Abstract
Plant diseases caused by pathogens lead to economic and agricultural losses, while plant resistance is defined by robustness and timing of defence response. Exposure to microbial-associated molecular patterns or specific chemical compounds can promote plants into a primed state with more robust defence responses. β-aminobutyric acid (BABA) is an endogenous stress metabolite that induces resistance, thereby protecting various plants’ diverse stresses by induction of non-canonical activity after binding into aspartyl-tRNA synthetase (AspRS). In this study, by integrating BABA-induced changes in selected metabolites and transcript data, we describe the molecular processes involved in BABA-induced resistance (BABA-IR) in tomatoes. BABA significantly restricted the growth of the pathogens P. syringae pv. tomato DC3000 and was related to the accumulation of transcripts for pathogenesis-related proteins and jasmonic acid signalling but not salicylic acid signalling in Arabidopsis. The resistance was considerably reduced by applying amino acids L-Asp and L-Gln when L-Gln prevents general amino acid inhibition in plants. Analysis of amino acid changes suggests that BABA-IR inhibition by L-Asp is due to its rapid metabolisation to L-Gln and not its competition with BABA for the aspartyl-tRNA synthetase (AspRS) binding site. Our results showed differences between the effect of BABA on tomatoes and other model plants. They highlighted the importance of comparative studies between plants of agronomic interest subjected to treatment with BABA.
1. Introduction
Due to the inability to move, plants are exposed to various environmental stresses, both biological and abiotic, without having the possibility to escape. As a result of numerous pathogens and pest attacks, plants have formed passive and active physical and biochemical barriers. Once innate immunity is insufficient, local and systemic inducible responses can be activated to fortify defences. Defence responses are activated upon recognising the pathogen when the robustness of early activation and response cannot prevent the pathogen’s spread. Often, plant defence mechanisms are ineffective, leading to extensive plant infections resulting in catastrophic harvest failures and causing considerable economic and social problems worldwide.
Even though plants do not possess mobile immune system cells, defence responses can be activated at both the local and systemic levels after pathogen recognition. In these cases, the timing and strength of the immune response are decisive. The state of induced resistance is then characterised by a faster and more potent immune response of the plant activated upon the pathogen perception. Induced resistance can be activated upon the stimulus of different origins and provides resistance against a broad spectrum of stresses either through systemic acquired resistance (SAR) or induced systemic resistance (ISR). Besides native intrinsic activation, induced resistance can be activated by exogenously applied chemical compounds. Today, managing plant defences is a promising approach, especially using priming agents that prepare plants to enhance their defence responses. Defence priming causes enhanced expression of genes related to stress and defence [1], including many defence regulatory transcription factors [2]. Defence priming is now considered an essential component of various types of systemic plant immunity.
One of the most influential and promising defence priming agents is the non-proteinaceous amino acid beta-aminobutyric acid (BABA), which induces broad-spectrum resistance to pathogens covering the oomycetes, fungi, bacteria, nematodes, and insects in many plant species [3]. BABA was considered a xenobiotic compound, but recent research showed that it accumulates in plants exposed to stress [4] and acts as an endogenous stress metabolite [5]. The molecular mechanism of BABA-induced resistance (BABA-IR) was partially elucidated in Arabidopsis plants via the binding of R-BABA to the aspartate binding site of the aspartyl-tRNA synthetase (AspRS) IBI-1 and induction of its non-canonical activity resulting in translocation to the cytoplasm [6]. Here, VOZ1 and VOZ2 transcription factors were identified as IBI1-interacting partners, and this interaction repressed the expression of ABA genes resulting in increased expression of pattern-triggered immunity (PTI) genes and callose-associated defence. An increased Asp level after R-BABA application further supported the proposed mechanism; however, direct evidence of the inhibition of the BABA effect on IBI-1 activity is missing. Interestingly, BABA-IR inhibition was observed in Arabidopsis after the L-glutamine treatment, which prevents a general amino acid stress inhibition in plants [7].
Tomato (Solanum lycopersicum) is one of the most important agricultural plants in the world, with an annual production of 186 million tons in 2020 [8]. Besides its agricultural importance, the tomato is widely used in research due to the presence of features such as compound leaves, fleshy fruit, and sympodial shoot. In contrast, other model plants, such as Arabidopsis and rice, lack these agronomically essential traits. In addition, the tomato is a member of the Solanaceae family and studies conducted on the tomato can be easily applied to members of this family, which also includes commercially important crops such as tobacco, potatoes, peppers, and eggplants, making it a vital research material [9]. Previous studies showed that BABA induced resistance to biotrophic and hemibiotrophic pathogens in tomato plants, but our study further advances our understanding of the detailed description of the molecular mechanism [3].
Here, we investigated the effect of BABA treatment in combination with amino acids Asp and Gln on BABA-IR in tomatoes. We used cultivars Solanum lycopersicum cv. Micro-Tom and cv. Amateur, representing a model tomato cultivar and a widely grown tomato cultivar, respectively, and the established virulent bacterial model pathogen Pseudomonas syringae pv. tomato DC3000. Analysis of resistance in conjunction with analysis of amino acid changes, transcripts, and phytohormones involved in resistance demonstrate that treatment with Asp and Gln reduces the rate of BABA-IR, which in tomato, appears to be mediated by JA rather than the SA signalling pathway.
