Transcriptome and Gene Regulatory Network Analyses Reveal New Transcription Factors in Mature Fruit Associated with Harvest Date in Prunus persica
Abstract
1. Introduction
2. Results
2.1. Fruit Quality Attributes Evaluation on Individuals with Contrasting Harvest Dates
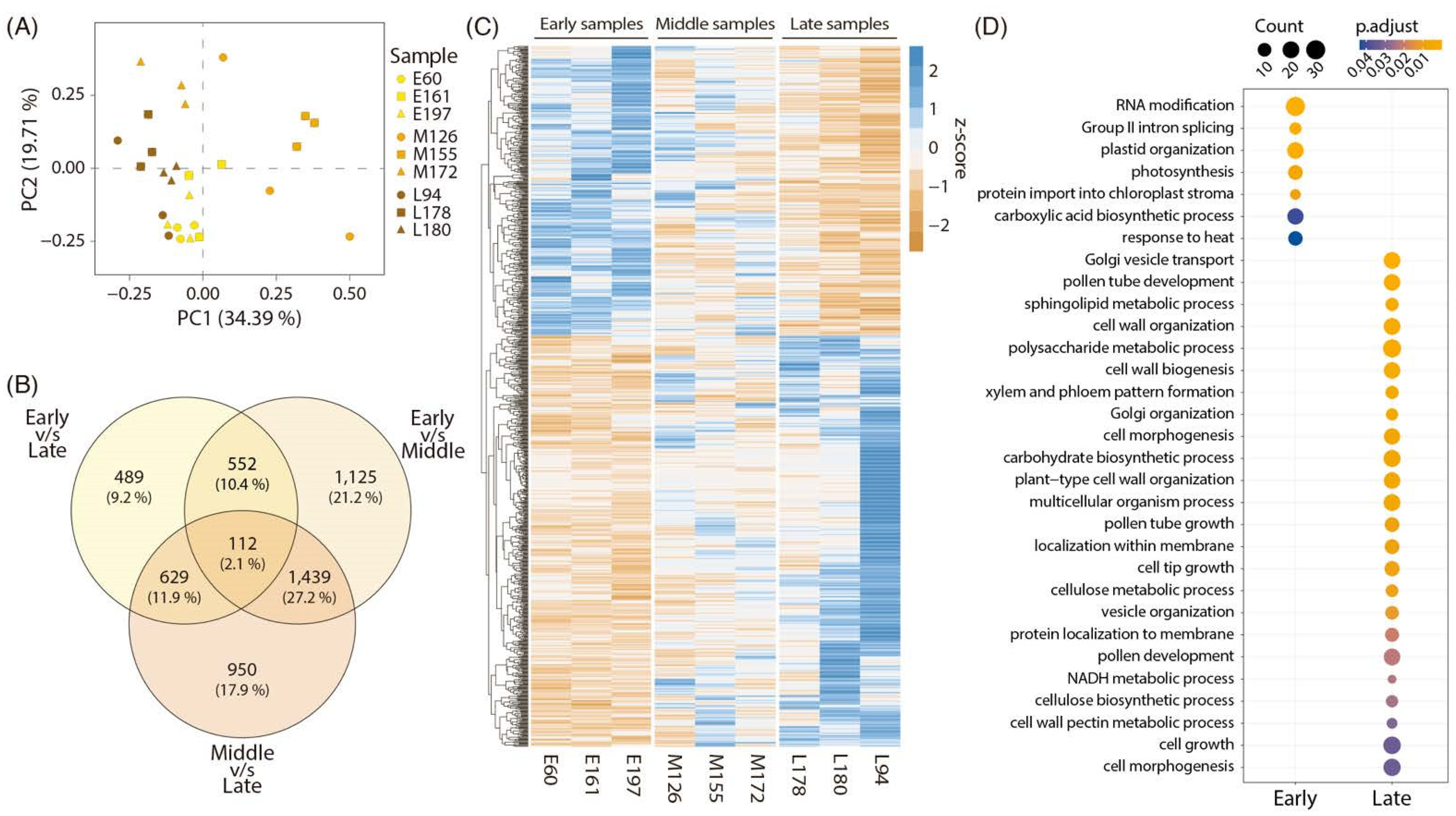
2.2. RNAseq Analysis and Bioinformatic Results
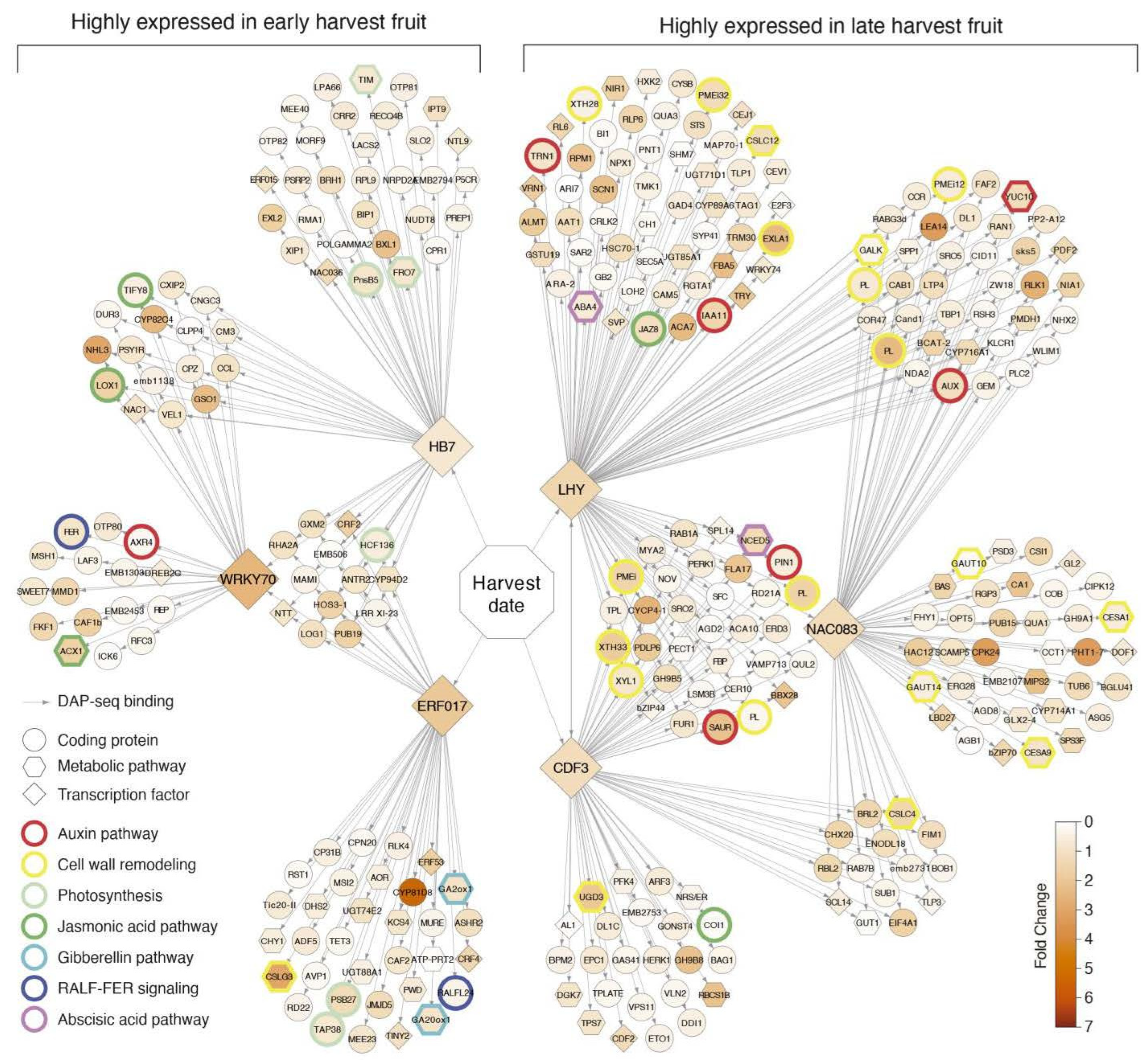
2.3. Network Analysis of Differentially Expressed Genes between Early and Late Harvest Samples
2.4. Candidate Gene Selection and Evaluation in Contrasting Harvest Date Peach Varieties
3. Discussion
3.1. ‘O×N’ Segregating Population Sequencing Evaluation and Bioinformatic Analysis for Harvest Date
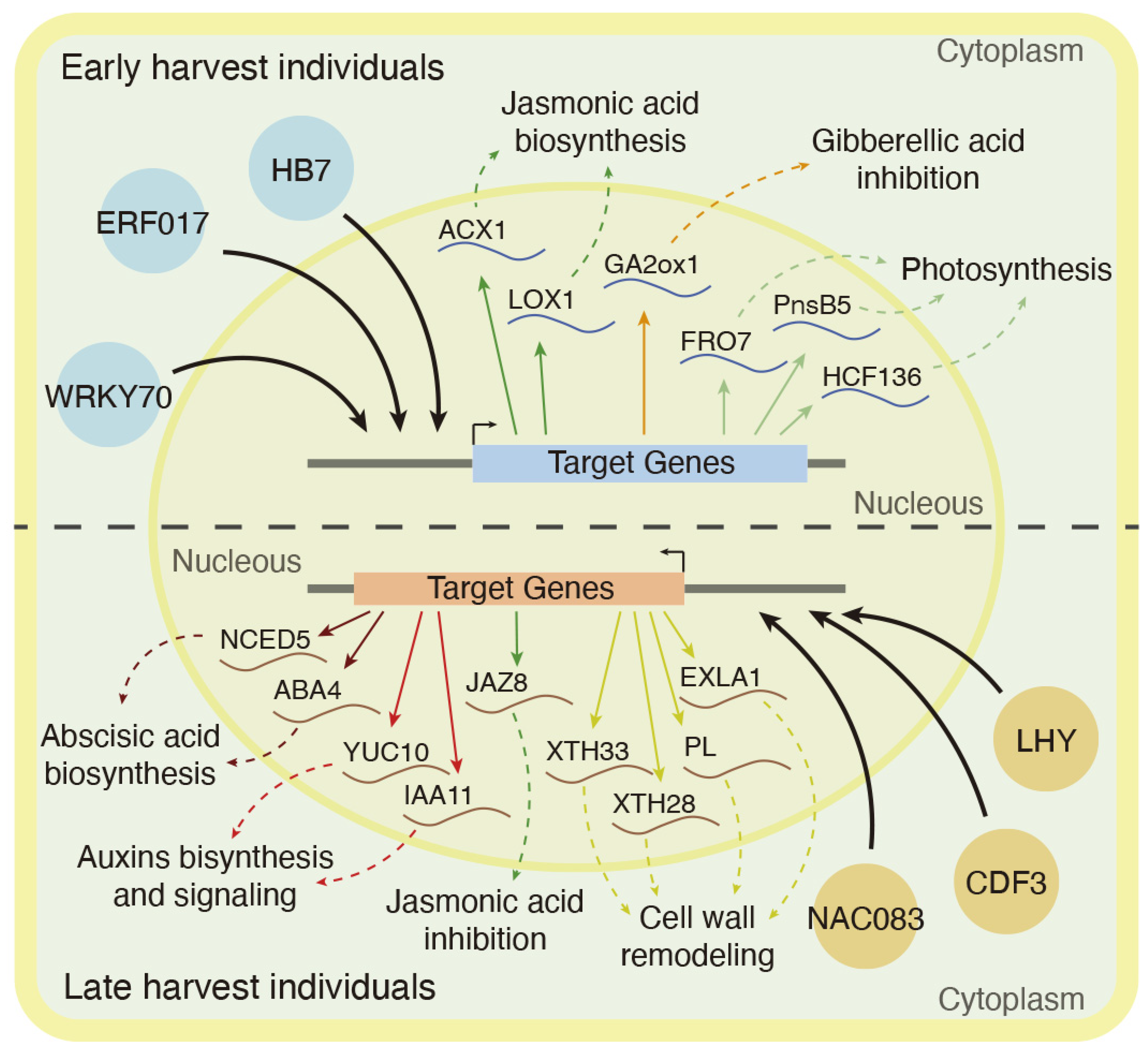
3.2. Hormonal Regulation Mechanisms Associated with Late Harvest Samples and Cell Wall Remodeling Enzymes
3.3. Jasmonic Acid Biosynthesis and Gibberellin Inhibition as Hormonal Signals Associated with Early Harvest Samples
3.4. Transcriptional Regulation of Harvest Date Phenotype
4. Materials and Methods
4.1. Plant Material
4.2. RNA Extraction and Library Construction
4.3. Sequencing Data Analysis and Network Construction
4.4. RT-qPCR Gene Evaluation in Peach Varieties
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arús, P.; Verde, I.; Sosinski, B.; Zhebentyayeva, T.; Abott, A.G. The peach genome. Tree Genet. Genomes 2012, 8, 531–547. [Google Scholar] [CrossRef]
- Monti, L.L.; Bustamante, C.A.; Osorio, S.; Gabilondo, J.; Borsani, J.; Lauxmann, M.A.; Maulión, E.; Valentini, G.; Budde, C.O.; Fernie, A.R.; et al. Metabolic profiling of a range of peach fruit varieties reveals high metabolic diversity and commonalities and differences during ripening. Food Chem. 2016, 190, 879–888. [Google Scholar] [CrossRef]
- Zhang, J.; Cheng, D.; Wang, B.; Khan, I.; Ni, Y. Ethylene control technologies in extending the postharvest shelf life of climacteric fruit. J. Agric. Food Chem. 2017, 65, 7308–7319. [Google Scholar] [CrossRef]
- Martínez-García, P.J.; Peace, C.P.; Parfitt, D.E.; Ogundiwin, E.A.; Fresnedo-Ramírez, J.; Dandekar, A.M.; Gradziel, T.M.; Crisosto, C.H. Influence of year and genetic factors on chilling injury susceptibility in peach (Prunus persica (L.) Batsch). Euphytica 2012, 185, 267–280. [Google Scholar] [CrossRef]
- Infante, R.; Meneses, C.; Byrne, D. Present situation of peach breeding programs: Postharvest and fruit quality assessment. Acta Hortic. 2006, 713, 121–124. [Google Scholar] [CrossRef]
