Coffea canephora: Heterotic Crosses Indicated by Molecular Approach
Abstract
1. Introduction
2. Results and Discussion
2.1. Diversity Parameters via SSR
2.2. Relative Kinship and Estimates of Genetic Dissimilarity in C. canephora
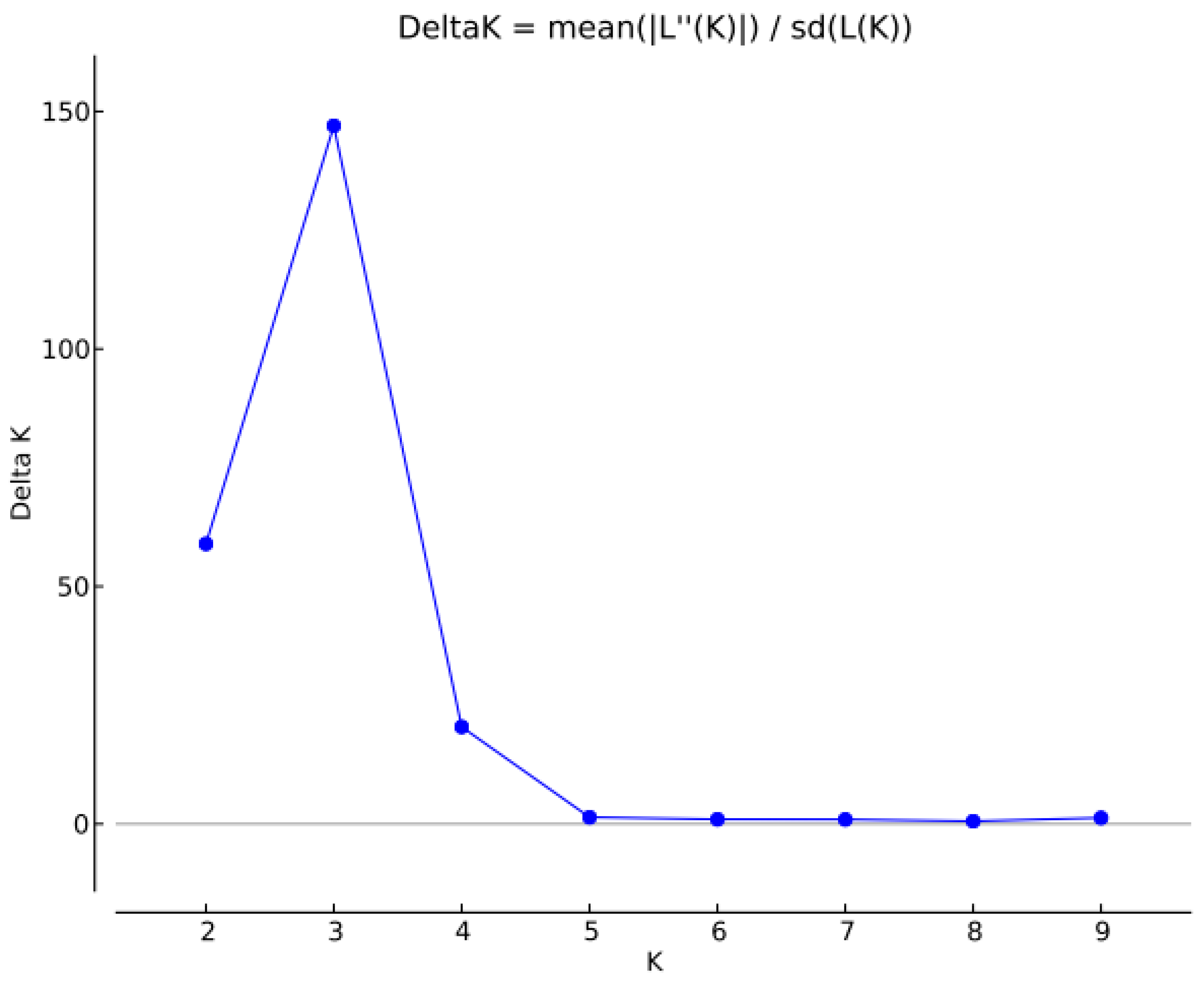
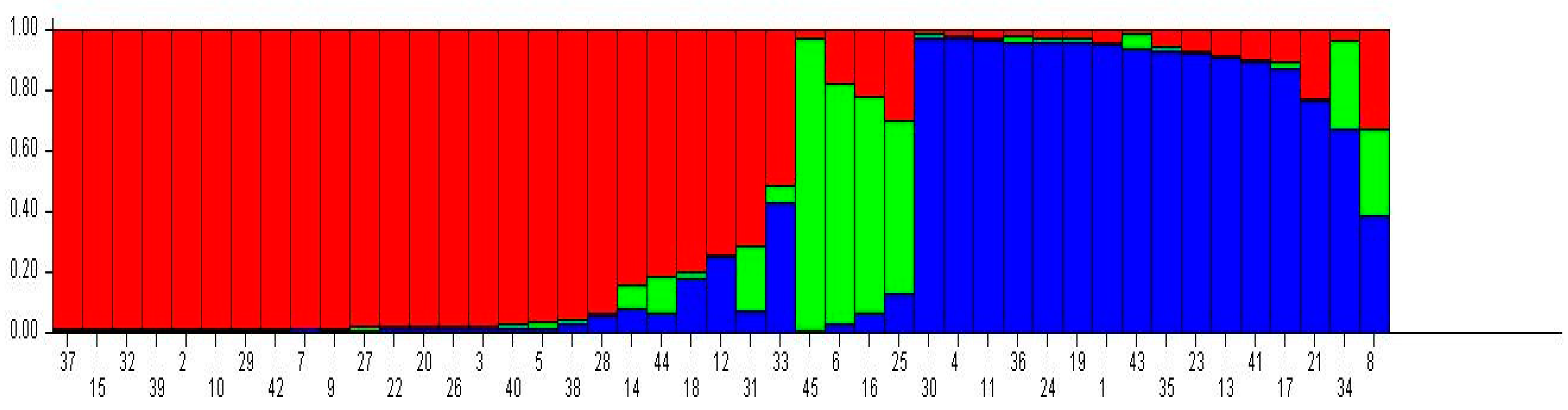
2.3. Analysis of Genetic Structure
3. Materials and Methods
3.1. Experimental Area
3.2. Sampling of Plant Material
3.3. DNA Extraction
3.4. Primer Screening
3.5. Polymerase Chain Reaction (PCR)
3.6. Statistical Analysis of Molecular Variables
3.7. Genetic Structure of the Population
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Companhia Nacional de Abastecimento—CONAB. Acompanhamento da Safra Brasileira de Café–Safra 2021. Brasília. 2021. Available online: https://www.conab.gov.br/info-agro/safras/cafe (accessed on 24 March 2022).
- Moraes, M.S.; Teixeira, A.L.; Ramalho, A.R.; Espíndula, M.C.; Ferrão, M.A.G.; Rocha, R.B. Characterization of gametophytic self-incompatibility of superior clones of Coffea canephora. Genet. Mol. Res. 2018, 17, 16039876. [Google Scholar] [CrossRef]
- Ramalho, A.R.; Rocha, R.B.; Souza, F.F.; Veneziano, W.; Teixeira, A.L. Genetic gain in the productivity of processed coffee from the selection of clones of ‘Conilon’ coffee. Rev. Cienc. Agron. 2016, 47, 516–523. [Google Scholar] [CrossRef]
- Bergo, C.L.L.; Miqueloni, D.P.; Lunz, A.M.P.; De Assis, G.M.L. Estimation of genetic parameters and selection of Coffea canephora progenies evaluated in Brazilian Western Amazon. Coffee Sci. 2020, 15, e151663. [Google Scholar] [CrossRef]
- Babova, O.; Occhipinti, A.; Maffei, M.E. Chemical partitioning and antioxidant capacity of green coffee (Coffea arabica and Coffea canephora) of different geographical origin. Phytochemistry 2016, 123, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Toppa, E.V.B.; Jadoski, C.J. O Uso dos Marcadores Moleculares no Melhoramento Genético de Plantas. Sci. Agrar. Parana. 2013, 12, 1–5. [Google Scholar] [CrossRef]
- Sousa, T.V.; Caixeta, E.T.; Alkimim, E.R.; de Oliveira, A.C.B.; Pereira, A.A.; Zambolim, L.; Sakiyama, N.S. Molecular markers useful to discriminate Coffea arabica cultivars with high genetic similarity. Euphytica 2017, 213, 75. [Google Scholar] [CrossRef]
