Identification and Analysis of TCP Transcription Factors Involved in the Formation of Leafy Head in Chinese Cabbage. Int. J. Mol. Sci. 2018, 19, 847. [Google Scholar] [CrossRef]
- Liu, C.; Liu, H.; Zhou, C.; Timko, M.P. Genome-Wide Identification of the VQ Protein Gene Family of Tobacco (Nicotiana tabacum L.) and Analysis of Its Expression in Response to Phytohormones and Abiotic and Biotic Stresses. Genes 2020, 11, 284. [Google Scholar] [CrossRef]
- Wang, A.; Garcia, D.; Zhang, H.; Feng, K.; Chaudhury, A.; Berger, F.; Peacock, W.J.; Dennis, E.S.; Luo, M. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 2010, 63, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, Y.; Sui, N. Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 2018, 503, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Jolma, A.; Yin, Y.; Nitta, K.R.; Dave, K.; Popov, A.; Taipale, M.; Enge, M.; Kivioja, T.; Morgunova, E.; Taipale, J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 2015, 527, 384–388. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.; Dolan, L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-B.; Bokowiec, M.T.; Rushton, P.J.; Han, S.-C.; Timko, M.P. Tobacco Transcription Factors NtMYC2a and NtMYC2b Form Nuclear Complexes with the NtJAZ1 Repressor and Regulate Multiple Jasmonate-Inducible Steps in Nicotine Biosynthesis. Mol. Plant 2012, 5, 73–84. [Google Scholar] [CrossRef]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.-M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Castelain, M.; Le Hir, R.; Bellini, C. The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiol. Plant 2012, 145, 450–460. [Google Scholar] [CrossRef]
- Friedrichsen, D.M.; Nemhauser, J.; Muramitsu, T.; Maloof, J.N.; Alonso, J.; Ecker, J.R.; Furuya, M.; Chory, J. Three Redundant Brassinosteroid Early Response Genes Encode Putative bHLH Transcription Factors Required for Normal Growth. Genetics 2002, 162, 1445–1456. [Google Scholar] [CrossRef]
- Garin, J.P.; Pellerat, J.; Maillard; Woehrle-Hezez, R. Theoretical basis of the prevention of congenital toxoplasmosis in pregnant women by spiramycin. Lyon Med. 1969, 221, 21–25. [Google Scholar] [PubMed]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef] [PubMed]
- Hetherington, A.M.; Woodward, F.I. The role of stomata in sensing and driving environmental change. Nature 2003, 424, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.E.; Kwon, Y.; Kim, J.H.; Noh, H.; Hong, S.-W.; Lee, H. A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol. Biol. 2011, 77, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.-J. Overexpression of AtMYB44 Enhances Stomatal Closure to Confer Abiotic Stress Tolerance in Transgenic Arabidopsis. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, M.R.; Feurtado, J.A.; Huang, D.; Lu, Y.; Cutler, A.J. Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 2013, 13, 192. [Google Scholar] [CrossRef]
- Li, D.; Li, Y.; Zhang, L.; Wang, X.; Zhao, Z.; Tao, Z.; Wang, J.; Wang, J.; Lin, M.; Li, X.; et al. Arabidopsis ABA Receptor RCAR1/PYL9 Interacts with an R2R3-Type MYB Transcription Factor, AtMYB44. Int. J. Mol. Sci. 2014, 15, 8473–8490. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.-J.; Gao, J.; Wang, P.; Duan, C.-G.; Zhu, X.; Zhu, J.-K. The ABA Receptor PYL8 Promotes Lateral Root Growth by Enhancing MYB77-Dependent Transcription of Auxin-Responsive Genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Wei, Q.; Li, B.; Zhong, X.; Hu, T.; Hu, H.; Bao, C. Transcriptome-Wide Identification and Characterization of Circular RNAs in Leaves of Chinese Cabbage (Brassica rapa L. ssp. pekinensis) in Response to Calcium Deficiency-Induced Tip-burn. Sci. Rep. 2019, 9, 14544. [Google Scholar] [CrossRef]