- Pirona, R.; Eduardo, I.; Pacheco, I.; Linge, C.D.; Miculan, M.; Verde, I.; Tartarini, S.; Dondini, L.; Pea, G.; Bassi, D.; et al. Fine mapping and identification of a candidate gene for a major locus controlling maturity date in peach. BMC Plant Biol. 2013, 13, 166–179. [Google Scholar] [CrossRef]
- Cantín, C.M.; Crisosto, C.H.; Ogundiwin, E.A.; Gradziel, T.; Torrents, J.; Moreno, M.A.; Gogorcena, Y. Chilling injury susceptibility in an intraspecific peach [Prunus persica (L.) Batsch] progeny. Postharvest Biol. Technol. 2010, 58, 79–87. [Google Scholar] [CrossRef]
- Romeu, J.F.; Monforte, A.J.; Sánchez, G.; Granell, A.; García-Brunton, J.; Badenes, M.L.; Ríos, G. Quantitative trait loci affecting reproductive phenology in peach. BMC Plant Biol. 2014, 14, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Nuñez-Lillo, G.; Cifuentes-Esquivel, A.; Troggio, M.; Micheletti, D.; Infante, R.; Campos-Vargas, R.; Orellana, A.; Blanco-Herrera, F.; Meneses, C. Identification of candidate genes associated with mealiness and maturity date in peach [Prunus persica (L.) Batsch] using QTL analysis and deep sequencing. Tree Genet. Genomes 2015, 11, 86–98. [Google Scholar] [CrossRef]
- Lurie, S.; Friedman, H.; Weksler, A.; Dagar, A.; Zerbini, P.E. Maturity assessment at harvest and prediction of softening in an early and late season melting peach. Postharvest Biol. Technol. 2013, 76, 10–16. [Google Scholar] [CrossRef]
- Calontuono, F.; Amodio, M.L.; Piazzolla, F.; Colelli, G. Influence of quality attributes of early, intermediate and late peach varieties on suitability as fresh-convenience products. Adv. Hortic. Sci. 2012, 26, 32–38. [Google Scholar] [CrossRef]
- Meneses, C.; Ulloa-Zepeda, L.; Cifuentes-Esquivel, A.; Infante, R.; Cantin, C.M.; Batlle, I.; Arús, P.; Eduardo, I. A codominant diagnostic marker for the slow ripening trait in peach. Mol. Breed. 2016, 36, 77. [Google Scholar] [CrossRef]
- Núñez-Lillo, G.; Ulloa-Zepeda, L.; Pavez, C.; Riveros, A.; Blanco-Herrera, F.; Campos-Vargas, R.; Pedreschi, R.; Meneses, C. Unravelling the molecular regulation mechanisms of slow ripening trait in Prunus persica. Plants 2021, 10, 2380. [Google Scholar] [CrossRef]
- Tonutti, P.; Bonghi, C.; Ruperti, B.; Tornielli, G.B.; Ramina, A. Ethylene evolution and 1-aminocyclopropane-1-carboxylate oxidase gene expression during early development and ripening of peach fruit. J. Am. Soc. Hortic. Sci. 1997, 122, 642–647. [Google Scholar] [CrossRef]
- Paul, V.; Pandey, R.; Srivastava, G.C. The fading distinctions between classical patterns of ripening in climacteric and non-climacteric fruit and the ubiquity of ethylene—An overview. J. Food Sci. Technol. 2012, 49, 1–21. [Google Scholar] [CrossRef]