- Alkimim, E.R.; Caixeta, E.T.; Sousa, T.V.; Pereira, A.A.; de Oliveira, A.C.B.; Zambolim, L.; Sakiyama, N.S. Marker-assisted selection provides arabica coffee with genes from other Coffea species targeting on multiple resistance to rust and coffee berry disease. Mol. Plant Breed. 2017, 37, 6. [Google Scholar] [CrossRef]
- Gartner, G.A.L.; McCouch, S.R.; Moncada, M.D.P. A genetic map of an interspecific diploid pseudo testcross population of coffee. Euphytica 2013, 192, 305–323. [Google Scholar] [CrossRef]
- Alkimim, E.R.; Caixeta, E.T.; Sousa, T.V.; da Silva, F.L.; Sakiyama, N.S.; Zambolim, L. High-throughput targeted genotyping using next-generation sequencing applied in Coffea canephora breeding. Euphytica 2018, 214, 50. [Google Scholar] [CrossRef]
- Anagbogu, C.F.; Bhattacharjee, R.; Ilori, C.; Tongyoo, P.; Dada, K.E.; Muyiwa, A.A.; Gepts, P.; Beckles, D.M. Genetic diversity and re-classification of coffee (Coffea canephora Pierre ex A. Froehner) from South Western Nigeria through geno-typing-by-sequencing-single nucleotide polymorphism analysis. Genet. Resour. Crop Evol. 2019, 66, 685–696. [Google Scholar] [CrossRef]
- Baltazar, M.D.; Fabella, J.M.A.O. Assessment of the genetic diversity of Philippine Arabica coffee (Coffea arabica L.) using SSR markers. Philipp. J. Sci. 2020, 149, 993–1003. [Google Scholar]
- da Silva, B.S.R.; Sant’Ana, G.C.; Chaves, C.L.; Androcioli, L.G.; Ferreira, R.V.; Sera, G.H.; Charmetant, P.; Leroy, T.; Pot, D.; Domingues, D.S.; et al. Population structure and genetic relationships between Ethiopian and Brazilian Coffea arabica genotypes revealed by SSR markers. Genetica 2019, 147, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Ferrão, M.A.G.; Ferrão, R.G.; Fonseca, A.F.A.; Verdin Filho, A.C.; Volpi, P.S. Origin, geographical dispersion, taxonomy and genetic diversity of Coffea canephora. In Conilon Coffee; Ferrão, R.G., Fonseca, A.F.A., Ferrão, M.A.G., De Muner, L.H., Eds.; Incaper: Vitória, Brazil, 2019; pp. 85–110. [Google Scholar]
- Zucchi, M.I.; Brondani, R.P.V.; Pinheiro, J.B.; Chaves, L.J.; Coelho, A.S.G.; Vencovsky, R. Genetic structure and gene flow in Eugenia dysenterica DC in the Brazilian Cerrado utilizing SSR markers. Genet. Mol. Biol. 2003, 26, 449–457. [Google Scholar] [CrossRef][Green Version]
