Phytotoxic Metabolites Produced by Fungi Involved in Grapevine Trunk Diseases: Progress, Challenges, and Opportunities
Abstract
1. Introduction
2. Phytotoxic Metabolites Produced by Fungi Involved in GTDs and Their Role in Grapevine Tissues
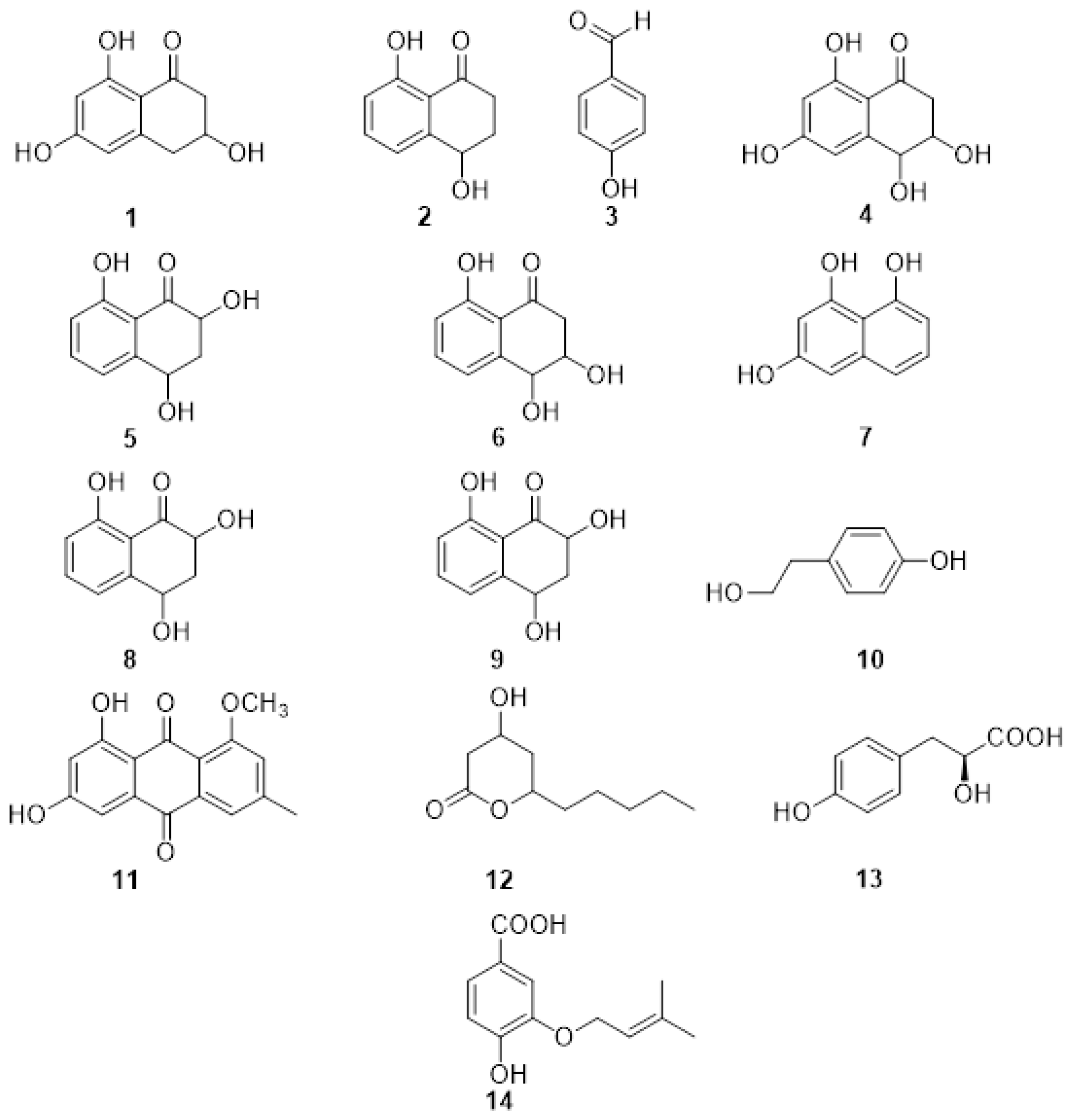
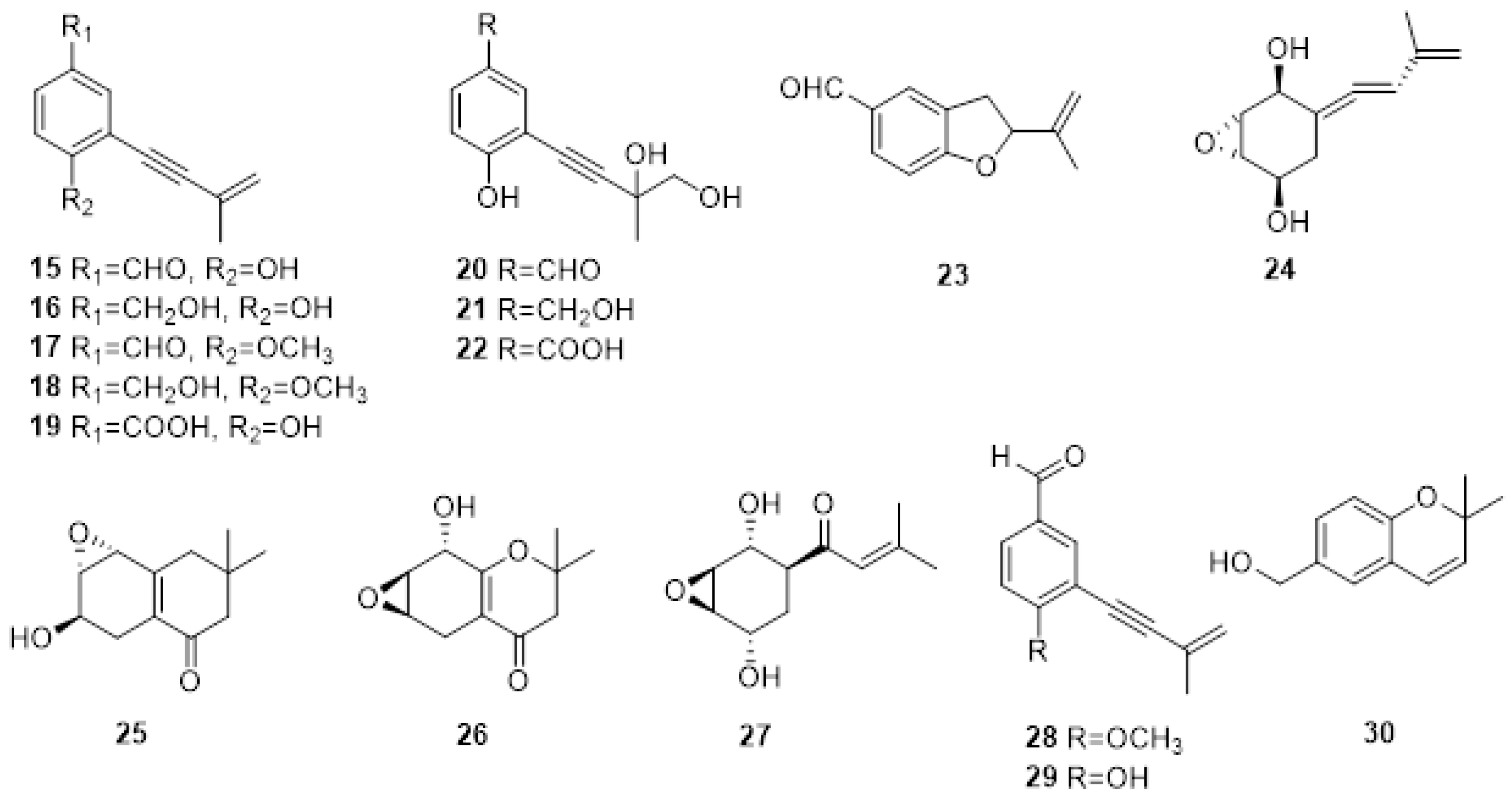
2.1. Phytotoxic Metabolites Produced by Fungi Involved in Esca Complex
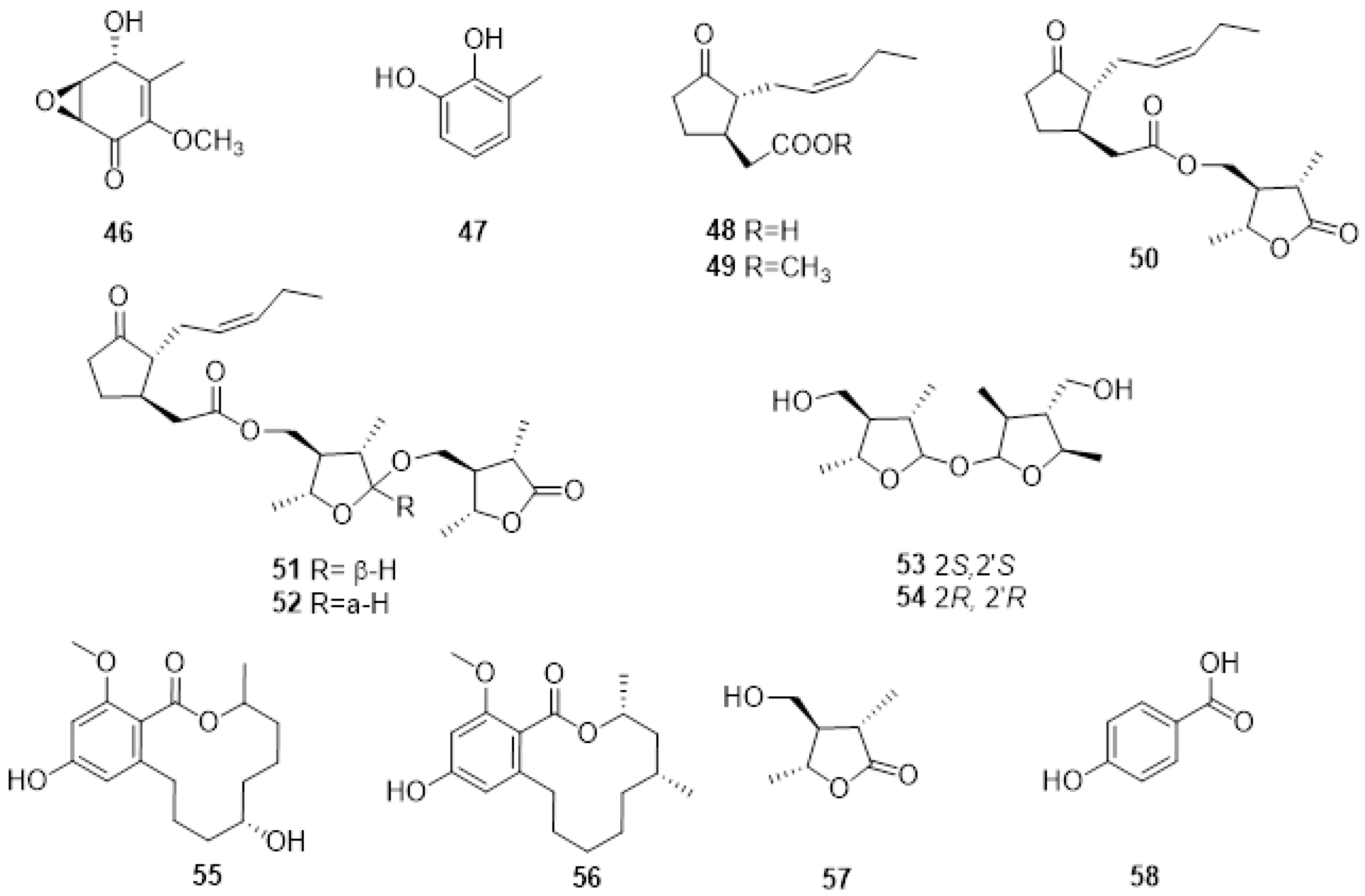
2.2. Phytotoxic Metabolites Produced by Fungi Involved in Eutypa Dieback
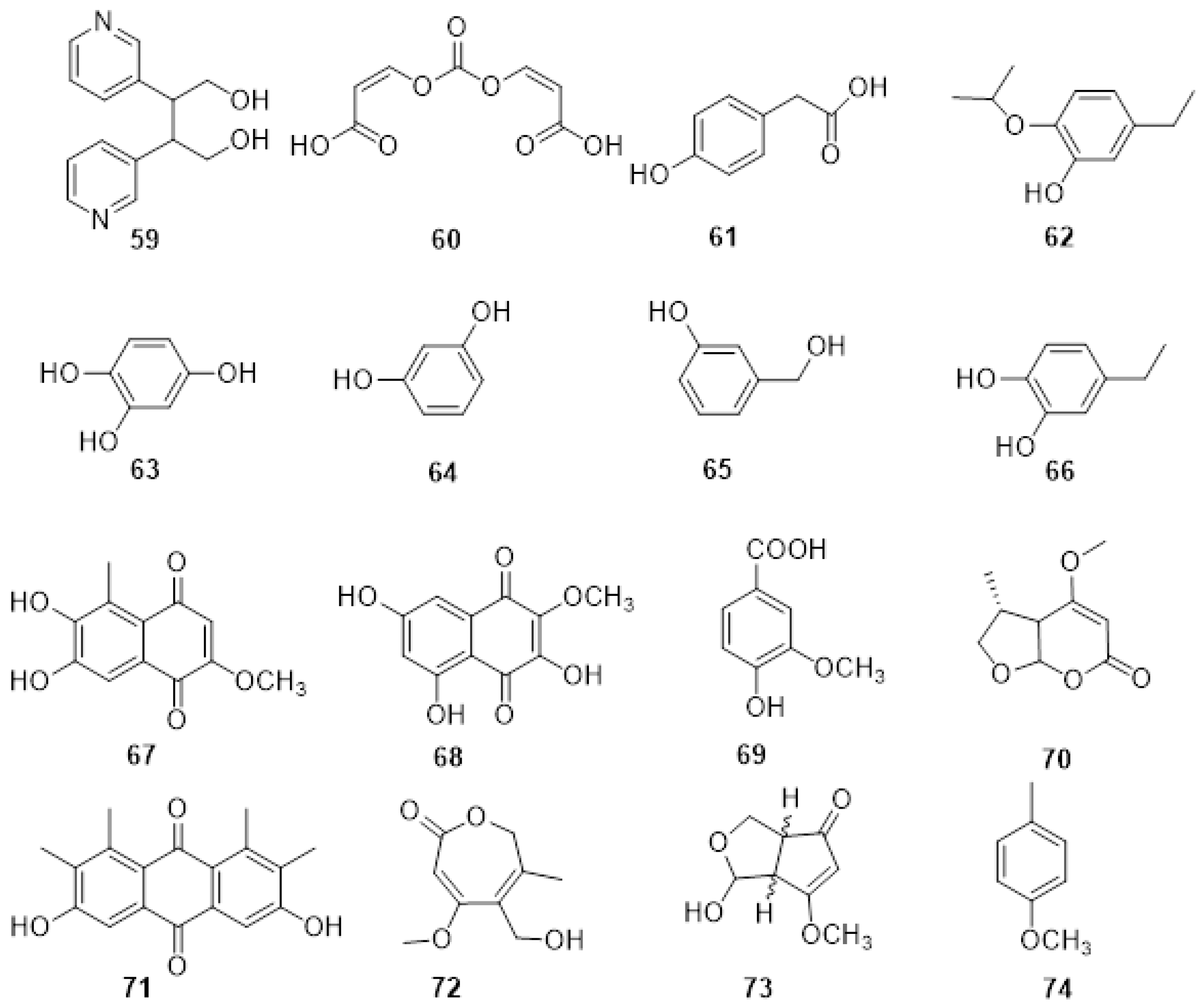
2.3. Phytotoxic Metabolites Produced by Fungi Involved in Botryosphaeria Dieback
3. Metabolomics Studies
4. Challenges and Perspective
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alston, J.M.; Sambucci, O. Grapes in the world economy. In The Grape Genome; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–24. [Google Scholar]
- Mondello, V.; Songy, A.; Battiston, E.; Pinto, C.; Coppin, C.; Trotel-Aziz, P.; Clément, C.; Mugnai, L.; Fontaine, F. Grapevine trunk diseases: A review of fifteen years of trials for their control with chemicals and biocontrol agents. Plant Dis. 2018, 102, 1189–1217. [Google Scholar] [CrossRef] [PubMed]