- Li, C.; Jia, H.; Chai, Y.; Shen, Y. Abscisic acid perception and signaling transduction in strawberry: A model for non-climacteric fruit ripening. Plant Signal. Behav. 2011, 6, 1950–1953. [Google Scholar] [CrossRef]
- Zaharah, S.S.; Singh, Z.; Symons, G.M.; Reid, J.B. Mode of action of abscisic acid in triggering ethylene biosynthesis and softening during ripening in mango fruit. Postharvest Biol. Technol. 2013, 75, 37–44. [Google Scholar] [CrossRef]
- Kou, X.; Yang, S.; Chai, L.; Wu, C.; Zhou, J.; Liu, Y.; Xue, Z. Abscisic acid and fruit ripening: Multifaceted analysis of the effect of abscisic acid on fleshy fruit ripening. Sci. Hortic. 2021, 281, 109999. [Google Scholar] [CrossRef]
- Ziosi, V.; Bonghi, C.; Bregoli, A.M.; Trainotti, L.; Biondi, S.; Sutthiwal, S.; Kondo, S.; Costa, G.; Torrigiani, P. Jasmonate-induced transcriptional changes suggest a negative interference with the ripening syndrome in peach fruit. J. Exp. Bot. 2008, 59, 563–573. [Google Scholar] [CrossRef]
- Soto, A.; Ruiz, K.B.; Ziosi, V.; Costa, G.; Torrigiani, P. Ethylene and auxin biosynthesis and signaling are impaired by methyl jasmonate leading to a transient slowing down of ripening in peach fruit. J. Plant Physiol. 2012, 18, 1858–1865. [Google Scholar] [CrossRef]
- Ruiz, K.B.; Trainotti, L.; Bonghi, C.; Ziosi, V.; Costa, G.; Torrigiani, P. Early methyl jasmonate application to peach delays fruit/seed development by altering the expression of multiple hormone-related genes. J. Plant Growth Regul. 2013, 32, 852–864. [Google Scholar] [CrossRef]
- Wei, J.; Wen, X.; Tang, L. Effect of methyl jasmonic acid on peach fruit ripening progress. Sci. Hortic. 2017, 220, 206–213. [Google Scholar] [CrossRef]
- Concha, C.M.; Figueroa, N.E.; Poblete, L.A.; Oñate, F.A.; Schwab, W.; Figueroa, C.R. Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in Fragaria chiloensis fruit. Plant Physiol. Biochem. 2013, 70, 433–444. [Google Scholar] [CrossRef]
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Trainotti, L.; Zanin, D.; Casadoro, G. A cell wall-oriented genomic approach reveals a new and unexpected complexity of the softening in peaches. J. Exp. Bot. 2003, 54, 1821–1832. [Google Scholar] [CrossRef]
- Brummell, D.A.; Dal Cin, V.; Crisosto, C.H.; Labavitch, J.M. Cell wall metabolism during maturation, ripening and senescence of peach fruit. J. Exp. Bot. 2004, 55, 2029–2039. [Google Scholar] [CrossRef]
- The International Peach Genome Initiative; Verde, I.; Abbott, A.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef]