- Caballero, A.; Rodríguez-Ramilo, S.T.; Ávila, V.; Fernández, J. Management of genetic diversity of subdivided populations in conservation programmes. Conserv. Genet. 2010, 11, 409–419. [Google Scholar] [CrossRef]
- Kiwuka, C.; Goudsmit, E.; Tournebize, R.; de Aquino, S.O.; Douma, J.C.; Bellanger, L.; Crouzillat, D.; Stoffelen, P.; Sumirat, U.; Legnaté, H.; et al. Genetic diversity of native and cultivated Ugandan Robusta coffee (Coffea canephora Pierre ex A. Froehner): Climate influences, breeding potential and diversity conservation. PLoS ONE 2021, 16, e0245965. [Google Scholar] [CrossRef]
- Sánchez, E.; Solano, W.; Gatica-Arias, A.; Chavarría, M.; Araya-Valverde, E. Microsatellite DNA fingerprinting of Coffea sp. germplasm conserved in Costa Rica through singleplex and multiplex PCR. Crop Breed. Appl. Biotechnol. 2020, 20, 1–9. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- De Moura, T.M.; Sebbenn, A.M.; Chaves, L.J.; Coelho, A.S.G.; Oliveira, G.C.X.; Kageyama, P.Y. Diversidade e estrutura genética espacial em populações fragmentadas de Solanum spp. do Cerrado, estimadas por meio de locos microssatélites. Sci. For. 2009, 37, 143–150. [Google Scholar]
- Labouisse, J.-P.; Cubry, P.; Austerlitz, F.; Rivallan, R.; Nguyen, H.A. New insights on spatial genetic structure and diversity of Coffea canephora (Rubiaceae) in Upper Guinea based on old herbaria. Plant Ecol. Evol. 2020, 153, 82–100. [Google Scholar] [CrossRef]
- Alves, A.L.; Leitão, A.E.B.; de Souza, P.E.N.; Santos, M.D.F.P.D.; Moscon, P.S.; Pessoa, M.S.; Pinheiro, C.A.; Morais, P.C.; Partelli, F.L. Antioxidant Activity and Stable Free Radicals in Robusta Green Coffee Genotypes/Atividade antioxidante e Radicais Estáveis Livres em Genótipos de Café Verde Robusta. Braz. J. Dev. 2021, 7, 37312–37330. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6, 5: Genetic analysis in Excel, Population genetic software for teaching and research an update. BMC Bioinform. 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.D.; Ferreira, F.M.; Pessoni, L.A. Biometria Aplicada ao Estudo da Diversidade Genética, 1st ed.; Produção Independente: Viçosa, Brazil, 2011. [Google Scholar]