2. Results
2.1. L-Glutamine and L-Aspartate Reduce the BABA-IR in Tomato
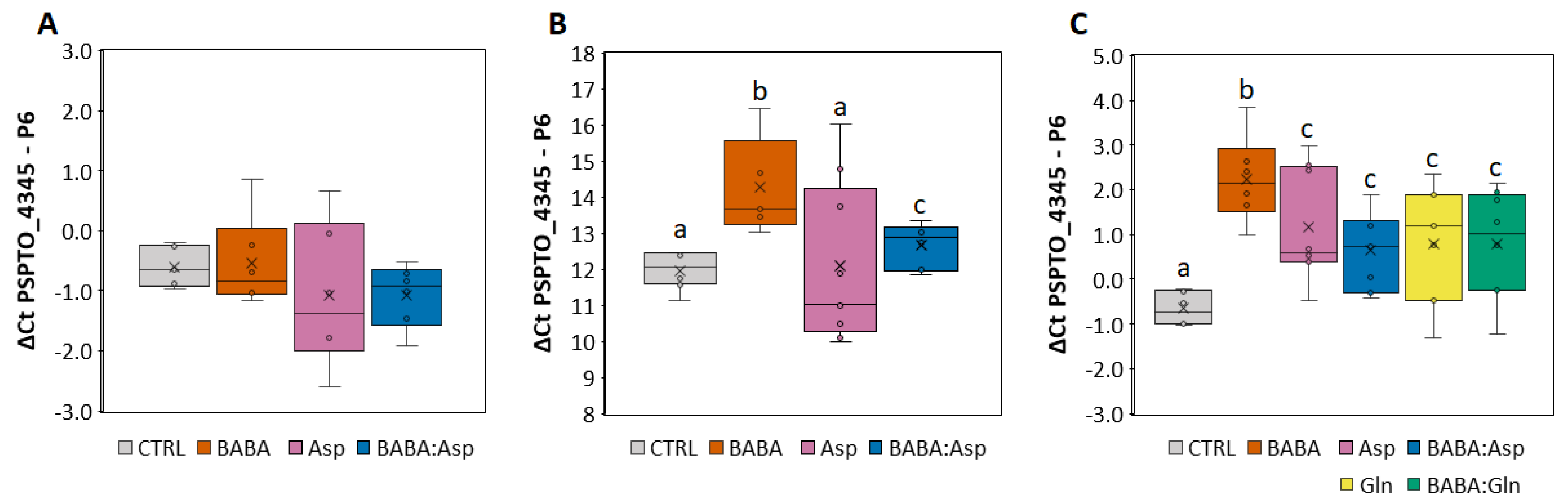
To determine the protective effect of BABA treatment against the pathogen P. syringae pv. tomato DC3000, we treated 6–7-week-old S. lycopersicum cv. MicroTom and cv. Amateur leaflets with 5 mM BABA, L-Asp, or equimolar mixtures BABA:Asp and BABA:Gln through petiole aspiration to avoid the formation of small necrotic lesions observed after BABA treatment by spraying [10,11]. In the case of S. lycopersicum cv. MicroTom, bacterial inoculation was performed by spraying the bacterial suspension on the leaflets or by direct syringe infiltration of bacteria to the apoplast of the leaflets 24 h after BABA treatment. Previous studies with diverse fungi and bacteria showed that BABA has no direct toxic effect. Thus, BABA-mediated resistance is most likely based on the activation of host resistance mechanisms [12]. The infection was evaluated 48 h later by the previously described qPCR method [13] using primers for the P6 gene (Solyc00g174340) from tomato and the PSPTO_4345 gene (GenBank: CP047072) for P. syringae pv. tomato DC3000. Noticeably, the resistance depended on the technique of the application. The application by syringe did not induce significant resistance after the BABA or Asp treatment (Figure 1A), but application by spraying induced considerable resistance after the BABA treatment (Figure 1B). Moreover, treatment with an equimolar mixture of BABA: Asp weakened this resistance to the basal level (Figure 1B).
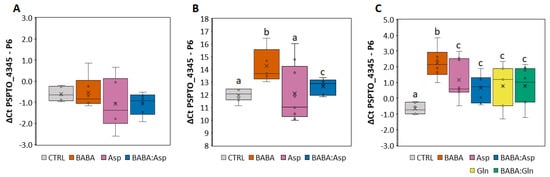
Figure 1.
Resistance levels of tomato against pathogen P. syringae. (A,B) Solanum lycopersicum cv. MicroTom leaflets were inoculated with Pseudomonas syringae pv. tomato DC3000 by direct syringe infiltration of bacteria into the apoplast (A) or by spraying the bacterial suspension on the leaflets (B) treated for 24 h with 5 mM BABA, Aspartate, or their equimolar mixture (BABA:Asp). (C) Solanum lycopersicum cv. Amateur leaflets were inoculated with Pseudomonas syringae pv. tomato DC3000 by spraying the bacterial suspension on the leaflets treated for 24 h with 5 mM BABA, Aspartate (Asp), Glutamine (Gln), or their equimolar mixture (BABA:Asp, BABA:Gln). The level of resistance was measured after 72 h by qPCR and evaluated by the ΔΔCt method [14] using the P6 gene (Solyc00g174340) from tomato and the PSPTO_4345 gene (GenBank: CP047072) from P. syringae pv. tomato DC3000. Significant differences compared with the water-treated control were determined using the ANOVA, and Duncan’s test is a post-hoc test; different lowercase letters indicate significant differences at p ≤ 0.05 (Duncan’s test).
For the results obtained in the case of the S. lycopersicum cv. Amateur, we inoculated the pathogen only using the spraying technique. Compared with the MicroTom cultivar, we also used 5 mM L-Gln and equimolar mixtures BABA:Gln. Not only BABA but also BABA enriched with amino acids Asp and Gln induced significant resistance in the Amateur cultivar but to a lesser extent (Figure 1C). Noticeably, treatment with an equimolar mixture of BABA:Asp or BABA:Gln resulted in the significant weakening of observed BABA-IR (Figure 1C).