- Núñez-Lillo, G.; Balladares, C.; Pavez, C.; Urra, C.; Sanhueza, D.; Vendramin, E.; Dettori, M.T.; Arús, P.; Verde, I.; Blanco-Herrera, F.; et al. High-density genetic map and QTL analysis of soluble solid content, maturity date and mealiness in peach using genotyping by sequencing. Sci. Hortic. 2019, 257, 108734. [Google Scholar] [CrossRef]
- Xiong, L.; Zhu, J.K. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003, 133, 29–36. [Google Scholar] [CrossRef]
- Perreau, F.; Frey, A.; Effroy-Cuzzi, D.; Savane, P.; Berger, A.; Gissot, L.; Marion-Poll, A. Abscisic acid-deficient 4 has an essential function in both cis-violaxanthin and cis-neoxanthin synthesis. Plant Physiol. 2020, 184, 1303–1316. [Google Scholar] [CrossRef]
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512. [Google Scholar] [CrossRef]
- Gavassi, M.A.; Silva, G.S.; da Silva, C.d.M.S.; Thompson, A.J.; Macleod, K.; Oliveira, P.M.R.; Cavalheiro, M.F.; Domingues, D.S.; Habermann, G. NCED expression is related to increased ABA biosynthesis and stomatal closure under aluminum stress. Environ. Exp. Bot. 2021, 185, 104404. [Google Scholar] [CrossRef]
- Hernández, I.; Uarrota, V.; Fuentealba, C.; Paredes, D.; Defilippi, B.G.; Campos-Vargas, R.; Nuñez, G.; Carrera, E.; Meneses, C.; Hertog, M.; et al. Transcriptome and hormone analysis reveals differences in physiological age of ‘Hass’ avocado fruit. Postharvest Biol. Technol. 2022, 185, 111806. [Google Scholar] [CrossRef]
- Soto, A.; Ruiz, K.B.; Ravaglia, D.; Costa, G.; Torrigiani, P. ABA may promote or delay peach fruit ripening through modulation of ripening- and hormone-related gene expression depending on the developmental stage. Plant Physiol. Biochem. 2013, 64, 11–24. [Google Scholar] [CrossRef]
- Diao, D.; Hu, X.; Guan, D.; Wang, W.; Yang, H.; Liu, Y. Genome-wide identification of the ARF (Auxin response factor) gene family in peach and their expression analysis. Mol. Biol. Rep. 2020, 47, 4331–4344. [Google Scholar] [CrossRef]
- Ohmiya, A. Effects of auxin on growth and ripening of mesocarp discs of peach fruit. Sci. Hortic. 2000, 84, 309–319. [Google Scholar] [CrossRef]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Function of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef]
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.S.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2021, 40, 1513–1541. [Google Scholar] [CrossRef]