- Morgante, M.; Hanafey, M.; Powell, W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002, 30, 194–200. [Google Scholar] [CrossRef]
- Mojena, R. Hierarchical grouping methods and stopping rules: An evaluation. Comput. J. 1977, 20, 359–363. [Google Scholar] [CrossRef]
- Partelli, F.L.; Oliosi, G.; Dalazen, J.R.; da Silva, C.A.; Vieira, H.D.; Espindula, M.C. Proportion of ripe fruit weight and volume to green coffee: Differences in 43 genotypes of Coffea canephora. Agron. J. 2021, 113, 1050–1057. [Google Scholar] [CrossRef]
- Fazuoli, L.C.; Mistro, J.C.; Braghini, M.T. Melhoramento de café robusta no Instituto Agronômico de Campinas. In Tecnologia para Produção do Café Conilon; Zambolim, L., Ed.; UFV: Viçosa, Brazil, 2009; Volume cap.8, pp. 201–248. [Google Scholar]
- Cruz, C.D.; Carneiro, P.C.S.; Regazzi, A.J. Modelos Biométricos Aplicados ao Melhoramento Genético, 3rd ed.; UFV: Viçosa, Brazil, 2014; 668p. [Google Scholar]
- Carmona, P.A.; Peixoto, J.R.; Amaro, G.B.; Mendonça, M.A. Divergência genética entre acessos de batata-doce utilizando descritores morfoagronômicos das raízes. Hortic. Bras. 2015, 33, 241–250. [Google Scholar] [CrossRef]
- Bragança, S.M.; Carvalho, C.H.S.; Fonseca, A.F.A.F.; Ferrão, R.G. Clonal varieties of Conilon coffee for the Espírito Santo State, Brazil. Pesq. Agropec. Bras. 2001, 36, 765–770. [Google Scholar] [CrossRef]
- Partelli, F.L.; Giles, J.A.D.; Oliosi, G.; Covre, A.M.; Ferreira, A.; Rodrigues, E.V.M. Tributun: A coffee cultivar developed in partnership with farmers. Crop Breed. Appl. Biotechnol. 2020, 20, 30002025. [Google Scholar] [CrossRef]
- Partelli, F.B.; Golynski, A.; Ferreira, A.; Martins, M.Q.; Mauri, A.L.; Ramalho, J.C.; Vieira, H.D. Andina—First clonal cultivar of high-altitude conilon coffee. Crop Breed. Appl. Biotechnol. 2019, 19, 476–480. [Google Scholar] [CrossRef]
- Diniz, L.E.C.; Sakiyama, N.S.; Lashermes, P.; Caixeta, E.T.; Oliveira, A.C.B.; Zambolim, E.; Loureiro, M.E.; Pereira, A.A.; Zambolim, L. Analysis of AFLP markers associated to the Mex-1 resistance locus in Icatu progenies. Crop Breed. Appl. Biotechnol. 2005, 5, 387–393. [Google Scholar] [CrossRef]
- Souza, F.D.F.; Caixeta, E.T.; Ferrão, L.F.V.; Pena, G.F.; Sakiyama, N.S.; Zambolim, E.M.; Zambolim, L.; Cruz, C.D. Molecular diversity in Coffea canephora germplasm conserved and cultivated in Brazil. Crop Breed. Appl. Biotechnol. 2013, 13, 221–227. [Google Scholar] [CrossRef]