2.2. BABA Did Not Induce Changes in L-Aspartate Levels in Tomato
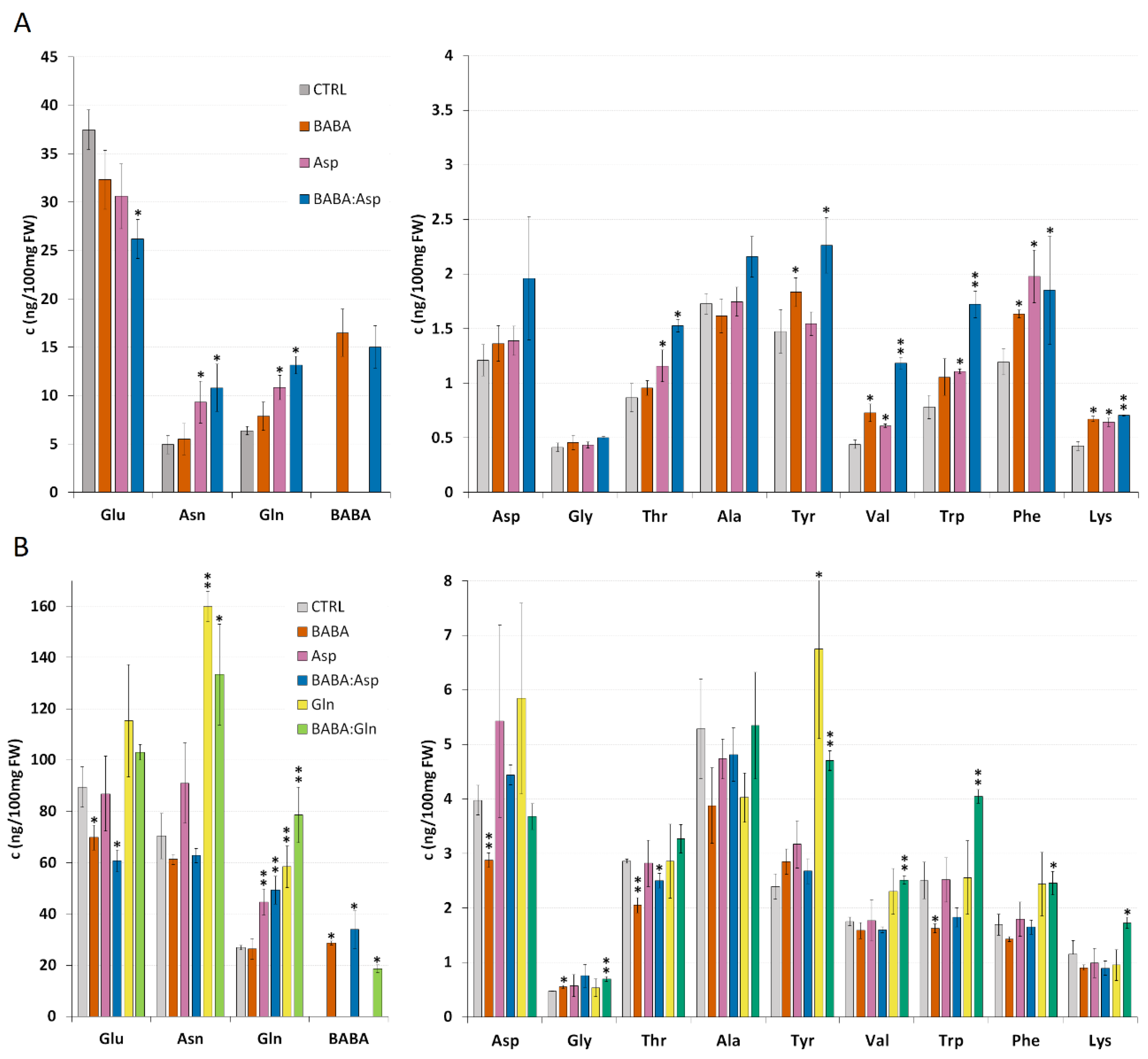
The result from the resistance experiments indicated the role of aspartic acid and glutamine on the level of BABA-IR. Hence, a detailed amino acids analysis was carried out in S. lycopersicum cv. MicroTom leaflets treated with 5 mM solutions of BABA, L-Asp, or equimolar mixture BABA:Asp and S. lycopersicum cv. Amateur leaflets treated with 5 mM solutions of BABA, L-Asp, L-Gln, or equimolar mixtures BABA:Asp and BABA:Gln through petiole aspiration. In all BABA-treated samples, we found a significantly increased level of BABA corresponding to the basal level of Gln, which greatly exceeded the basal level of aspartate (Figure 2A,B).
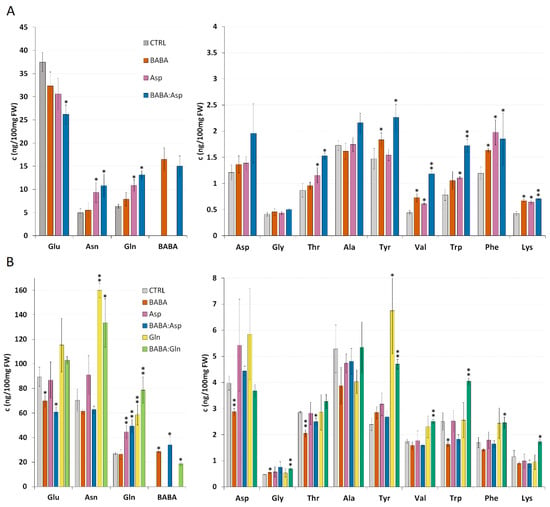
Figure 2.
Amino acid levels in tomato plants. Amino acid levels of 8-week-old leaflets of Solanum lycopersicum cv. MicroTom (A) and Solanum lycopersicum cv. Amateur (B) plants. Leaflets were treated for 24 h with BABA or amino acids solutions (BABA, L-Asp, BABA: Asp, L-Gln, BABA: Gln), and amino acid levels were measured by the HPLC method. Data are means from free biological replicates for each time; the errors represent standard errors of means. Statistically, significant differences recorded for each amino acid as determined by t-tests are annotated with different asterisks (* p < 0.05, ** p < 0.01).