- Wasternack, C.; Goetz, S.; Hellwege, A.; Forner, S.; Strnad, M.; Hause, B. Another JA/COI1-independent role of OPDA detected in tomato embryo development. Plant Signal. Behav. 2012, 7, 1349–1353. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, C.; Pollmann, S. Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 2009, 276, 4693–4704. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Chakrabarty, S.K. Gibberellic acid in plant. Plant Signal. Behav. 2013, 8, e25504. [Google Scholar] [CrossRef] [PubMed]
- Pergoraro, C.; Zanuzo, M.R.; Chaves, F.C.; Brackmann, A.; Girardi, C.L.; Lucchetta, L.; Nora, L.; Silva, J.A.; Rombaldi, C.V. Physiological and molecular changes associated with prevention of woolliness in peach following pre-harvest application of gibberellic acid. Postharvest Biol. Technol. 2010, 57, 19–26. [Google Scholar] [CrossRef]
- Dagar, A.; Weksler, A.; Friedman, H.; Lurie, S. Gibberellic acid (GA3) application at the end of pit ripening: Effect on ripening and storage of two harvests of ‘September snow’ peach. Sci. Hortic. 2012, 140, 125–130. [Google Scholar] [CrossRef]
- Rodrigues, C.; Vandenbergue, L.P.d.S.; de Oliveira, J.; Soccol, C.R. New perspectives of gibberellic acid production: A review. Crit. Rev. Biotechnol. 2012, 32, 263–273. [Google Scholar] [CrossRef]
- Ré, D.A.; Capella, M.; Bonaventure, G.; Chan, R.L. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 2014, 14, 150. [Google Scholar] [CrossRef]
- Valdés, A.E.; Overnäs, E.; Johansson, H.; Rada-Iglesias, A.; Engström, P. The homeodomain-leucine zipper (HB-Zip) class-I transcription factor ATHB7 and ATHB12 modulate abscisic acid signaling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 2012, 80, 405–418. [Google Scholar] [CrossRef]
- Yin, X.R.; Xie, X.L.; Xia, X.J.; Yu, J.Q.; Ferguson, I.B.; Giovannoni, J.J.; Chen, K.S. Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening. Plant J. 2016, 86, 403–412. [Google Scholar] [CrossRef]
- Han, Z.; Hu, Y.; Lv, Y.; Rose, J.K.C.; Sun, Y.; Shen, F.; Wang, Y.; Zhang, X.; Xu, X.; Wu, T.; et al. Natural variation underlies differences in ETHYLENE RESPONSE FACTOR17 activity in fruit peel degreening. Plant Physiol. 2018, 176, 2292–2304. [Google Scholar] [CrossRef]
- Shim, J.S.; Jung, C.; Lee, S.; Min, K.; Lee, Y.W.; Choi, Y.; Lee, J.S.; Song, J.T.; Kim, J.K.; Choi, Y.D. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 2012, 73, 483–495. [Google Scholar] [CrossRef]
- Domínguez-Figueroa, J.; Carrillo, L.; Renau-Morata, B.; Yang, L.; Molina, R.V.; Marino, D.; Canales, J.; Weih, M.; Vicente-Carbajosa, J.; Nebauer, S.G.; et al. The Arabidopsis transcription factor DOF3 is involved in nitrogen response and improves nitrogen use efficiency in tomato. Front. Plant Sci. 2020, 11, 601558. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef] [PubMed]