- Cruz, C.D. Genes: A Software Package for Analysis in Experimental Statistics and Quantitative Genetics. Acta Sci. 2013, 35, 271–276. [Google Scholar]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Hardy, O.J.; Vekemans, X. spagedi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Loiselle, B.A.; Spork, V.L.; Nason, J.; Graham, C. Spatial genetic structure of a tropical understory shrub. Psychotria officinalis (Rubiaceae). Am. J. Bot. 1995, 82, 1420–1425. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]


| Locus | NA | I | Ho | He | F |
|---|---|---|---|---|---|
| CaEST-010 | 2 | 0.413 | 0.289 | 0.247 | −0.169 |
| SSR-081 | 3 | 0.987 | 0.5 | 0.588 | 0.15 |
| SSR-303 | 4 | 0.45 | 0.211 | 0.196 | −0.076 |
| SSR-343 | 2 | 0.474 | 0.273 | 0.298 | 0.083 |
| SSR-353 | 2 | 0.65 | 0.024 | 0.457 | 0.947 |
| SSR-373 | 3 | 0.625 | 0.394 | 0.363 | −0.085 |
| SSR-433 | 2 | 0.625 | 0.636 | 0.434 | −0.467 |
| SSR-463 | 5 | 0.882 | 0.512 | 0.487 | −0.05 |
| SSR-482 | 4 | 0.869 | 0.571 | 0.477 | −0.198 |
| SSR-493 | 3 | 0.516 | 0.2 | 0.284 | 0.296 |
| SSR-552 | 3 | 0.184 | 0.075 | 0.073 | −0.03 |
| SSR-593 | 2 | 0.264 | 0.148 | 0.137 | −0.08 |
| SSR-704 | 2 | 0.528 | 0.395 | 0.344 | −0.148 |
| SSR-714 | 2 | 0.253 | 0.14 | 0.13 | −0.075 |
| SSR-744 | 3 | 1.003 | 1 | 0.609 | −0.643 |
| SSR-845 | 4 | 1.038 | 0.432 | 0.553 | 0.219 |
| SSR-875 | 3 | 0.482 | 0.25 | 0.257 | 0.029 |
| SSR-1005 | 5 | 0.462 | 0.093 | 0.195 | 0.522 |
| SSR-1061 | 2 | 0.611 | 0.6 | 0.42 | −0.429 |
| SSR-1146 | 3 | 0.333 | 0.178 | 0.164 | −0.086 |
| SSR-1193 | 2 | 0.619 | 0.619 | 0.427 | −0.448 |
| Mean | 2.904 | 0.584 | 0.359 | 0.34 | −0.035 |
| Groups | Genotypes | Yield | Productivity |
|---|---|---|---|
| Liter/Bag | Bag/ha | ||
| I | Emcapa 143 | 312.26 | 94.65 |
| Tardio V | 329.95 | 69.84 | |
| Imbigudinho | 324.82 | 79.67 | |
| Z29 | 366.61 | 70.15 | |
| 18 | 422.67 | 49.30 | |
| Clementino | 355.48 | 97.86 | |
| Bicudo | 366.21 | 93.09 | |
| Z37 | 363.78 | 86.84 | |
| Ouro Negro | 346.39 | 75.24 | |
| CH1 | 359.87 | 81.40 | |
| Beira rio 8 | 439.72 | 61.81 | |
| Z35 | 402.23 | 72.48 | |
| Bamburral | 357.23 | 86.60 | |