In contrast to previous data on Arabidopsis plants [15], BABA treatment of both tomato cultivars through petiole aspiration did not induce a significant increase in L-Asp level (Figure 2A), while in the case of S. lycopersicum cv. Amateur, we even detected a decrease in the L-Asp level (Figure 2B). This result could be a consequence of metabolic disbalance in susceptible cultivars after BABA treatment because we observed a decrease in many other amino acids such as Glu, Asn, Ala, Trp, Phe, or Lys. This phenomenon will need to be confirmed and described in more detail in further studies.
Surprisingly, in both tomato cultivars, the treatment with Asp did not result in its significantly elevated level but we observed a substantial increase in the level of Gln together with Asn, the common nitrogen carriers playing the primary role in the recycling, storage, and transport of nitrogen in plants (Figure 2A,B) [16,17]. In the case of treatment with equimolar mixtures of BABA:Asp, we observed an additive effect of both substances. In S. lycopersicum cv. MicroTom, treatment resulted in elevated levels of aromatic amino acids Tyr, Trp, and Phe and amino acids Val and Lys, similar to BABA and Asp treatment. S. lycopersicum cv. Amateur treatment decreased Glu and Thr levels, identical to the BABA treatment (Figure 2A,B). In both tomato cultivars, we observed a significant increase in Gln level similar to the Asp treatment (Figure 2A,B). The treatment of S. lycopersicum cv. Amateur plants with Gln and equimolar mixtures BABA:Gln led to a significant increase in Asn, Gln, and Tyr levels when, in the case of the BABA:Gln treatment, we found increased levels of the aromatic amino acids Trp and Phe (Figure 2B).
2.3. BABA up-Regulated Transcripts and Signalling in the Tomato
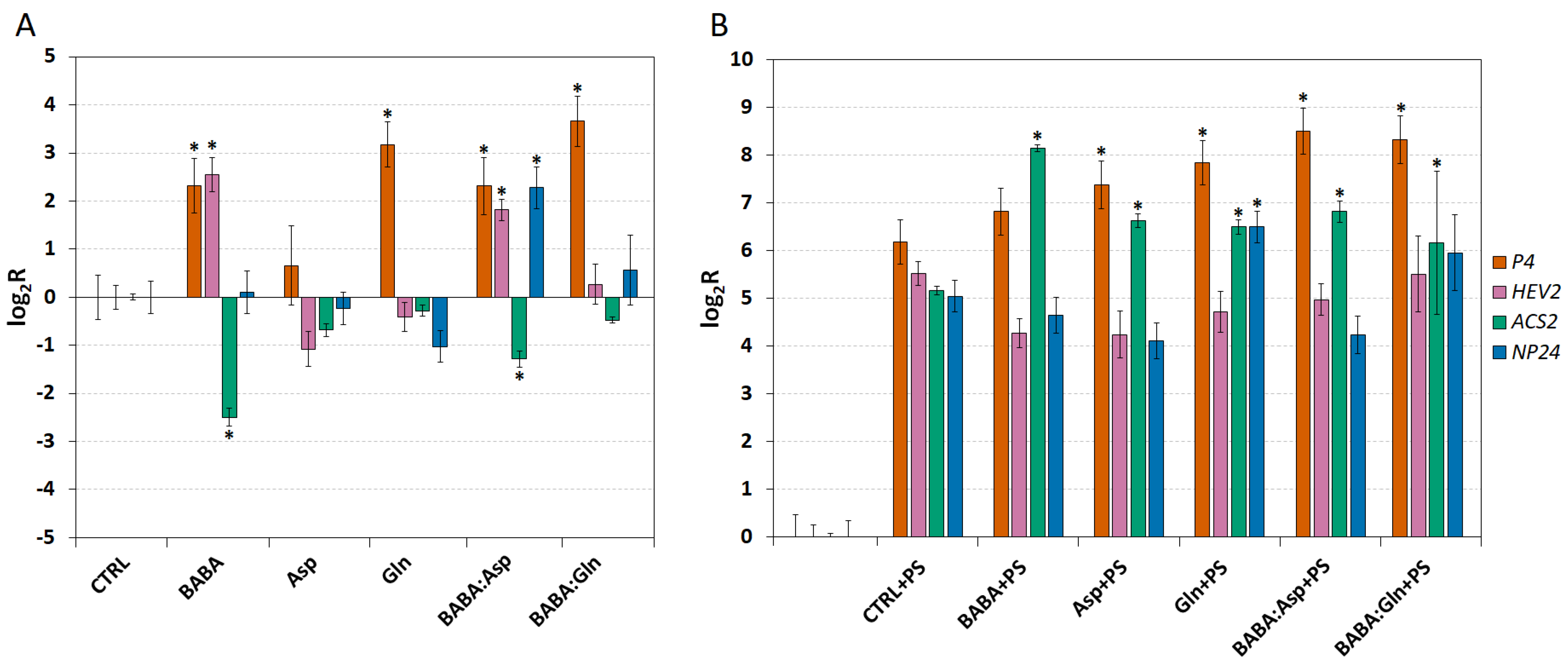
To demonstrate the role of defence genes in the resistance of S. lycopersicum cv. Amateur leaflets induced by BABA and amino acids treatment, we measured the expression of four defence genes, P4 (PR1 protein), HEV2 (hevein 2), NP24 (osmotin 1), and ACS2 (1-aminocyclopropane-1-carboxylic acid synthase 2), whose increased expression was previously demonstrated in BABA-IR [11]. The biological function of gene P4 is unclear, HEV2 gene coding protein acting as a chitinase, NP24 gene coding thaumatin-like protein PR-NP24, and ACS2 gene coding enzyme e ACC synthase regulating the ethylene (ET) synthesis [18].