- Min, K.; Yi, G.; Lee, J.G.; Kim, H.S.; Hong, Y.; Choi, J.H.; Lim, S.; Lee, E.J. Comparative transcriptome and metabolome analyses of two strawberry cultivars with different storability. PLoS ONE 2020, 15, e0242556. [Google Scholar] [CrossRef]
- Carré, I.A.; Kim, J.Y. MYB-transcription factors in the Arabidopsis circadian clock. J. Exp. Bot. 2002, 53, 1551–1557. [Google Scholar] [CrossRef]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.L.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Grundy, J.; Veflingstad, S.R.; Dyer, N.P.; Hannah, M.A.; Ott, S.; Carré, I.A. Circadian control of abscisic acid biosynthesis and signaling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 2018, 220, 893–907. [Google Scholar] [CrossRef]
- Xue, X.; Sun, K.; Zhu, Z. CIRCADIAN CLOCK ASSOCIATED 1 gates morning phased auxin response in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2020, 527, 935–940. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Horikoshi, M.; Li, W. ggfortify: Unified Interface to Visualize Statistical Result of Popular R Packages. R J. 2016, 8, 478–489. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Brooks, M.D.; Juang, C.L.; Katari, M.S.; Alvarez, J.M.; Pasquino, A.; Shih, H.J.; Huang, J.; Shanks, C.; Cirrone, J.; Coruzzi, G.M. ConnecTF: A platform to integrate transcription factor-gene interactions and validate regulatory networks. Plant Physiol. 2021, 185, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]

| Quality Trait | ‘O×N’ Population | Varieties | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Early Harvest | Middle Harvest | Late Harvest | BB | LRJ | |||||||
| E60 | E161 | E197 | M126 | M155 | M172 | L94 | L178 | L180 | |||
| IAD | 1.1 a | 1.1 a | 1.0 a | 1.1 a | 1.1 a | 1.1 a | 1.1 a | 1.1 a | 0.9 a | 0.8 A | 1.2 A |
| Harvest date (DAB) | 127.7 ab | 123.7 a | 132.3 ab | 136.0 abc | 134.7 abc | 139.7 bcd | 157.3 e | 149.3 cde | 153.3 de | 104.5 A | 170.0 B |
| Weight (g) | 153.6 ab | 170.9 ab | 154.1 ab | 133.3 a | 160.7 ab | 160.7 ab | 164.9 ab | 275.2 c | 217.5 bc | 112.9 A | 207.6 B |
| SSC (ºBrix) | 12.4 ab | 13.0 b | 11.0 ab | 12.8 b | 12.3 ab | 8.8 a | 14.4 b | 11.2 ab | 13.9 b | 15.4 A | 15.8 A |
| Firmness (N) | 7.0 ab | 11.1 b | 10.3 ab | 10.4 ab | 11.2 b | 9.1 ab | 6.4 a | 8.2 ab | 6.4 a | 14.1 A | 15.1 A |
| PpersicaID | AthalianaID | Symbol | Description | Normalized Expression * | ||
|---|---|---|---|---|---|---|
| Early | Middle | Late | ||||