- Hrycan, J.; Hart, M.; Bowen, P.; Forge, T.; Urbez-Torres, J.R. Grapevine trunk disease fungi: Their roles as latent pathogens and stress factors that favour disease develop-ment and symptom expression. Phytopathol. Mediterr. 2020, 59, 395–424. [Google Scholar]
- Gramaje, D.; Úrbez-Torres, J.R.; Sosnowski, M.R. Managing grapevine trunk diseases with respect to etiology and epidemiology: Current strategies and future prospects. Plant Dis. 2018, 102, 12–39. [Google Scholar] [CrossRef] [PubMed]
- Bertsch, C.; Ramirez-Suero, M.; Magnin-Robert, M.; Larignon, P.; Chong, J.; Abou-Mansour, E.; Spagnolo, A.; Clement, C.; Fontaine, F. Grapevine trunk diseases: Complex and still poorly understood. Plant Pathol. 2013, 62, 243–265. [Google Scholar] [CrossRef]
- Billones-Baaijens, R.; Savocchia, S. A review of Botryosphaeriaceae species associated with grapevine trunk diseases in Australia and New Zealand. Australas. Plant Pathol. 2018, 48, 3–18. [Google Scholar] [CrossRef]
- Sosnowski, M.R.; Ayres, M.; Billones-Baaijens, R.; Savocchia, S. Practical Management of Grapevine Trunk Diseases. Final Report to Wine Australia, Project SAR1205, February 2017. Wine Australia 2017. Available online: https://www.wineaustralia.com/research/search/completed-projects/sar-1205 (accessed on 23 October 2022).