| LB1 | 312.21 | 118.08 | |
| Verdim R | 398.32 | 82.35 | |
| Z21 | 294.01 | 102.49 | |
| II | Tardio C | 345.15 | 74.73 |
| B01 | 418.21 | 43.11 | |
| Z39 | 339.92 | 95.48 | |
| Ouro Negro 2 | 316.46 | 74.70 | |
| A1 | 365.44 | 108.19 | |
| Alecrim | 341.46 | 54.37 | |
| Z36 | 316.62 | 91.45 | |
| Verdim D | 336.65 | 90.43 | |
| Ouro Negro 1 | 359.87 | 72.23 | |
| Clone 1 | - | - | |
| Valcir P | 341.11 | 87.93 | |
| Graudão HP | 317.59 | 86.13 | |
| Z38 | 372.53 | 74.94 | |
| III | AP | 313.53 | 86.28 |
| 122 | 330.03 | 81.07 | |
| P2 | 316.7 | 92.32 | |
| P1 | 332.5 | 93.47 | |
| 700 | 303.23 | 89.19 | |
| Clone 2 | - | - | |
| Pirata | 379.63 | 76.22 | |
| L80 | 364.68 | 105.53 | |
| IV | Z18 | 330.2 | 80.05 |
| Z40 | 356.18 | 74.55 | |
| Emcapa 153 | 322.45 | 85.52 | |
| Peneirão | 326.6 | 99.22 | |
| AT | 303.49 | 128.93 | |
| Emcapa 02 | 323.18 | 97.10 | |
| V | Sementes | 348.06 | 70.32 |
| VI | Arabica | - | - |
| Sample Number | Name | Sample Number | Name |
|---|---|---|---|
| 1 | AP | 24 | Clone 1 |
| 2 | Ouro negro 2 | 25 | Z40 |
| 3 | 18 | 26 | CH1 |
| 4 | Ouro negro 1 | 27 | Emcapa 143 |
| 5 | Graudão HP | 28 | Verdim R |
| 6 | Peneirão | 29 | Tardio C |
| 7 | Ouro Negro | 30 | Clone 2 |
| 8 | Z18 | 31 | 700 |
| 9 | Clementino | 32 | Z29 |
| 10 | Beira Rio 8 | 33 | Emcapa 02 |
| 11 | Verdim D | 34 | Pirata |
| 12 | Z39 | 35 | A1 |
| 13 | Bamburral | 36 | Z36 |
| 14 | P2 | 37 | Z37 |
| 15 | Imbigudinho | 38 | B01 |
| 16 | AT | 39 | Tardio V |
| 17 | Emcapa 153 | 40 | Z21 |
| 18 | Bicudo | 41 | Z35 |
| 19 | Alecrim | 42 | LB1 |
| 20 | Z38 | 43 | L80 |
| 21 | 122 | 44 | P1 |
| 22 | Sementes | 45 | Arabica |
| 23 | Valcir P |
| Locus | Sequence (5′3′) | at | Reference |
|---|---|---|---|
| CaEST-010 | F: CTTCTTCATCCAACAACACG | 54 °C | [35] |
| R: TGCCATTCCACTGTGTCACT | |||
| SSR-081 | F: CACTGGCATTAGAAAGCACC | 55 °C | [35] |
| R: GGCAAAGTCAATGATGACTC | |||
| SSR-303 | F: ATGGGGCCAACTTGAATATG | 55 °C | [35] |
| R: CAGGGCATCTATCTACTTCTCTTT | |||
| SSR-343 | F: GGAGACGCAGGTGGTAGAAG | 55 °C | [35] |
| R: TCGAGAAGTCTTGGGGTGTT | |||
| SSR-353 | F: CTGGCATTAGAAAGCACCTTG | 54 °C | [35] |
| R: GCTTGGCTCACTGTAGGACTG | |||
| SSR-373 | F: CAACACTATCTCTTGATTTTTCACT | 53 °C | [35] |
| R: CGTGCAAGTCACATACTTTACTAC | |||