| Prupe.2G316600 | AT2G46680 | HB7 | Homeobox 7 | 172 | 96 | 86 |
| Prupe.7G194400 | AT1G19210 | ERF017 | Ethylene-responsive transcription factor ERF017 | 231 | 197 | 50 |
| Prupe.2G265000 | AT3G56400 | WRKY70 | WRKY DNA-binding protein 70 | 87 | 18 | 12 |
| Prupe.2G200400 | AT1G01060 | LHY | MYB-related transcription factor LHY | 122 | 153 | 404 |
| Prupe.5G194600 | AT3G47500 | CDF3 | Cycling DOF factor 3 | 58 | 103 | 157 |
| Prupe.1G220400 | AT5G13180 | NAC083 | NAC domain containing protein 83 | 2234 | 2459 | 3404 |
| Prupe.1G478600 | AT4G32410 | CESA1 | Cellulose synthase A1 | 1308 | 1814 | 2255 |
| Prupe.8G035100 | AT2G21770 | CESA9 | Cellulose synthase A9 | 2104 | 3247 | 4602 |
| Prupe.1G418400 | AT4G07960 | CSLC12 | Cellulose-synthase-like C12 | 596 | 840 | 1575 |
| Prupe.3G280100 | AT3G28180 | CSLC4 | Cellulose-synthase-like C4 | 75 | 162 | 241 |
| Prupe.8G174500 | AT3G45970 | EXLA1 | Expansin-like A1 | 226 | 511 | 1407 |
| Prupe.3G258200 | AT5G15470 | GAUT14 | Galacturonosyltransferase 14 | 793 | 1003 | 1339 |
| Prupe.2G206100 | AT5G04310 | PL | Pectin lyase-like superfamily protein | 37 | 42 | 137 |
| Prupe.1G129300 | AT3G16850 | PL | Pectin lyase-like superfamily protein | 202 | 280 | 393 |
| Prupe.4G271300 | AT4G33440 | PL | Pectin lyase-like superfamily protein | 185 | 211 | 264 |
| Prupe.1G114500 | AT1G47960 | PMEi | Pectin methylesterase inhibitor superfamily | 1153 | 2908 | 3911 |
| Prupe.7G190400 | AT2G26440 | PMEi12 | Pectin methylesterase inhibitor superfamily | 853 | 1082 | 1552 |
| Prupe.7G190300 | AT3G43270 | PMEi32 | Pectin methylesterase inhibitor superfamily | 1512 | 3079 | 4303 |
| Prupe.5G202800 | AT5G15490 | UGD3 | UDP-glucose 6-dehydrogenase family protein | 55 | 113 | 264 |
| Prupe.1G337000 | AT1G14720 | XTH28 | Xyloglucan endotransglucosylase/hydrolase 28 | 1819 | 2545 | 2868 |
| Prupe.1G255100 | AT1G10550 | XTH33 | Xyloglucan endotransglucosylase/hydrolase 33 | 327 | 380 | 1447 |
| Prupe.1G309900 | AT1G68560 | XYL1 | Alpha-xylosidase 1 | 439 | 753 | 895 |
| Prupe.1G165400 | AT3G23805 | RALFL24 | Ralf-like 24 | 533 | 475 | 360 |
| Prupe.4G150200 | AT1G78440 | GA2ox1 | Gibberellin 2-β-dioxygenase | 917 | 862 | 391 |
| Prupe.8G192500 | AT3G25290 | AUX | Auxin-responsive family protein | 161 | 165 | 412 |
| Prupe.7G247500 | AT4G28640 | IAA11 | Indole-3-acetic acid inducible 11 | 563 | 1409 | 1906 |
| Prupe.5G233100 | AT1G73590 | PIN1 | Auxin efflux carrier family protein | 1553 | 2326 | 2743 |
| Prupe.4G231800 | AT5G55540 | TRN1 | Tornado 1 | 108 | 158 | 239 |