- Tripathi, R.K.; Wilkins, O. Single cell gene regulatory networks in plants: Opportunities for enhancing climate change stress resilience. Plant Cell Environ. 2021, 44, 2006–2017. [Google Scholar] [CrossRef]
- Martinez-Pastor, M.; Tonner, P.D.; Darnell, C.L.; Schmid, A.K. Transcriptional Regulation in Archaea: From Individual Genes to Global Regulatory Networks. Annu. Rev. Genet. 2017, 51, 143–170. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasulu, N.; Wobus, U. Seed-Development Programs: A Systems Biology–Based Comparison Between Dicots and Monocots. Annu. Rev. Plant Biol. 2013, 64, 189–217. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.G.; Hsu, S.-W.; Harada, J.J.; Belmonte, M.F. Genomic dissection of the seed. Front. Plant Sci. 2014, 5, 464. [Google Scholar]
- Haas, K.T.; Wightman, R.; Peaucelle, A.; Höfte, H. The role of pectin phase separation in plant cell wall assembly and growth. Cell Surf. 2021, 7, 100054. [Google Scholar] [CrossRef]
- Su, T.; Yu, S.; Yu, R.; Zhang, F.; Yu, Y.; Zhang, D.; Zhao, X.; Wang, W. Effects of Endogenous Salicylic Acid During Calcium Deficiency-Induced Tipburn in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Plant Mol. Biol. Rep. 2016, 34, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, L.; Gao, H.; Song, Q.; Gao, J. First Report of a New Bacterial Leaf Blight of Chinese Cabbage (Brassica rapa pekinensis) Caused by Pantoea ananatis in China. Plant Dis. 2019, 103, 2942. [Google Scholar] [CrossRef]
- Galant, A.; Preuss, M.L.; Cameron, J.C.; Jez, J.M. Plant Glutathione Biosynthesis: Diversity in Biochemical Regulation and Reaction Products. Front. Plant Sci. 2011, 2, 45. [Google Scholar] [CrossRef]
- Zhang, H.; Forman, H.J. Glutathione synthesis and its role in redox signaling. Semin. Cell Dev. Biol. 2012, 23, 722–728. [Google Scholar] [CrossRef]
- Gallé, A.; Czékus, Z.; Bela, K.; Horváth, E.; Ördög, A.; Csiszár, J.; Poór, P. Plant Glutathione Transferases and Light. Front. Plant Sci. 2019, 9, 1944. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Cho, Y.-G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar] [CrossRef]
- Tian, Q.; Sun, P.; Zhang, W. Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. New Phytol. 2009, 184, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.-L.; Fan, Z.-Q.; Shan, W.; Yin, X.-R.; Kuang, J.-F.; Lu, W.-J.; Chen, J.-Y. Association of BrERF72 with methyl jasmonate-induced leaf senescence of Chinese flowering cabbage through activating JA biosynthesis-related genes. Hortic. Res. 2018, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Ludwig, A.A.; Saitoh, H.; Felix, G.; Freymark, G.; Miersch, O.; Wasternack, C.; Boller, T.; Jones, J.D.G.; Romeis, T. Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl. Acad. Sci. USA 2005, 102, 10736–10741. [Google Scholar] [CrossRef]
- Liu, Q.; Wen, C.-K. Arabidopsis ETR1 and ERS1 Differentially Repress the Ethylene Response in Combination with Other Ethylene Receptor Genes. Plant Physiol. 2012, 158, 1193–1207. [Google Scholar] [CrossRef]
- Datta, R.; Kumar, D.; Sultana, A.; Hazra, S.; Bhattacharyya, D.; Chattopadhyay, S. Glutathione regulates ACC synthase transcription via WRKY33 and ACC oxidase by modulating mRNA stability to induce ethylene synthesis during stress. Plant Physiol. 2015, 169, 2963–2981. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Araki, T.; Endo, M. Circadian clock during plant development. J. Plant Res. 2018, 131, 59–66. [Google Scholar] [CrossRef]