- Sosnowski, M.R. Best Practice Management Guide-Grapevine Trunk Disease, Version 2.1 Wine Australia 2021. Available online: www.wineaustralia.com (accessed on 23 October 2022).
- Pitt, W.M.; Sosnowski, M.R.; Huang, R.; Qiu, Y.; Steel, C.C.; Savocchia, S. Evaluation of fungicides for the management of Botryosphaeria canker of grapevines. Plant Dis. 2012, 96, 1303–1308. [Google Scholar] [CrossRef]
- Ayres, M.; Wicks, T.; Scott, E.S.; Sosnowski, M. Developing pruning wound protection strategies for managing Eutypa dieback. Austral. J. Grape Wine Res. 2017, 23, 103–111. [Google Scholar] [CrossRef]
- Ayres, M.; Billones-Baaijens, R.; Savocchia, S.; Scott, E.; Sosnowski, M. Critical timing of fungicide application for pruning wound protection to control grapevine trunk diseases. Austral. J. Grape Wine Res. 2022, 28, 70–74. [Google Scholar] [CrossRef]
- Kotze, C.; Van Niekerk, J.; Mostert, L.; Halleen, F.; Fourie, P. Evaluation of biocontrol agents for grapevine pruning wound protection against trunk pathogen infection. Phytopathol. Mediterr. 2011, 50, S247–S263. [Google Scholar]
- Mutawila, C.; Fourie, P.H.; Halleen, F.; Mostert, L. Grapevine cultivar variation to pruning wound protection by Trichoderma species against trunk pathogens. Phytopathol. Mediterr. 2011, 50, S264–S276. [Google Scholar]
- Leal, C.; Richet, N.; Guise, J.-F.; Gramaje, D.; Armengol, J.; Fontaine, F.; Trotel-Aziz, P. Cultivar contributes to the beneficial effects of Bacillus subtilis PTA-271 and Trichoderma atroviride SC1 to protect grapevine against Neofusicoccum parvum. Front. Microbiol. 2021, 12, 2889. [Google Scholar] [CrossRef] [PubMed]
- Pollard-Flamand, J.; Boulé, J.; Hart, M.; Úrbez-Torres, J.R. Biocontrol Activity of Trichoderma Species Isolated from Grapevines in British Columbia against Botryosphaeria Dieback Fungal Pathogens. J. Fungi 2022, 8, 409. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Kassemeyer, H.-H. Fungi associated with Esca disease of grapevine in Germany. Vitis-J. Grapevine Res. 2015, 42, 109. [Google Scholar]
- Scharf, D.H.; Heinekamp, T.; Brakhage, A.A. Human and plant fungal pathogens: The role of secondary metabolites. PLoS Pathog. 2014, 10, e1003859. [Google Scholar] [CrossRef] [PubMed]
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef] [PubMed]
- Kusari, S.; Pandey, S.P.; Spiteller, M. Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites. Phytochemistry 2013, 91, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Cimmino, A.; Masi, M.; Reveglia, P.; Nocera, P.; Solano, R.; Evidente, A. The fungal phytotoxin Lasiojasmonate A activates the plant jasmonic acid pathway. J. Exp. Bot. 2018, 69, 3095–3102. [Google Scholar] [CrossRef]
- Fonseca, S.; Radhakrishnan, D.; Prasad, K.; Chini, A. Fungal production and manipulation of plant hormones. Curr. Med. Chem. 2018, 25, 253–267. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Gen. 2010, 11, 539. [Google Scholar] [CrossRef]
- Kusari, S.; Hertweck, C.; Spiteller, M. Chemical ecology of endophytic fungi: Origins of secondary metabolites. Chem. Biol. 2012, 19, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Peyraud, R.; Dubiella, U.; Barbacci, A.; Genin, S.; Raffaele, S.; Roby, D. Advances on plant–pathogen interactions from molecular toward systems biology perspectives. Plant J. 2017, 90, 720–737. [Google Scholar] [CrossRef] [PubMed]
- Evidente, A.; Kornienko, A.; Cimmino, A.; Andolfi, A.; Lefranc, F.; Mathieu, V.; Kiss, R. Fungal metabolites with anticancer activity. Nat. Prod. Rep. 2014, 31, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Masi, M.; Evidente, M.; Superchi, S.; Evidente, A. Fungal phytotoxins with potential herbicidal activity: Chemical and biological characterization. Nat. Prod. Rep. 2015, 32, 1629–1653. [Google Scholar] [CrossRef] [PubMed]
- Masi, M.; Nocera, P.; Reveglia, P.; Cimmino, A.; Evidente, A. Fungal Metabolites Antagonists towards Plant Pests and Human Pathogens: Structure-Activity Relationship Studies. Molecules 2018, 23, 834. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.J. Polyketides, proteins and genes in fungi: Programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem. 2007, 5, 2010–2026. [Google Scholar] [CrossRef] [PubMed]
- Chooi, Y.-H.; Krill, C.; Barrow, R.A.; Chen, S.; Trengove, R.; Oliver, R.P.; Solomon, P.S. An in planta-expressed polyketide synthase produces (R)-mellein in the wheat pathogen Parastagonospora nodorum. App. Env. Microbiol. 2015, 81, 177–186. [Google Scholar] [CrossRef]
- Li, H.; Wei, H.; Hu, J.; Lacey, E.; Sobolev, A.N.; Stubbs, K.A.; Solomon, P.S.; Chooi, Y.-H. Genomics-driven discovery of phytotoxic cytochalasans involved in the virulence of the wheat pathogen Parastagonospora nodorum. ACS Chem. Biol. 2019, 15, 226–233. [Google Scholar] [CrossRef]
- Idnurm, A.; Howlett, B.J. Pathogenicity genes of phytopathogenic fungi. Mol. Plant Pathol. 2001, 2, 241–255. [Google Scholar] [CrossRef]
- Lim, F.Y.; Sanchez, J.F.; Wang, C.C.; Keller, N.P. Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Methods Enzymol. 2012, 517, 303. [Google Scholar]
- Yaegashi, J.; Oakley, B.R.; Wang, C.C. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans. J. Ind. Microbiol. Biotec. 2014, 41, 433–442. [Google Scholar] [CrossRef]
- Cacho, R.A.; Tang, Y.; Chooi, Y.-H. Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi. Front. Microbiol. 2015, 5, 774. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.-R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef] [PubMed]
- Bielska, E.; Higuchi, Y.; Schuster, M.; Steinberg, N.; Kilaru, S.; Talbot, N.J.; Steinberg, G. Long-distance endosome trafficking drives fungal effector production during plant infection. Nat. Comm. 2014, 5, 5097. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, I.; Steinberg, G.; Gurr, S. The role of the fungal cell wall in the infection of plants. Trends Microbiol. 2017, 25, 957–967. [Google Scholar] [CrossRef]
- Fontaine, F.; Gramaje, D.; Armengol, J.; Smart, R.; Nagy, Z.A.; Borgo, M.; Rego, C.; Corio-Costet, M.-F. Grapevine Trunk Diseases. A review; OIV Publications: Paris, France, 2016; Volume 24, ISBN 979-10-91799-60-7. [Google Scholar]
- Tey-Rulh, P.; Philippe, I.; Renaud, J.-M.; Tsoupras, G.; de Angelis, P.; Fallot, J.; Tabacchi, R. Eutypine, a phytotoxin produced by Eutypa lata the causal agent of dying-arm disease of grapevine. Phytochemistry 1991, 30, 471–473. [Google Scholar] [CrossRef]