The transcript levels were analysed in leaves collected 24 h post-treatment through petiole aspiration with 5 mM solutions of BABA, L-Asp, L-Gln, or equimolar mixtures BABA:Asp and BABA:Gln and two days after infection with P. syringae pv. tomato DC3000. In all treatments except the Asp treatment, we observed a strong up-regulation of the P4 transcript after 24 h. Interestingly, the Asp treatment did not up-regulate any other measured genes when only the tBABA and BABA:Asp treatment up-regulated the HEV2 transcript and down-regulated the ACS2 transcript. Finally, the NP24 transcript was up-regulated only in the BABA:Asp treatment (Figure 3A).
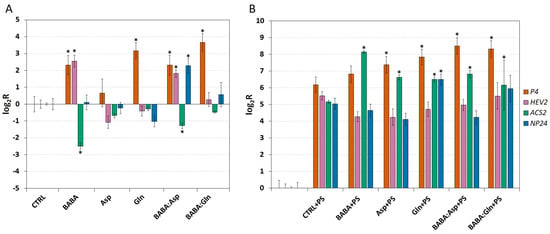
Figure 3.
The expression of defence genes inSolanum lycopersicum cv. Amateur plants. The transcript levels of four defence genes (P4, HEV2, ACS2 and NP24) were monitored in 8-week-old leaflets (n = 6) 24 h after treatment with 5 mM BABA and/or amino acids solutions (BABA, L-Asp, BABA:Asp, L-Gln, BABA:Gln) (A) or 48 h after their inoculation with P. syringae pv. tomato DC3000 (B). Transcript levels were measured by the RT-qPCR method and evaluated by the ΔΔCt method [14]. The control tissue (CTRL) was a water-treated control sample. Each bar represents the mean ± SE. Asterisks denote mean values that differ significantly from that of the control tissue based on Dunnett’s test (* p < 0.05).
On the other hand, two days after infection, we observed strong up-regulation of all measured genes in all treatment conditions and inoculated control leaflets treated with MgCl2 buffer containing 0.02% Silwet L77 as a mock inoculation (Figure 3B). Two days after inoculating leaflets with the pathogen P. syringae, compared to the inoculated control, we observed a significantly higher ACS2 transcript accumulation, coding a key regulatory enzyme in the ET synthesis pathway in all treatments (Figure 3B). Moreover, the P4 transcript was significantly accumulated in all treatments except BABA and the NP24 transcript was significantly increased only in the L-Gln treatment (Figure 3B).
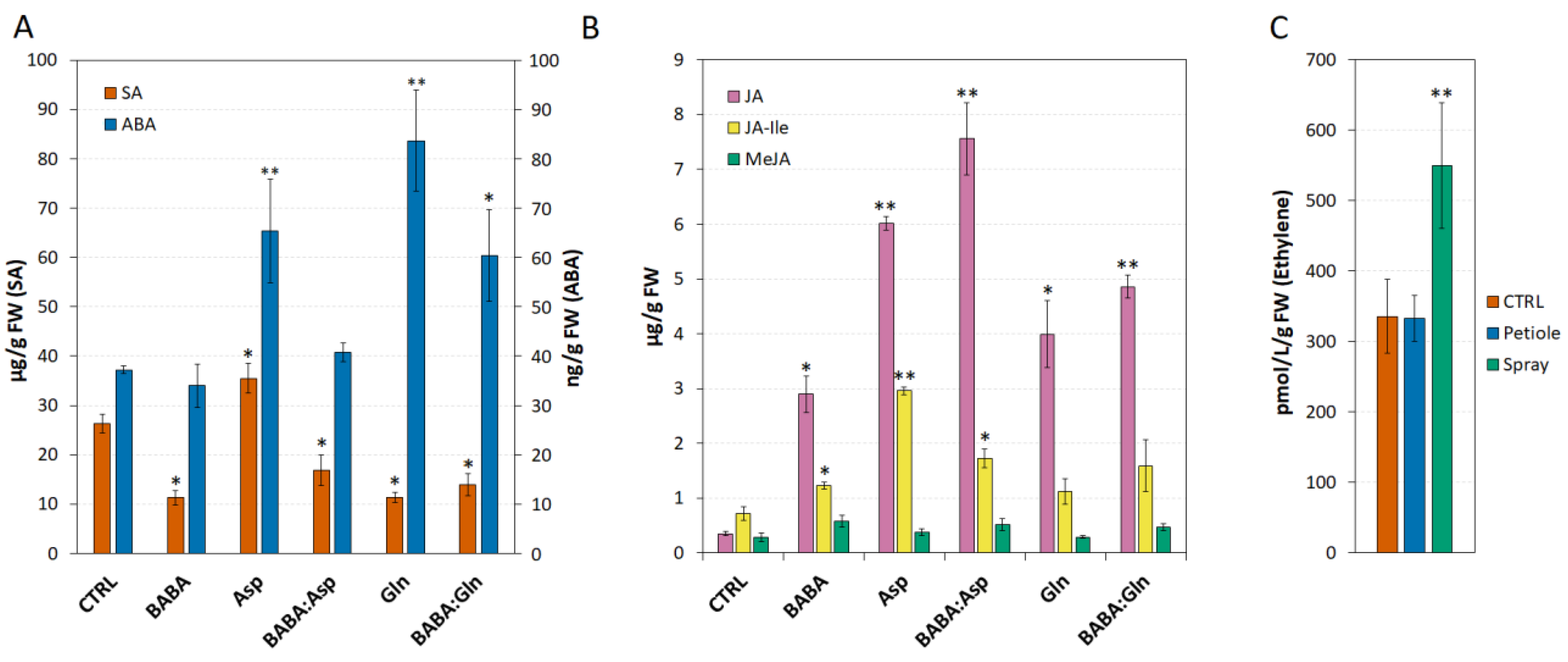
BABA induces resistance via several hormones, including SA [19], JA [20], abscisic acid [21] or ET [12]. In both potato and Arabidopsis, BABA potentiates SA-dependent defence against pathogens [2]. Indeed, in Arabidopsis, BABA-induced priming seems to be also ABA-dependent [6]. Surprisingly, here in S. lycopersicum cv. Amateur leaflets, BABA treatment alone or in combination with amino acids L-Asp and L-Gln led to a significant decrease in SA compared to its higher level after L-Asp treatment alone (Figure 4A). In addition, the application of BABA to tomato leaflets did not change the level of ABA and its combination with amino acids L-Asp and L-Glu reduced the amplitude of the increase in the ABA level caused by the amino acids themselves. On the other hand, treatment with 5 mM solutions of BABA, L-Asp, L-Gln, or equimolar mixtures BABA:Asp and BABA:Gln significantly induced the accumulation of JA and its endogenous bioactive conjugate JA-Ile but to a lesser extent (Figure 4B). Consistent with the transcriptomic data for the ACS2 enzyme, one of the key enzymes of the ET synthesis pathway in tomato and Arabidopsis [11,22], we did not observe an increase in ET concentration in leaflets treated with 5 mM BABA through petiole aspiration, but only after the spray treatment with 5 mM BABA, as described earlier [11] (Figure 4C).