| SSR-433 | F: TTTTCTGGGTTTTCTGTGTTCTC | 50 °C | [35] |
| R: TAACTCTCCATTCCCGCATT | |||
| SSR-463 | F: AATGAAGAAGAGGGTGGTG | 53 °C | [35] |
| R: CGAGGGTATTGTTTTCCAG | |||
| SSR-482 | F: AGCAAGTGGAGCAGAAGAAG | 56 °C | [35] |
| R: CGGTGAATAAGTCGCAGTC | |||
| SSR-493 | F: TGGAGAAGGCTGTTGAAACC | 56 °C | [35] |
| R: GGCGTGAAGCAAAAAGGTAT | |||
| SSR-552 | F: GCAGGTATTTGAAGGATGAACC | 56 °C | [35] |
| R: GTGTAGGTGGTGCGATGTGT | |||
| SSR-593 | F: CCAGCTCTCCTCACTCTTTTCA | 58 °C | [35] |
| R: GGTGGTGGAGGGGTAATAGG | |||
| SSR-704 | F: GTAACCACCACCTCCTCTGC | 59 °C | [35] |
| R: TGGAGGTAACGGAAGCTCTG | |||
| SSR-714 | F: GCTAAGTTCAATTGCCCCTGT | 55 °C | [35] |
| R: GGGTTAATTTGATTGCGTGA | |||
| SSR-744 | F: TGGGGAAAAGAAGGATATAGACAAGAG | 59 °C | [35] |
| R: GAGGGGGGCTAAGGGAATAACATA | |||
| SSR-845 | F: AAGTAGATTGGTGAAAGGGAAGC | 57 °C | [35] |
| R: TCCTTCATTTTCTCCTCTTGGTT | |||
| SSR-875 | F: ATTCGACGACTCCAAAGCATA | 58 °C | [35] |
| R: CCTTGCTGGCCCTTCCTT | |||
| SSR-1005 | F: ACCCTTTACTACTTATTTACTCTC | 50 °C | [35] |
| R: ACATCCCCTTGCCATTTCTTC | |||
| SSR-1061 | F: CCCTCCCTCTTTCTCCTCTC | 56 °C | [35] |
| R: TCTGGGTTTTCTGTGTTCTCG | |||
| SSR-1146 | F: TAACAGAAGCACCAAAACC | 53 °C | [35] |
| R: TCTAAACCCACCTCACAAC | |||
| SSR-1193 | F: TTGCCATCATCGTTCATTCT | 56 °C | [35] |
| R: GCATAGTGTCGGTTGTGTTGTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa, P.; Vieira, H.; Santos, E.; Viana, A.; Boaechat, M.; Partelli, F. Coffea canephora: Heterotic Crosses Indicated by Molecular Approach. Plants 2022, 11, 3023. https://doi.org/10.3390/plants11223023
Sousa P, Vieira H, Santos E, Viana A, Boaechat M, Partelli F. Coffea canephora: Heterotic Crosses Indicated by Molecular Approach. Plants. 2022; 11(22):3023. https://doi.org/10.3390/plants11223023
Chicago/Turabian StyleSousa, Priscila, Henrique Vieira, Eileen Santos, Alexandre Viana, Marcela Boaechat, and Fábio Partelli. 2022. "Coffea canephora: Heterotic Crosses Indicated by Molecular Approach" Plants 11, no. 22: 3023. https://doi.org/10.3390/plants11223023
APA StyleSousa, P., Vieira, H., Santos, E., Viana, A., Boaechat, M., & Partelli, F. (2022). Coffea canephora: Heterotic Crosses Indicated by Molecular Approach. Plants, 11(22), 3023. https://doi.org/10.3390/plants11223023