- Sosnowski, M.; Shtienberg, D.; Creaser, M.; Wicks, T.; Lardner, R.; Scott, E. The influence of climate on foliar symptoms of eutypa dieback in grapevines. Phytopathology 2007, 97, 1284–1289. [Google Scholar] [CrossRef]
- Rolshausen, P.; Greve, L.; Labavitch, J.; Mahoney, N.; Molyneux, R.; Gubler, W. Pathogenesis of Eutypa lata in grapevine: Identification of virulence factors and biochemical characterization of cordon dieback. Phytopathology 2008, 98, 222–229. [Google Scholar] [CrossRef]
- Gramaje, D.; Mostert, L.; Groenewald, J.Z.; Crous, P.W. Phaeoacremonium: From esca disease to phaeohyphomycosis. Fungal Biol. 2015, 119, 759–783. [Google Scholar] [CrossRef]
- Raimondo, M.L.; Carlucci, A.; Ciccarone, C.; Saddallah, A.; Francesco, L. Identification and pathogenicity of lignicolous fungi associated with grapevine trunk diseases in southern Italy. Phytopathol. Mediter. 2019, 58, 639–662. [Google Scholar]
- Aigoun-Mouhous, W.; Mahamedi, A.E.; León, M.; Chaouia, C.; Zitouni, A.; Barankova, K.; Eichmeier, A.; Armengol, J.; Gramaje, D.; Berraf-Tebbal, A. Cadophora sabaouae sp. nov. and Phaeoacremonium species associated with Petri disease on grapevine propagation material and young grapevines in Algeria. Plant Dis. 2021, 105, 3657–3668. [Google Scholar] [CrossRef] [PubMed]
- Mondello, V.; Piccolo, S.L.; Conigliaro, G.; Alfonzo, A.; Torta, L.; Burruano, S. First report of Neofusiccoccum vitifusiforme and presence of other Botryosphaeriaceae species associated with Botryosphaeria dieback of grapevine in Sicily (Italy). Phytopathol. Mediterr. 2013, 52, 388–396. [Google Scholar]
- Raimondo, M.L.; Lops, F.; Carlucci, A. Phaeoacremonium italicum sp. nov., associated with esca of grapevine in southern Italy. Mycologia 2014, 106, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Surico, G.; Mugnai, L.; Marchi, G. The esca disease complex. In Integrated Management of Diseases Caused by Fungi, Phytoplasma and Bacteria; Springer: Berlin/Heidelberg, Germany, 2008; pp. 119–136. [Google Scholar]
- Guerin-Dubrana, L.; Fontaine, F.; Mugnai, L. Grapevine trunk disease in European and Mediterranean vineyards: Occurrence, distribution and associated disease-affecting cultural factors. Phytopathol. Mediterr. 2019, 58, 49–71. [Google Scholar]
- Mundy, D.C.; Brown, A.; Jacobo, F.; Tennakoon, K.; Woolley, R.H.; Vanga, B.; Tyson, J.; Johnston, P.; Ridgway, H.J.; Bulman, S. Pathogenic fungi isolated in association with grapevine trunk diseases in New Zealand. N. Z. J. Crop. Hortic. Sci. 2020, 48, 84–96. [Google Scholar] [CrossRef]
- Fischer, M. Grapevine trunk diseases in German viticulture. III. Biodiversity and spatial distribution of fungal pathogens in rootstock mother plants and possible relation to leaf symptoms. Vitis- J. Grapevine Res. 2019, 58, 141–149. [Google Scholar]
- Urbez-Torres, J.R.; Peduto, F.; Striegler, R.; Urrea-Romero, K.; Rupe, J.; Cartwright, R.; Gubler, W. Characterization of fungal pathogens associated with grapevine trunk diseases in Arkansas and Missouri. Fungal Div. 2012, 52, 169–189. [Google Scholar] [CrossRef]
- Blanco-Ulate, B.; Rolshausen, P.; Cantu, D. Draft genome sequence of the ascomycete Phaeoacremonium aleophilum strain UCR-PA7, a causal agent of the esca disease complex in grapevines. Genome Announc. 2013, 1, e00390–e00413. [Google Scholar] [CrossRef]
- Antonielli, L.; Compant, S.; Strauss, J.; Sessitsch, A.; Berger, H. Draft genome sequence of Phaeomoniella chlamydospora strain RR-HG1, a grapevine trunk disease (Esca)-related member of the Ascomycota. Genome Announc. 2014, 2, e00098–e00114. [Google Scholar] [CrossRef]
- Massonnet, M.; Morales-Cruz, A.; Minio, A.; Figueroa-Balderas, R.; Lawrence, D.P.; Travadon, R.; Rolshausen, P.E.; Baumgartner, K.; Cantu, D. Whole-genome resequencing and pan-transcriptome reconstruction highlight the impact of genomic structural variation on secondary metabolism gene clusters in the grapevine Esca pathogen Phaeoacremonium minimum. bioRxiv 2018, 9, 252221. [Google Scholar]
- Yacoub, A.; Magnin, N.; Gerbore, J.; Haidar, R.; Bruez, E.; Compant, S.; Guyoneaud, R.; Rey, P. The biocontrol root-oomycete, Pythium oligandrum, triggers grapevine resistance and shifts in the transcriptome of the trunk pathogenic fungus, Phaeomoniella chlamydospora. Int. J. Mol. Sci. 2020, 21, 6876. [Google Scholar] [CrossRef] [PubMed]
- Evidente, A.; Bruno, G.; Andolfi, A.; Sparapano, L. Two Naphthalenone Pentakides from Liquid Cultures of Phaeoacremonium aleophilum, a Fungus Associated with Esca of Grapevine. Phytopathol. Mediterr. 2000, 39, 1000–1007. [Google Scholar] [CrossRef]
- Tabacchi, R.; Fkyerat, A.; Poliart, C.; Dubin, G. Phytotoxins from fungi of esca grapevine [Vitis vinifera L.]. Phytopathol. Mediterr. 2000, 1000–1006. [Google Scholar] [CrossRef]
- Abou-Mansour, A.; Tabacchi, R.; Couché, E. Do fungal naphthalenones have a role in the development of esca symptoms? Phytopathol. Mediterr. 2004, 1000–1008. [Google Scholar] [CrossRef]
- Andolfi, A.; Mugnai, L.; Luque, J.; Surico, G.; Cimmino, A.; Evidente, A. Phytotoxins produced by fungi associated with grapevine trunk diseases. Toxins 2011, 3, 1569–1605. [Google Scholar] [CrossRef]
- Medentsev, A.; Akimenko, V. Naphthoquinone metabolites of the fungi. Phytochemistry 1998, 47, 935–959. [Google Scholar] [CrossRef]
- De Gara, L.; de Pinto, M.C.; Tommasi, F. The antioxidant systems vis-à-vis reactive oxygen species during plant–pathogen interaction. Plant Physiol. Biochem. 2003, 41, 863–870. [Google Scholar] [CrossRef]
- Trouillas, F.P.; Urbez-Torres, J.R.; Gubler, W.D. Diversity of diatrypaceous fungi associated with grapevine canker diseases in California. Mycologia 2010, 102, 319–336. [Google Scholar] [CrossRef]
- Trouillas, F.P.; Pitt, W.M.; Sosnowski, M.R.; Huang, R.; Peduto, F.; Loschiavo, A.; Savocchia, S.; Scott, E.S.; Gubler, W.D. Taxonomy and DNA phylogeny of Diatrypaceae associated with Vitis vinifera and other woody plants in Australia. Fungal Div. 2011, 49, 203–223. [Google Scholar] [CrossRef]
- Pitt, W.M.; Trouillas, F.P.; Gubler, W.D.; Savocchia, S.; Sosnowski, M.R. Pathogenicity of diatrypaceous fungi on grapevines in Australia. Plant Dis. 2013, 97, 749–756. [Google Scholar] [CrossRef]
- Trouillas, F.; Sosnowski, M.; Gubler, W. Two new species of Diatrypaceae from coastal wattle in Coorong National Park, South Australia. Mycosphere 2010, 1, 183–188. [Google Scholar]