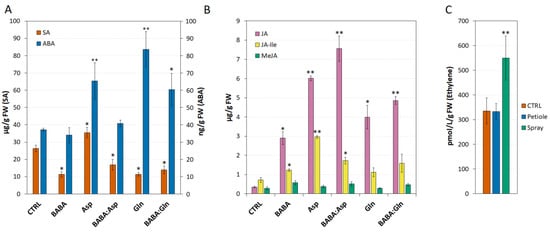
Figure 4.
Phytohormones levels in Solanum lycopersicum cv. Amateur plants. (A,B) The levels of salicylic acid (SA), abscisic acid (ABA), jasmonic acid (JA), jasmonate isoleucine (JA-Ile) and methyl jasmonate (MeJA) were measured by LC–MS 24 h after 5 mM BABA and/or amino acids solutions (BABA, L-Asp, BABA:Asp, L-Gln, BABA:Gln) of the 8-week-old leaflets through petiole aspiration (n = 6). (C) ET accumulation was measured 24 h after BABA treatment (5 mM) of the 8-week-old leaflets through petiole aspiration (n = 6) or spraying (n = 6) by gas chromatography. The control tissue (CTRL) was a water-treated control sample. Each bar represents the mean ± SE. Asterisks denote mean values that differ significantly from the control tissue based on Dunnett’s test (* p < 0.05, ** p < 0.01).
3. Discussion
The phenomena of BABA-IR in different plants were proven by various studies [23,24]. Here, we tested the effectiveness of BABA, L-Asp, and L-Gln or their equimolar mixture treatments on the resistance of the dwarf cultivar Solanum lycopersicum cv. Micro-Tom [25] and Solanum lycopersicum cv. Amateur against the pathogenic bacterium Pseudomonas syringae pv. tomato DC3000 [26]. Solanum lycopersicum cv. Micro-Tom represents a preferred variety for molecular research in tomatoes [27] and Solanum lycopersicum cv. Amateur is a good model system for an agricultural crop. The L-Asp was selected due to the previous finding that aspartyl-tRNA synthetase serves as an R-BABA receptor in Arabidopsis and L-Asp accumulates after BABA application [15]. On the other hand, the L-Gln treatment of the Arabidopsis plant inhibited BABA-IR previously [7].
A BABA-IR against pathogenic bacteria P. syringae pv. tomato DC3000 depended on the technique of pathogen application when, in the case of S. lycopersicum cv. Micro-Tom, we observed a high basal resistance level to P. syringae pv. tomato DC3000 with limited symptoms corresponding with the previous finding [28]. In the case of syringe infiltration, where the bacterial pathogen is delivered directly to the apoplastic space, we observed no induction of resistance compared to the application of bacterial pathogen on leaves. This result suggests that BABA application-mediated priming of tomato defence mechanisms involved in stomata closure restricts pathogen penetration to the apoplast as a part of the pattern-triggered immunity (PTI). PTI controls the production of long-distance signals that systemically prime plants against future attacks, resulting in systemic acquired resistance [29,30,31]. Noticeably, in the case of S. lycopersicum cv. Amateur, the significantly increased resistance, even lower than BABA, was observed after applying 5 mM L-Asp and L-Gln. This finding agrees with the study of Kadotani et al., 2016, in which strong rice resistance against rice blast fungus Magnaporthe grisea was observed after applying 10 mM proteinaceous amino acids L-Glu, L-Asn, L-Met, and L-Asp [32].
In Arabidopsis thaliana, the molecular mechanism described by Luna et al., 2014 suggests induction of the non-canonical activity of AspRS mediated by the binding of R-BABA to a catalytic site of the enzyme and increase in L-Asp level [15]. Here, this suggestion seems to be supported by the observed effect of L-Asp on reducing BABA-IR resistance. However, the measured level of L-Asp showed no increase 24 h after L-Asp or BABA treatment, and the BABA level in leaflets was approximately 10-times higher compared to L-Asp due to its persistence in plants. On the other hand, we detected an increased level of L-Gln and L-Asn after L-Asp or BABA/L-Asp treatment, indicating its rapid metabolisation to these typical amino acids used by the nitrophilous plants for nitrogen storage [33]. Indeed, we observed a significant reduction of BABA-IR after L-Gln treatment, in agreement with previous studies on Arabidopsis [34]. Therefore, it seems probable that the observed decrease in BABA-IR after L-Asp treatment could be somewhat due to an increased level of L-Gln than due to competition of L-Asp with BABA regarding the binding site. In the case of S. lycopersicum cv. Micro-Tom, a significant increase in the level of precursor amino acids of the phenylpropanoid pathway (Phe, Tyr, and Trp) involved in increased resistance was observed [35] after BABA treatment. In addition, an increased level of amino acid Lys, representing a precursor of pipecolic acid taking a role in defence reaction [36], may also play a role in this priming mechanism. Interestingly, in the case of S. lycopersicum cv. Amateur, we found an increased level of these amino acids only after BABA/L-Gln treatment, showing different mechanisms operating in tomato cultivars with different resistance mechanisms, as reported earlier [11].