- Xu, C.; Zhu, Q.; Chen, J.; Shen, L.; Yi, X.; Huang, Z.; Wang, W.; Chen, M.; Kuang, T.; Shen, J.; et al. A unique photosystem I reaction center from a chlorophyll d -containing cyanobacterium Acaryochloris marina. J. Integr. Plant Biol. 2021, 63, 1740–1752. [Google Scholar] [CrossRef]
- Zou, Z. Mining Gene Families in the Castor Bean Genome. In The Castor Bean Genome; Kole, C., Rabinowicz, P., Eds.; Compendium of Plant Genomes; Springer: Cham, Switzerland, 2018; Chapter 8; pp. 135–173. [Google Scholar]
- Zou, Z.; Li, M.; Jia, R.; Zhao, H.; He, P.; Zhang, Y.; Guo, A. Genes encoding light-harvesting chlorophyll a/b-binding proteins in papaya (Carica papaya L.) and insight into lineage-specific evolution in Brassicaceae. Gene 2020, 748, 144685. [Google Scholar] [CrossRef]
- Engelken, J.; Brinkmann, H.; Adamska, I. Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol. Biol. 2010, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. Understanding the Complexity of Iron Sensing and Signaling Cascades in Plants. Plant Cell Physiol. 2019, 60, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yin, F.; Bao, Y. Genome-wide identification and characterization of basic helix-loop-helix genes in nine molluscs. Gene 2021, 785, 145604. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Feng, R.; Ma, R.; Shen, Z.; Cai, Z.; Song, Z.; Peng, B.; Yu, M. Genome-wide analysis of basic helix-loop-helix superfamily members in peach. PLoS ONE 2018, 13, e0195974. [Google Scholar] [CrossRef]
- Mao, T.-Y.; Liu, Y.-Y.; Zhu, H.-H.; Zhang, J.; Yang, J.-X.; Fu, Q.; Wang, N.; Wang, Z. Genome-wide analyses of the bHLH gene family reveals structural and functional characteristics in the aquatic plant Nelumbo nucifera. PeerJ 2019, 7, e7153. [Google Scholar] [CrossRef]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and Calcineurin B–like Proteins. Plant Cell 2002, 14, S389–S400. [Google Scholar] [CrossRef]
- Bouché, N.; Yellin, A.; Snedden, W.A.; Fromm, H. Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 2005, 56, 435–466. [Google Scholar] [CrossRef]
- Zhou, Y.; Cai, H.; Xiao, J.; Li, X.; Zhang, Q.; Lian, X. Over-expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor. Appl. Genet. 2009, 118, 1381–1390. [Google Scholar] [CrossRef]
- Yu, G.-H.; Zou, J.; Feng, J.; Peng, X.-B.; Wu, J.-Y.; Wu, Y.-L.; Palanivelu, R.; Sun, M.-X. Exogenous γ-aminobutyric acid (GABA) affects pollen tube growth via modulating putative Ca2+-permeable membrane channels and is coupled to negative regulation on glutamate decarboxylase. J. Exp. Bot. 2014, 65, 3235–3248. [Google Scholar] [CrossRef]
- van Doorn, W.G. Classes of programmed cell death in plants, compared to those in animals. J. Exp. Bot. 2011, 62, 4749–4761. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, Z.; Kong, X.; Chen, Y.; Li, J. Exogenous tryptophan application improves cadmium tolerance and inhibits cadmium upward transport in broccoli (Brassica oleracea var. italica). Front. Plant Sci. 2022, 13, 969675. [Google Scholar] [CrossRef]
- Arnao, M.B.; Ruiz, J.H. Melatonin: A New Plant Hormone and/or a Plant Master Regulator? Trends Plant Sci. 2019, 24, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kang, Y.; Zhong, M.; Zhang, L.; Chai, X.; Jiang, X.; Yang, X. Effects of Iron Deficiency Stress on Plant Growth and Quality in Flowering Chinese Cabbage and Its Adaptive Response. Agronomy 2022, 12, 875. [Google Scholar] [CrossRef]