- Moyo, P.; Mostert, L.; Spies, C.F.; Damm, U.; Halleen, F. Diversity of Diatrypaceae species associated with dieback of grapevines in South Africa, with the description of Eutypa cremea sp. nov. Plant Dis. 2018, 102, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Catal, M.; Jordan, S.; Butterworth, S.; Schilder, A. detection of Eutypa lata and Eutypella vitis in grapevine by nested multiplex polymerase chain reaction. Phytopathology 2007, 97, 737–747. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Trouillas, F.P.; Hand, F.P.; Inderbitzin, P.; Gubler, W.D. The genus Cryptosphaeria in the western United States: Taxonomy, multilocus phylogeny and a new species, C. multicontinentalis. Mycologia 2015, 107, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Luque, J.; Garcia, F.; Torres, E.; Sierra, D. “Cryptovalsa ampelina” on grapevines in NE Spain: Identification and pathogenicity. Phytopatol. Mediterr. 2006, 1000–1009. [Google Scholar] [CrossRef]
- Bruez, E.; Baumgartner, K.; Bastien, S.; Travadon, R.; Guérin-Dubrana, L.; Rey, P. Various fungal communities colonise the functional wood tissues of old grapevines externally free from grapevine trunk disease symptoms. Aus. J. Grape Wine Res. 2016, 22, 288–295. [Google Scholar] [CrossRef]
- Blanco-Ulate, B.; Rolshausen, P.E.; Cantu, D. Draft genome sequence of the grapevine dieback fungus Eutypa lata UCR-EL1. Genome Announc. 2013, 1, e00228–e00313. [Google Scholar] [CrossRef]
- Morales-Cruz, A.; Amrine, K.C.; Blanco-Ulate, B.; Lawrence, D.P.; Travadon, R.; Rolshausen, P.E.; Baumgartner, K.; Cantu, D. Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens. BMC genom. 2015, 16, 1–22. [Google Scholar] [CrossRef]
- Onetto, C.A.; Sosnowski, M.R.; Van Den Heuvel, S.; Borneman, A.R. Population genomics of the grapevine pathogen Eutypa lata reveals evidence for population expansion and intraspecific differences in secondary metabolite gene clusters. PLoS Genet. 2022, 18, e1010153. [Google Scholar] [CrossRef]
- Renaud, J.M.; Tsoupras, G.; Stoeckli-Evans, H.; Tabacchi, R. A novel allenic epoxycyclohexane and related compounds from Eutypa lata (Pers: F.) Tul. Helv. Chim. Acta 1989, 72, 1262–1267. [Google Scholar] [CrossRef]
- Renaud, J.M.; Tsoupras, G.; Tabacchi, R. Biologically active natural acetylenic compounds from Eutypa lata (Pers: F.) TUL. Helv. Chim. Acta 1989, 72, 929–932. [Google Scholar] [CrossRef]
- Defrancq, E.; Gordon, J.; Brodard, A.; Tabacchi, R. The synthesis of a novel epoxycyclohexane from the fungus Eutypa lata (Pers: F.) TUL. Helv. Chim. Acta 1992, 75, 276–281. [Google Scholar] [CrossRef]
- Molyneux, R.J.; Mahoney, N.; Bayman, P.; Wong, R.Y.; Meyer, K.; Irelan, N. Eutypa dieback in grapevines: Differential production of acetylenic phenol metabolites by strains of Eutypa lata. J. Agr. Food Chemi. 2002, 50, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, N.; Molyneux, R.J.; Smith, L.R.; Schoch, T.K.; Rolshausen, P.E.; Gubler, W.D. Dying-arm disease in grapevines: Diagnosis of infection with Eutypa lata by metabolite analysis. J. Agr. Food Chem. 2005, 53, 8148–8155. [Google Scholar] [CrossRef] [PubMed]
- Lardner, R.; Mahoney, N.; Zanker, T.P.; Molyneux, R.J.; Scott, E.S. Secondary metabolite production by the fungal pathogen Eutypa lata: Analysis of extracts from grapevine cultures and detection of those metabolites in planta. Aus. J. Grape Wine Res. 2006, 12, 107. [Google Scholar] [CrossRef]
- Crous, P.W.; Slippers, B.; Wingfield, M.J.; Rheeder, J.; Marasas, W.F.; Philips, A.J.; Alves, A.; Burgess, T.; Barber, P.; Groenewald, J.Z. Phylogenetic lineages in the Botryosphaeriaceae. Stud. Mycol. 2006, 55, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Jones, E.G.; Liu, J.-K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.-Q. Families of dothideomycetes. Fungal Div. 2013, 63, 1–313. [Google Scholar]
- Slippers, B.; Crous, P.W.; Denman, S.; Coutinho, T.A.; Wingfield, B.D.; Wingfield, M.J. Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia 2004, 96, 83–101. [Google Scholar] [CrossRef]
- Sakalidis, M.L.; Hardy, G.E.S.; Burgess, T.I. Class III endophytes, clandestine movement amongst hosts and habitats and their potential for disease; a focus on Neofusicoccum australe. Australas. Plant Pathol. 2011, 40, 510–521. [Google Scholar] [CrossRef]
- Urbez-Torres, J.R. The status of Botryosphaeriaceae species infecting grapevines. Phytopathol. Mediterr. 2011, 50, 5–45. [Google Scholar]
- Úrbez-Torres, J.; Leavitt, G.; Voegel, T.; Gubler, W. Identification and distribution of Botryosphaeria spp. associated with grapevine cankers in California. Plant Dis. 2006, 90, 1490–1503. [Google Scholar] [CrossRef] [PubMed]
- Pitt, W.M.; Huang, R.; Steel, C.; Savocchia, S. Identification, distribution and current taxonomy of Botryosphaeriaceae species associated with grapevine decline in New South Wales and South Australia. Aus. J. Grape Wine Res. 2010, 16, 258–271. [Google Scholar] [CrossRef]
- Linaldeddu, B.T.; Deidda, A.; Scanu, B.; Franceschini, A.; Serra, S.; Berraf-Tebbal, A.; Boutiti, M.Z.; Jamâa, M.B.; Phillips, A. Diversity of Botryosphaeriaceae species associated with grapevine and other woody hosts in Italy, Algeria and Tunisia, with descriptions of Lasiodiplodia exigua and Lasiodiplodia mediterranea sp. nov. Fungal Div. 2015, 71, 201–214. [Google Scholar] [CrossRef]
- Pitt, W.M.; Urbez-Torres, J.R.; Trouillas, F.P. Dothiorella and Spencermartinsia, new species and records from grapevines in Australia. Australas. Plant Pathol. 2015, 44, 43–56. [Google Scholar] [CrossRef]
- Comont, G.; Mayet, V.; Corio-Costet, M. First report of Lasiodiplodia viticola, Spencermartinsia viticola and Diplodia intermedia associated with Vitis vinifera grapevine decline in French vineyards. Plant Dis. 2016, 100, 2328. [Google Scholar] [CrossRef]
- Correia, K.C.; Silva, M.A.; de Morais Jr, M.A.; Armengol, J.; Phillips, A.J.; Cômara, M.; Michereff, S.J. Phylogeny, distribution and pathogenicity of Lasiodiplodia species associated with dieback of table grape in the main Brazilian exporting region. Plant Pathol. 2016, 65, 92–103. [Google Scholar] [CrossRef]
- Correia, K.; Silva, M.; Netto, M.; Vieira, W.; Câmara, M.; Michereff, S. First report of grapevine dieback caused by Neoscytalidium hyalinum in Brazil. Plant Dis. 2016, 100, 213. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.; Gubler, W. Pathogenicity of Botryosphaeriaceae species isolated from grapevine cankers in California. Plant Dis. 2009, 93, 584–592. [Google Scholar] [CrossRef]
- Pitt, W.M.; Huang, R.; Steel, C.; Savocchia, S. Pathogenicity and epidemiology of Botryosphaeriaceae species isolated from grapevines in Australia. Australas. Plant Pathol. 2013, 42, 573–582. [Google Scholar] [CrossRef]