| Prupe.6G157500 | AT1G48910 | YUC10 | Flavin-containing monooxygenase family | 81 | 179 | 220 |
| Prupe.5G053500 | AT1G67080 | ABA4 | Abscisic acid (aba)-deficient 4 | 169 | 181 | 302 |
| Prupe.4G082000 | AT1G30100 | NCED5 | Nine-cis-epoxycarotenoid dioxygenase 5 | 175 | 2677 | 5153 |
| Prupe.7G067100 | AT4G16760 | ACX1 | Acyl-CoA oxidase 1 | 54 | 18 | 14 |
| Prupe.4G082500 | AT1G30135 | JAZ8 | Jasmonate-zim-domain protein 8 | 201 | 355 | 515 |
| Prupe.1G467500 | AT4G32570 | TIFY8 | TIFY domain protein 8 | 68 | 63 | 41 |
| Prupe.1G587800 | AT5G49740 | FRO7 | Ferric reduction oxidase 7 | 75 | 69 | 36 |
| Prupe.1G347100 | AT5G23120 | HCF136 | Photosystem II stability/assembly factor | 175 | 134 | 101 |
| Prupe.1G026900 | AT5G43750 | PnsB5 | NAD(P)H dehydrogenase 18 | 40 | 20 | 19 |
| Prupe.2G313700 | AT4G27800 | TAP38 | Thylakoid-associated phosphatase 38 | 57 | 35 | 29 |
| Prupe.8G160500 | AT2G21170 | TIM | Triosephosphate isomerase | 392 | 219 | 197 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Núñez-Lillo, G.; Pérez-Reyes, W.; Riveros, A.; Lillo-Carmona, V.; Rothkegel, K.; Álvarez, J.M.; Blanco-Herrera, F.; Pedreschi, R.; Campos-Vargas, R.; Meneses, C. Transcriptome and Gene Regulatory Network Analyses Reveal New Transcription Factors in Mature Fruit Associated with Harvest Date in Prunus persica. Plants 2022, 11, 3473. https://doi.org/10.3390/plants11243473
Núñez-Lillo G, Pérez-Reyes W, Riveros A, Lillo-Carmona V, Rothkegel K, Álvarez JM, Blanco-Herrera F, Pedreschi R, Campos-Vargas R, Meneses C. Transcriptome and Gene Regulatory Network Analyses Reveal New Transcription Factors in Mature Fruit Associated with Harvest Date in Prunus persica. Plants. 2022; 11(24):3473. https://doi.org/10.3390/plants11243473
Chicago/Turabian StyleNúñez-Lillo, Gerardo, Wellasmin Pérez-Reyes, Anibal Riveros, Victoria Lillo-Carmona, Karin Rothkegel, José Miguel Álvarez, Francisca Blanco-Herrera, Romina Pedreschi, Reinaldo Campos-Vargas, and Claudio Meneses. 2022. "Transcriptome and Gene Regulatory Network Analyses Reveal New Transcription Factors in Mature Fruit Associated with Harvest Date in Prunus persica" Plants 11, no. 24: 3473. https://doi.org/10.3390/plants11243473
APA StyleNúñez-Lillo, G., Pérez-Reyes, W., Riveros, A., Lillo-Carmona, V., Rothkegel, K., Álvarez, J. M., Blanco-Herrera, F., Pedreschi, R., Campos-Vargas, R., & Meneses, C. (2022). Transcriptome and Gene Regulatory Network Analyses Reveal New Transcription Factors in Mature Fruit Associated with Harvest Date in Prunus persica. Plants, 11(24), 3473. https://doi.org/10.3390/plants11243473