- Hoagland, R.D.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil. Calif. Agr. Exp. Sta. Circ. 1950, 347, 357–359. [Google Scholar]
- Floros, K.V.; Jacob, S.; Kurupi, R.; Fairchild, C.K.; Hu, B.; Puchalapalli, M.; Koblinski, J.E.; Dozmorov, M.G.; Boikos, S.A.; Scaltriti, M.; et al. Targeting transcription of MCL-1 sensitizes HER2-amplified breast cancers to HER2 inhibitors. Cell Death Dis. 2021, 12, 179. [Google Scholar] [CrossRef]
- Zhan, J.; Thakare, D.; Ma, C.; Lloyd, A.; Nixon, N.M.; Arakaki, A.M.; Burnett, W.J.; Logan, K.O.; Wang, D.; Wang, X.; et al. RNA Sequencing of Laser-Capture Microdissected Compartments of the Maize Kernel Identifies Regulatory Modules Associated with Endosperm Cell Differentiation. Plant Cell 2015, 27, 513–531. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, C.; Yan, X.; Zhang, J.; Xu, J. Simultaneous analysis of ten phytohormones in Sargassum horneri by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J. Sep. Sci. 2016, 39, 1804–1813. [Google Scholar] [CrossRef] [PubMed]
- Floková, K.; Tarkowská, D.; Miersch, O.; Strnad, M.; Wasternack, C.; Novák, O. UHPLC–MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 2014, 105, 147–157. [Google Scholar] [CrossRef]
- Cai, B.-D.; Zhu, J.-X.; Gao, Q.; Luo, D.; Yuan, B.-F.; Feng, Y.-Q. Rapid and high-throughput determination of endogenous cytokinins in Oryza sativa by bare Fe3O4 nanoparticles-based magnetic solid-phase extraction. J. Chromatogr. 2014, 1340, 146–150. [Google Scholar] [CrossRef]
- Xiao, H.-M.; Cai, W.-J.; Ye, T.-T.; Ding, J.; Feng, Y.-Q. Spatio-temporal profiling of abscisic acid, indoleacetic acid and jasmonic acid in single rice seed during seed germination. Anal. Chim. Acta 2018, 1031, 119–127. [Google Scholar] [CrossRef]
- Niu, Q.; Zong, Y.; Qian, M.; Yang, F.; Teng, Y. Simultaneous quantitative determination of major plant hormones in pear flowers and fruit by UPLC/ESI-MS/MS. Anal. Methods 2014, 6, 1766–1773. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Gao, H.; Wang, L.; Zhang, Y.; Zhou, D.; Anwar, A.; Li, J.; Wang, F.; Li, C.; Zhang, Y.; et al. Comparative Transcriptome and Co-Expression Network Analyses Reveal the Molecular Mechanism of Calcium-Deficiency-Triggered Tipburn in Chinese Cabbage (Brassica rapa L. ssp. Pekinensis). Plants 2022, 11, 3555. https://doi.org/10.3390/plants11243555
Zhang S, Gao H, Wang L, Zhang Y, Zhou D, Anwar A, Li J, Wang F, Li C, Zhang Y, et al. Comparative Transcriptome and Co-Expression Network Analyses Reveal the Molecular Mechanism of Calcium-Deficiency-Triggered Tipburn in Chinese Cabbage (Brassica rapa L. ssp. Pekinensis). Plants. 2022; 11(24):3555. https://doi.org/10.3390/plants11243555
Chicago/Turabian StyleZhang, Shu, Hanzhong Gao, Lixia Wang, Yihui Zhang, Dandan Zhou, Ali Anwar, Jingjuan Li, Fengde Wang, Cheng Li, Ye Zhang, and et al. 2022. "Comparative Transcriptome and Co-Expression Network Analyses Reveal the Molecular Mechanism of Calcium-Deficiency-Triggered Tipburn in Chinese Cabbage (Brassica rapa L. ssp. Pekinensis)" Plants 11, no. 24: 3555. https://doi.org/10.3390/plants11243555
APA StyleZhang, S., Gao, H., Wang, L., Zhang, Y., Zhou, D., Anwar, A., Li, J., Wang, F., Li, C., Zhang, Y., & Gao, J. (2022). Comparative Transcriptome and Co-Expression Network Analyses Reveal the Molecular Mechanism of Calcium-Deficiency-Triggered Tipburn in Chinese Cabbage (Brassica rapa L. ssp. Pekinensis). Plants, 11(24), 3555. https://doi.org/10.3390/plants11243555