- Pitt, W.M.; Úrbez-Torres, J.R.; Trouillas, F.P. Dothiorella vidmadera, a novel species from grapevines in Australia and notes on Spencermartinsia. Fungal Div. 2013, 61, 209–219. [Google Scholar] [CrossRef]
- Rangel Montoya, E.A.; Paolinelli, M.; Rolshausen, P.E.; Valenzuela Solano, C.; Hernandez Martinez, R. Characterization of Lasiodiplodia species associated with grapevines in Mexico. Mediterr. Phytopathol. Union 2021, 60, 177–385. [Google Scholar] [CrossRef]
- Al-Saadoon, A.H.; Ameen, M.K.; Hameed, M.A.; Al-Badran, A.; Ali, Z. First report of grapevine dieback caused by Lasiodiplodia theobromae and Neoscytalidium dimidiatum in Basrah, Southern Iraq. Afr. J. Adv. Biotechnol. 2012, 11, 16165–16171. [Google Scholar]
- Rolshausen, P.; Akgül, D.; Perez, R.; Eskalen, A.; Gispert, C. First report of wood canker caused by Neoscytalidium dimidiatum on grapevine in California. Plant Dis. 2013, 97, 1511. [Google Scholar] [CrossRef] [PubMed]
- Akgül, D.S.; Savaş, N.; Özarslandan, M. First report96 of wood canker caused by Lasiodiplodia exigua and Neoscytalidium novaehollandiae on grapevine in Turkey. Plant Dis. 2019, 103, 1036. [Google Scholar] [CrossRef]
- Billones-Baaijens, R.; Ridgway, H.; Jones, E.; Cruickshank, R.; Jaspers, M. Prevalence and distribution of Botryosphaeriaceae species in New Zealand grapevine nurseries. Eur. J. Plant Pathol. 2013, 135, 175–185. [Google Scholar] [CrossRef]
- Baskarathevan, J.; Jaspers, M.V.; Jones, E.E.; Cruickshank, R.H.; Ridgway, H.J. Genetic and pathogenic diversity of Neofusicoccum parvum in New Zealand vineyards. Fungal Biol. 2012, 116, 276–288. [Google Scholar] [CrossRef]
- Garcia, J.F.; Lawrence, D.P.; Morales-Cruz, A.; Travadon, R.; Minio, A.; Hernandez-Martinez, R.; Rolshausen, P.E.; Baumgartner, K.; Cantu, D. Phylogenomics of plant-associated Botryosphaeriaceae species. Front. Microbiol. 2021, 12, 587. [Google Scholar] [CrossRef] [PubMed]
- Chethana, K.T.; Li, X.; Zhang, W.; Hyde, K.D.; Yan, J. Trail of decryption of molecular research on Botryosphaeriaceae in woody plants. Phytopathol. Mediterr. 2016, 55, 147. [Google Scholar]
- Robert-Siegwald, G.; Vallet, J.; Abou-Mansour, E.; Xu, J.; Rey, P.; Bertsch, C.; Rego, C.; Larignon, P.; Fontaine, F.; Lebrun, M.-H. Draft Genome Sequence of Diplodia seriata F98. 1, a Fungal Species Involved in Grapevine Trunk Diseases. Genome Announc. 2017, 5, e00061–e00117. [Google Scholar] [CrossRef]
- Billones-Baaijens, R.; Jones, E.; Ridgway, H.; Jaspers, M. Virulence affected by assay parameters during grapevine pathogenicity studies with Botryosphaeriaceae nursery isolates. Plant Pathol. 2013, 62, 1214–1225. [Google Scholar] [CrossRef]
- Martos, S.; Andolfi, A.; Luque, J.; Mugnai, L.; Surico, G.; Evidente, A. Production of phytotoxic metabolites by five species of Botryosphaeriaceae causing decline on grapevines, with special interest in the species Neofusicoccum luteum and N. parvum. Eur. J. Plant Pathol. 2008, 121, 451–461. [Google Scholar] [CrossRef]
- Djoukeng, J.D.; Polli, S.; Larignon, P.; Abou-Mansour, E. Identification of phytotoxins from Botryosphaeria obtusa, a pathogen of black dead arm disease of grapevine. Eur. J. Plant Pathol. 2009, 124, 303–308. [Google Scholar] [CrossRef]
- Evidente, A.; Punzo, B.; Andolfi, A.; Cimmino, A.; Melck, D.; Luque, J. Lipophilic phytotoxins produced by Neofusicoccum parvum, a grapevine canker agent. Phytopathol. Mediterr. 2010, 49, 74–79. [Google Scholar]
- Abou-Mansour, E.; Débieux, J.-L.; Ramírez-Suero, M.; Bénard-Gellon, M.; Magnin-Robert, M.; Spagnolo, A.; Chong, J.; Farine, S.; Bertsch, C.; L’Haridon, F. Phytotoxic metabolites from Neofusicoccum parvum, a pathogen of Botryosphaeria dieback of grapevine. Phytochemistry 2015, 115, 207–215. [Google Scholar] [CrossRef]
- Reveglia, P.; Masi, M.; Evidente, A. Melleins—Intriguing Natural Compounds. Biomolecules 2020, 10, 772. [Google Scholar] [CrossRef]
- Sun, H.; Ho, C.L.; Ding, F.; Soehano, I.; Liu, X.-W.; Liang, Z.-X. Synthesis of (R)-mellein by a partially reducing iterative polyketide synthase. J. Am. Chem. Soc. 2012, 134, 11924–11927. [Google Scholar] [CrossRef]
- Ramírez-Suero, M.; Bénard-Gellon, M.; Chong, J.; Laloue, H.; Stempien, E.; Abou-Mansour, E.; Fontaine, F.; Larignon, P.; Mazet-Kieffer, F.; Farine, S. Extracellular compounds produced by fungi associated with Botryosphaeria dieback induce differential defence gene expression patterns and necrosis in Vitis vinifera cv. Chardonnay cells. Protoplasma 2014, 251, 1417–1426. [Google Scholar] [CrossRef]
- Cimmino, A.; Cinelli, T.; Evidente, M.; Masi, M.; Mugnai, L.; Silva, M.A.; Michereff, S.J.; Surico, G.; Evidente, A. Phytotoxic fungal exopolysaccharides produced by fungi involved in grapevine trunk diseases. Nat. Prod. Comm. 2016, 11, 1481–1484. [Google Scholar] [CrossRef]
- Stempien, E.; Goddard, M.-L.; Leva, Y.; Bénard-Gellon, M.; Laloue, H.; Farine, S.; Kieffer-Mazet, F.; Tarnus, C.; Bertsch, C.; Chong, J. Secreted proteins produced by fungi associated with Botryosphaeria dieback trigger distinct defense responses in Vitis vinifera and Vitis rupestris cells. Protoplasma 2018, 255, 613–628. [Google Scholar] [CrossRef]
- Andolfi, A.; Maddau, L.; Cimmino, A.; Linaldeddu, B.T.; Franceschini, A.; Serra, S.; Basso, S.; Melck, D.; Evidente, A. Cyclobotryoxide, a phytotoxic metabolite produced by the plurivorous pathogen Neofusicoccum australe. J. Nat. Prod. 2012, 75, 1785–1791. [Google Scholar] [CrossRef]
- Andolfi, A.; Maddau, L.; Cimmino, A.; Linaldeddu, B.T.; Basso, S.; Deidda, A.; Serra, S.; Evidente, A. Lasiojasmonates A–C, three jasmonic acid esters produced by Lasiodiplodia sp., a grapevine pathogen. Phytochemistry 2014, 103, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Reveglia, P.; Chini, A.; Mandoli, A.; Masi, M.; Cimmino, A.; Pescitelli, G.; Evidente, A. Synthesis and mode of action studies of N-[(-)-jasmonyl]-S-tyrosin and ester seiridin jasmonate. Phytochemistry 2018, 147, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Andolfi, A.; Basso, S.; Giambra, S.; Conigliaro, G.; Lo Piccolo, S.; Alves, A.; Burruano, S. Lasiolactols A and B produced by the grapevine fungal pathogen Lasiodiplodia mediterranea. Chem. Biodivers. 2016, 13, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Cinelli, T.; Masi, M.; Reveglia, P.; da Silva, M.A.; Mugnai, L.; Michereff, S.J.; Surico, G.; Evidente, A. Phytotoxic lipophylic metabolites produced by grapevine strains of Lasiodiplodia species in Brazil. J. Agr. Food Chem. 2017, 65, 1102–1107. [Google Scholar] [CrossRef]