BABA treatment strongly induced the expression of the P4 and HEV2 genes, which corresponds well with our previous study on S. lycopersicum cv. Amateur [11]. On the other hand, compared to that study, we did not observe any increase in the NP24 and ACS2 transcripts. We suggest that this discrepancy is a result of a different application method. Applying BABA by foliar spraying caused the formation of hypersensitive reaction-like lesions on the leaves followed by ET synthesis [11,37], which could be related to stress responses triggered by high BABA concentrations in the drying droplets (white deposits) after spraying. This observation corresponds with the well-demonstrated involvement of ET in the regulation of the degree of PCD during plant–pathogen interactions [38], where the initiation of hypersensitive reaction results in a large burst of ET [39]. However, the BABA was aspirated into the leaflet through the petiole, causing no formation of hypersensitive reaction-like lesions and, thus, no ET production. In keeping with this hypothesis, applying high BABA concentrations (>20 mM) to the petiole of detached tomato leaflets induces the formation of HR-like lesions. Pathogen inoculation led to strong up-regulation of all selected genes; however, in the case of BABA pretreatment, there was no visible defence priming effect demonstrated by the more intense expression [40]. Only in the case of the ACS2, the key enzyme of ET synthesis, transcript was significantly higher than the control, supporting the suggested ET-dependent mechanisms in BABA-induced resistance in tomatoes [11,41].
In Arabidopsis, BABA treatment was accompanied by the accumulation of SA and caused significant changes in the abundance of isochorismate synthase (ICS), which is directly involved in SA biosynthesis [12,42]. Here, in S. lycopersicum cv. Amateur, we observed no accumulation of SA after BABA treatment. On the other hand, treatment of S. lycopersicum cv. Amateur with BABA and/or amino acids L-Asp and L-Gln increased the content of JA and JA-Ile in S. lycopersicum cv. Amateur leaflets in a manner similar to BABA-IR towards P. infestans in the tomato def mutant, which is defective in JA accumulation [43]. Noticeably, BABA treatment reduced the accumulation of abscisic acid (ABA) after L-Asp and L-Gln treatment, which corresponds to the recently observed suppression of ABA-inducible abiotic stress genes during BABA-IR against the biotrophic oomycete Hyaloperonospora arabidopsidis [6].
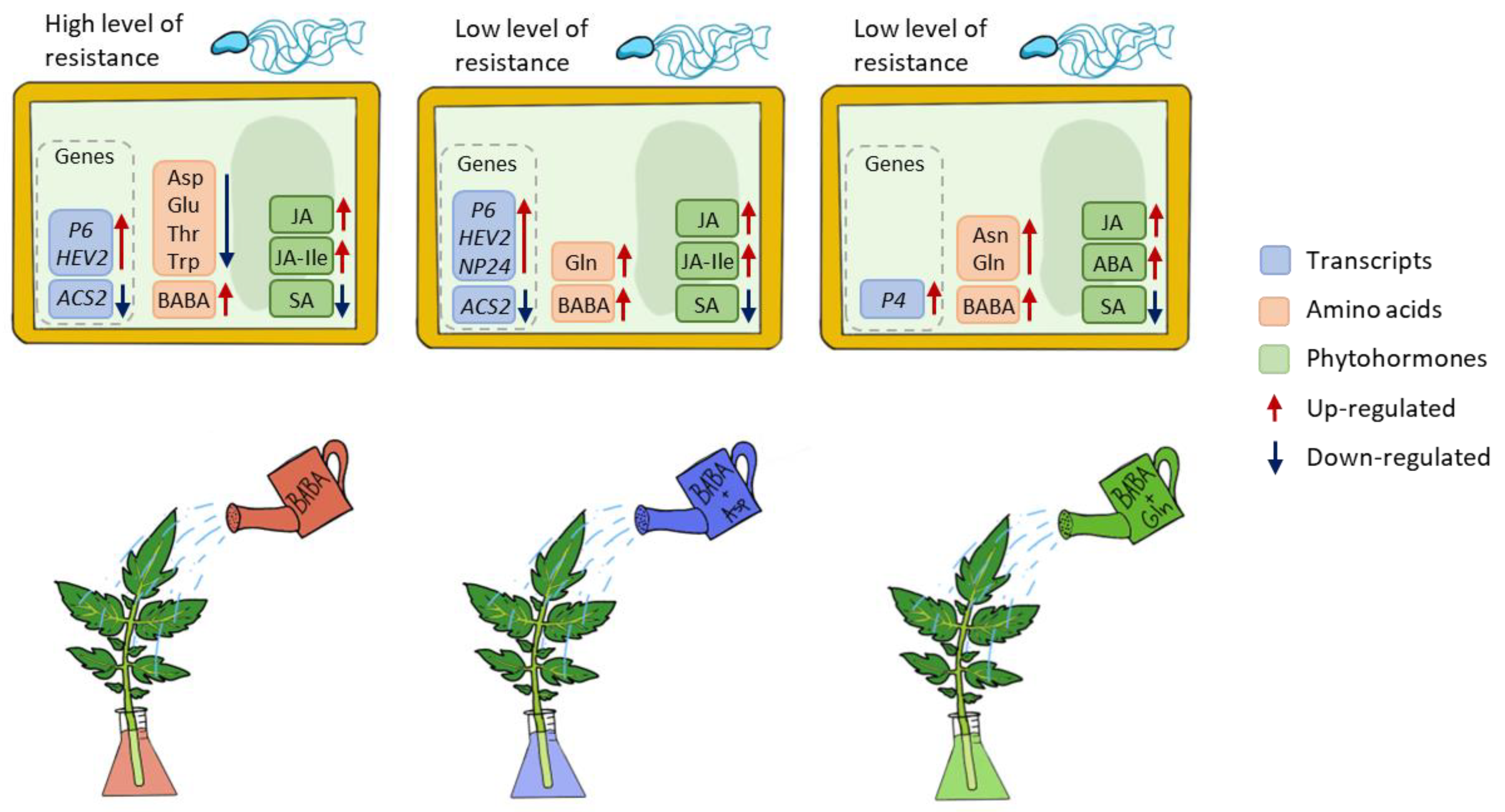
This work shows that the signalling processes and immune response activated during BABA-IR in tomatoes differed from those in Arabidopsis in response to P. syringae pv. tomato DC3000 in S. lycopersicum cv. Amateur. In BABA-treated tomato plants, the defence reaction is controlled by jasmonic acid signalling but not salicylic acid signalling. Moreover, BABA treatment of tomato caused no change in Asp levels; however, Asp- and Gln-treatment of tomato reduced BABA-IR (Figure 5). In conclusion, comparative studies between BABA-treated plants of agronomic interest must be carefully interpreted and expanded to other plant systems in general.