- Reveglia, P.; Marco, M.; Cimmino, a.; Michereff, S.; Cinelli, T.; Mugnai, L.; Evidente, A. Phytotoxins produced by Lasiodiplodia laeliocattleyae involved in Botryosphaeria dieback of grapevines in Brazil. Phytopathol. Mediterr. 2019, 58, 207–211. [Google Scholar]
- Reveglia, P.; Savocchia, S.; Billones-Baaijens, R.; Masi, M.; Cimmino, A.; Evidente, A. Phytotoxic metabolites by nine species of Botryosphaeriaceae involved in grapevine dieback in Australia and identification of those produced by Diplodia mutila, Diplodia seriata, Neofusicoccum australe and Neofusicoccum luteum. Nat. Prod. Res. 2019, 33, 2223–2229. [Google Scholar] [CrossRef] [PubMed]
- Reveglia, P.; Savocchia, S.; Billones-Baaijens, R.; Masi, M.; Evidente, A. Spencertoxin and spencer acid, new phytotoxic derivatives of diacrylic acid and dipyridinbutan-1, 4-diol produced by Spencermartinsia viticola, a causal agent of grapevine Botryosphaeria dieback in Australia. Arab. J. Chem. 2020, 13, 1803–1808. [Google Scholar] [CrossRef]
- Reveglia, P.; Savocchia, S.; Billones-Baaijens, R.; Cimmino, A.; Evidente, A. Isolation of Phytotoxic Phenols and Characterization of a New 5-Hydroxymethyl-2-isopropoxyphenol from Dothiorella vidmadera, a Causal Agent of Grapevine Trunk Disease. J. Agr. Food Chem. 2018, 66, 1760–1764. [Google Scholar] [CrossRef]
- Reveglia, P.; Savocchia, S.; Billones-Baaijens, R.; Masi, M.; Cimmino, A.; Evidente, A. Diploquinones A and B, two new phytotoxic tetrasubstituted 1, 4-naphthoquinones from Diplodia mutila, a causal agent of grapevine trunk disease. J. Agr. Food Chem. 2018, 66, 11968–11973. [Google Scholar] [CrossRef]
- Masi, M.; Reveglia, P.; Baaijens-Billones, R.; Górecki, M.; Pescitelli, G.; Savocchia, S.; Evidente, A. Phytotoxic metabolites from three Neofusicoccum species causal agents of Botryosphaeria Dieback in Australia, Luteopyroxin, Neoanthraquinone, and Luteoxepinone, a Disubstituted Furo-α-pyrone, a Hexasubstituted Anthraquinone, and a Trisubstituted Oxepi-2-one from Neofusicoccum luteum. J. Nat. Prod. 2020, 83, 453–460. [Google Scholar]
- Reis, P.; Magnin-Robert, M.; Nascimento, T.; Spagnolo, A.; Abou-Mansour, E.; Fioretti, C.; Clement, C.; Rego, C.; Fontaine, F. Reproducing Botryosphaeria Dieback Foliar Symptoms in a Simple Model System. Plant Dis. 2016, 100, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Spagnolo, A.; Magnin-Robert, M.; Alayi, T.D.; Cilindre, C.; Schaeffer-Reiss, C.; Van Dorsselaer, A.; Clément, C.; Larignon, P.; Ramirez-Suero, M.; Chong, J. Differential responses of three grapevine cultivars to Botryosphaeria dieback. Phytopathology 2014, 104, 1021–1035. [Google Scholar] [CrossRef] [PubMed]
- Bellée, A.; Comont, G.; Nivault, A.; Abou-Mansour, E.; Coppin, C.; Dufour, M.; Corio-Costet, M. Life traits of four Botryosphaeriaceae species and molecular responses of different grapevine cultivars or hybrids. Plant Pathol. 2017, 66, 763–776. [Google Scholar] [CrossRef]
- Reveglia, P.; Billones-Baaijens, R.; Millera Niem, J.; Masi, M.; Cimmino, A.; Evidente, A.; Savocchia, S. Production of Phytotoxic Metabolites by Botryosphaeriaceae in Naturally Infected and Artificially Inoculated Grapevines. Plants 2021, 10, 802. [Google Scholar] [CrossRef] [PubMed]
- Trotel-Aziz, P.; Robert-Siegwald, G.; Fernandez, O.; Leal, C.; Villaume, S.; Guise, J.-F.; Abou-Mansour, E.; Lebrun, M.-H.; Fontaine, F. Diversity of Neofusicoccum parvum for the Production of the Phytotoxic Metabolites (-)-Terremutin and (R)-Mellein. J. Fungi 2022, 8, 319. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. In Functional Genomics; Springer: Berlin/Heidelberg, Germany, 2002; pp. 155–171. [Google Scholar]
- Schauer, N.; Fernie, A.R. Plant metabolomics: Towards biological function and mechanism. Trends Plant Sci. 2006, 11, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Vasilev, N.; Boccard, J.; Lang, G.; Grömping, U.; Fischer, R.; Goepfert, S.; Rudaz, S.; Schillberg, S. Structured plant metabolomics for the simultaneous exploration of multiple factors. Sci. Rep. 2016, 6, 37390. [Google Scholar] [CrossRef]
- Tugizimana, F.; Mhlongo, M.; Piater, L.; Dubery, I. Metabolomics in Plant Priming Research: The Way Forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Metabolomics in plant stress physiology. In Plant Genetics and Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 187–236. [Google Scholar]
- Jan, S.; Ahmad, P. Ecometabolomics: Metabolic Fluxes Versus Environmental Stoichiometry; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Labois, C.; Wilhelm, K.; Laloue, H.; Tarnus, C.; Bertsch, C.; Goddard, M.-L.; Chong, J. Wood metabolomic responses of wild and cultivated grapevine to infection with Neofusicoccum parvum, a trunk disease pathogen. Metabolites 2020, 10, 232. [Google Scholar] [CrossRef]
- Moret, F.; Clement, G.; Grosjean, C.; Lemaitre-Guillier, C.; Morvan, G.; Trouvelot, S.; Adrian, M.; Fontaine, F. Metabolite fingerprints of Chardonnay grapevine leaves affected by esca is both clone-and year-dependent. Phytopathol. Mediterr. 2020, 59, 595–603. [Google Scholar]
- Galarneau, E.R.; Lawrence, D.P.; Wallis, C.M.; Baumgartner, K. A comparison of the metabolomic response of grapevine to infection with ascomycete wood-infecting fungi. Physiol. Mol. Plant Pathol. 2021, 113, 101596. [Google Scholar] [CrossRef]
- Azzollini, A.; Boggia, L.; Boccard, J.; Sgorbini, B.; Lecoultre, N.; Allard, P.-M.; Rubiolo, P.; Rudaz, S.; Gindro, K.; Bicchi, C. Dynamics of metabolite induction in fungal co-cultures by metabolomics at both volatile and non-volatile levels. Front. Microbiol. 2018, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Reveglia, P.; Raimondo, M.L.; Masi, M.; Cimmino, A.; Nuzzo, G.; Corso, G.; Fontana, A.; Carlucci, A.; Evidente, A. Untargeted and Targeted LC-MS/MS Based Metabolomics Study on In Vitro Culture of Phaeoacremonium Species. J. Fungi 2022, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R.; Broadhurst, D.; Smilde, A.K.; Kristal, B.S.; Baker, J.D.; Beger, R.; Bessant, C.; Connor, S.; Capuani, G.; Craig, A. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics 2007, 3, 231–241. [Google Scholar] [CrossRef]
- Burns, D.C.; Mazzola, E.P.; Reynolds, W.F. The role of computer-assisted structure elucidation (CASE) programs in the structure elucidation of complex natural products. Nat. Prod. Rep. 2019, 36, 919–933. [Google Scholar] [CrossRef]
- Ramos, A.E.F.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: Different manners, one goal. Nat. Prod. Rep. 2019, 36, 960–980. [Google Scholar] [CrossRef]
- Oberlies, N.H.; Knowles, S.L.; Amrine, C.S.M.; Kao, D.; Kertesz, V.; Raja, H.A. Droplet probe: Coupling chromatography to the in situ evaluation of the chemistry of nature. Nat. Prod. Rep. 2019, 36, 944–959. [Google Scholar] [CrossRef]
- Locatelli, M.; Epifano, F.; Genovese, S.; Carlucci, G.; Končić, M.Z.; Kosalec, I.; Kremer, D. Anthraquinone profile, antioxidant and antimicrobial properties of bark extracts of Rhamnus catharticus and R. orbiculatus. Nat. Prod. Comm. 2011, 6, 1934578X1100600917. [Google Scholar] [CrossRef]
- Gessler, N.; Egorova, A.; Belozerskaya, T. Fungal anthraquinones. App. Biochem. Microbiol. 2013, 49, 85–99. [Google Scholar] [CrossRef]
- Songy, A.; Fernandez, O.; Clément, C.; Larignon, P.; Fontaine, F. Grapevine trunk diseases under thermal and water stresses. Planta 2019, 249, 1655–1679. [Google Scholar] [CrossRef]
- Patanita, M.; Albuquerque, A.; Campos, M.D.; Materatski, P.; Varanda, C.M.; Ribeiro, J.A.; Félix, M.d.R. Metagenomic Assessment Unravels Fungal Microbiota Associated to Grapevine Trunk Diseases. Sci. Hort. 2022, 8, 288. [Google Scholar] [CrossRef]
- Félix, C.; Meneses, R.; Gonçalves, M.F.; Tilleman, L.; Duarte, A.S.; Jorrín-Novo, J.V.; Van de Peer, Y.; Deforce, D.; Van Nieuwerburgh, F.; Esteves, A.C. A multi-omics analysis of the grapevine pathogen Lasiodiplodia theobromae reveals that temperature affects the expression of virulence-and pathogenicity-related genes. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.F.; Nunes, R.B.; Tilleman, L.; Van de Peer, Y.; Deforce, D.; Van Nieuwerburgh, F.; Esteves, A.C.; Alves, A. Dual RNA sequencing of Vitis vinifera during Lasiodiplodia theobromae infection unveils host–pathogen interactions. Int. J. Mol. Sci. 2019, 20, 6083. [Google Scholar] [CrossRef] [PubMed]
- Ball, B.; Langille, M.; Geddes-McAlister, J. Fun (gi) omics: Advanced and diverse technologies to explore emerging fungal pathogens and define mechanisms of antifungal resistance. Mbio 2020, 11, e01020. [Google Scholar] [CrossRef] [PubMed]
- Crandall, S.G.; Gold, K.M.; Jiménez-Gasco, M.d.M.; Filgueiras, C.C.; Willett, D.S. A multi-omics approach to solving problems in plant disease ecology. PLoS ONE 2020, 15, e0237975. [Google Scholar] [CrossRef] [PubMed]
- Worrall, E.A.; Hamid, A.; Mody, K.T.; Mitter, N.; Pappu, H.R. Nanotechnology for plant disease management. Agronomy 2018, 8, 285. [Google Scholar] [CrossRef]
- Elmer, W.; White, J.C. The future of nanotechnology in plant pathology. Ann. Rev. Phytopathol. 2018, 56, 111–133. [Google Scholar] [CrossRef]
- Fletcher, S.J.; Reeves, P.T.; Hoang, B.T.; Mitter, N. A perspective on RNAi-based biopesticides. Front. Plant Sci. 2020, 11, 51. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, M.; Wang, N. Precision agriculture-a worldwide overview. Comput. Electron. Agric. 2002, 36, 113–132. [Google Scholar] [CrossRef]
- Keswani, C.; Mishra, S.; Sarma, B.K.; Singh, S.P.; Singh, H.B. Unraveling the efficient applications of secondary metabolites of various Trichoderma spp. App. Microbiol. Biotechnol. 2014, 98, 533–544. [Google Scholar] [CrossRef]
- Traversari, S.; Cacini, S.; Galieni, A.; Nesi, B.; Nicastro, N.; Pane, C. Precision agriculture digital technologies for sustainable fungal disease management of ornamental plants. Sustainability 2021, 13, 3707. [Google Scholar] [CrossRef]



| Genus | Species | References |
|---|---|---|
| Phaeoacremonium | P. angustius, P. alvesii, P. argentiniense, P. armeniacum, P. australiense, P. austroafricanum, P. canadense, P. cinereum, P. croatiense, P.globosum, P. hispanicum, P. hungaricum, P. inflatipes, P. italicum, P. iranianum, P. krajdenii, P. minimum, P. mortoniae, P. nordesticola, P. occidentale, P. roseum, P. scolyti, P. sicilianum, P. subulatum, P. tuscanum, P. venezuelense, P. viticola | [16,42,43,45,46,47,48,49,50] |
| Cadophora | Cad. luteo-olivacea, Cad. malorum, Cad. melinii, Cad. meredithiae, Cad. novi-eboraci, Cad. orchidicola, Cad. orientoamericana, Cad. spadicis, Cad. sabaouae, Cad. viticola | [4,43,44,49,51] |
| Genus | Species | References |
|---|---|---|
| Eutypa | E. laevata, E. consobrina, E. Cremea, E. lata, E. leptoplaca | [65,66] |
| Eutypella | Eu. Australiensis, Eu. Citricola, Eu. cryptovalsoidea, Eu. microtheca, Eutypella spp., Eu. vitis | [4,63,66,67] |
| Cryptosphaeria | C. lignyota, C. pullmanensis | [68] |
| Cryptovalsa | Cr. ampelina, Cr. rabenhorstii | [68,69] |
| Diatrype | D. brunneospora, D. oregonensis, D. stigma, Diatrype sp., D. whitmanensis | [62,68] |
| Diatrypella | Di. verrucaeformis, Di. vulgaris | [62,63] |
| Genus | Species | References |
|---|---|---|
| Botryosphaeria | B. dothidea | [84,92] |
| Diplodia | D. corticola, D. mutila, D. seriata, D.intermedia | [4,84,89] |
| Dothiorella | Do. americana, Do. neclivorem, Do. sarmentorum, Do.vidmadera, Do.vineagemmae | [6,88,93,94] |
| Lasiodiplodia | L. brasiliensis, L. citricola, L. crassipora, L. euphorbicola, L. exigua, L. hormozganensis, L. iraniensis, L.jatrophicola, L.laeliocattleyae, L.mahajangana L. mediterranea, L. missouriana, L. parva, L. pseudotheobromae, L. theobromae, L. viticola | [84,89,90,95,96] |
| Neoscytalidium | N. dimidiatum, N. hyalinum N. novaehollandiae | [91,97,98] |
| Neofusicoccum | N. australe, N. luteum, N. macroclavatum, N. mediterraneum, N. parvum, N. ribis, N. viticlavatum, N. vitifusiforme | [51,84,92,93,99,100,101] |
| Spencermartinsia | S. plurivora, S. viticola, S. westrale | [6,88,89] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reveglia, P.; Billones-Baaijens, R.; Savocchia, S. Phytotoxic Metabolites Produced by Fungi Involved in Grapevine Trunk Diseases: Progress, Challenges, and Opportunities. Plants 2022, 11, 3382. https://doi.org/10.3390/plants11233382
Reveglia P, Billones-Baaijens R, Savocchia S. Phytotoxic Metabolites Produced by Fungi Involved in Grapevine Trunk Diseases: Progress, Challenges, and Opportunities. Plants. 2022; 11(23):3382. https://doi.org/10.3390/plants11233382
Chicago/Turabian StyleReveglia, Pierluigi, Regina Billones-Baaijens, and Sandra Savocchia. 2022. "Phytotoxic Metabolites Produced by Fungi Involved in Grapevine Trunk Diseases: Progress, Challenges, and Opportunities" Plants 11, no. 23: 3382. https://doi.org/10.3390/plants11233382
APA StyleReveglia, P., Billones-Baaijens, R., & Savocchia, S. (2022). Phytotoxic Metabolites Produced by Fungi Involved in Grapevine Trunk Diseases: Progress, Challenges, and Opportunities. Plants, 11(23), 3382. https://doi.org/10.3390/plants11233382