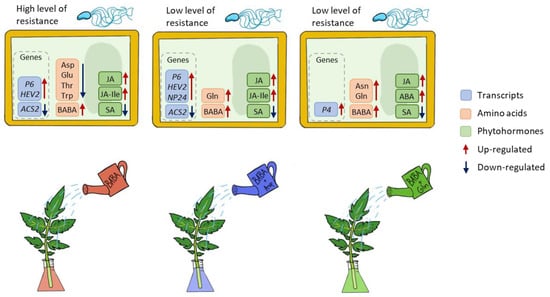
Figure 5.
Schematic representation of processes operated in tomato plants after treatment with BABA or BABA + amino acids Asp and Gln.
4. Materials and Methods
4.1. Plant Material
Solanum lycopersicum cv. Micro-Tom and Amateur were grown at 65% humidity, 24 °C with a 16 h daily light period (light intensity 100 µmol/m2). The detached leaflets from 6–7-week-old plants were immersed for 24 h in aqueous solutions of 5 mM BABA, 5 mM BABA with 5 mM Asp, 5 mM BABA with 5 mM Gln, 5 mM Asp and 5 mM Gln adjusted to pH = 7. Subsequently, the leaflets were inoculated with the pathogenic bacteria Pseudomonas syringae pv. tomato DC3000 and plants were incubated in a climatic chamber at 24 °C for 72 h. Leaves were collected before pathogen inoculation for amino acid and phytohormone level analysis and gene expression analysis and 72 h after the pathogen inoculation for resistance and gene expression analysis.
4.2. Plant Resistance Analysis against Pseudomonas Syringae
P. syringae pv. tomato DC3000 was inoculated into King’s B medium and cultured on a shaker at 225 RPM at 28 °C for 24 h. Subsequently, the culture was centrifuged at 4000× g for 10 min. The bacterial culture was diluted with 10 mM MgCl2 to an A620 of 0.2, corresponding to 108 CFU/mL concentration. Next, the bacteria were diluted to a concentration of 107 CFU/mL with 10 mM MgCl2 containing 0.1% Silwet L77 (AgroBio, Opava, Czechia). This was followed by applying bacteria into the leaves using direct infiltration into the leaf apoplast using a 1 mL needleless syringe or a brush to avoid wounding tissue until both the abaxial and adaxial surfaces were uniformly wet according to the method described previously [44]. The MgCl2 buffer containing 0.02% Silwet L77 was applied as a mock inoculation. The leaves were incubated in a climatic chamber for 72 h with water supplied by wet cotton at 23 °C, 85% humidity (16 h photoperiod), and then frozen in liquid nitrogen and stored at −70 °C until DNA isolation.
DNA was isolated according to the CTAB protocol [45]. Isolated DNA was analysed by qPCR method using Luna® Universal qPCR Master Mix (NEB, Ipswich, MA, USA) in a Light Cycler 480 (Roche, Mannheim, Germany). Primers for P6 (cv. Amateur) or Tip41 (cv. Micro-Tom) genes were used for tomato quantification and primers for PSPTO_4345 gene (GenBank: CP047072) for pathogen quantification. Data were evaluated by the ΔCt method [14]. PCR mixtures were prepared in a total volume of 15 µL and contained 4 µL of template DNA, 7.5 µL of Luna® Universal qPCR Master Mix (2×), and forward and reverse primer (Table S1) in a final concentration of 0.3 µM. qPCR temperature profile consisted of a 150-sec denaturation step at 95 °C followed by 45 cycles consisting of a 10-sec denaturation step at 95 °C and a 30-sec annealing extension step at 60 °C.
4.3. Amino Acid Levels Quantification
Amino acids were extracted 24 h after the solution treatment (BABA, L-Asp, L-Gln, BABA:Asp, and BABA:Gln). A 1 mL of extraction buffer consisting of 0.1 M HCl with the addition of 4.6 μg/mL 2-aminoadipic acid as an internal standard was added to 100 mg of leaf powder, mixed thoroughly, and incubated on ice for 5 min. Samples were centrifuged for 10 min at 4 °C and 15,000× g. A total of 500 μL of supernatant was diluted with 100 μL of methanol. Subsequently, solid-phase extraction was performed using Macherey-Nagel Chromabond® C18, 100 mg of solid-phase per 5 mL colony. The colony was washed with 1 mL of methanol and then with 1 mL of equilibration buffer (20% MeOH in 0.1 M HCl). Samples (600 μL) were applied to the column, and the column was washed with 400 μL of equilibration buffer, resulting in a volume of 1 mL of extracted amino acids. Extracted amino acids were derivatised and analysed, as described previously [46].
4.4. Transcription of Pathogenesis-Related Genes
Plant material before and after 72 h post-pathogen inoculation was used. Plant RNA was isolated by TRI REAGENT (Merck, Darmstadt, Germany) and treated by RapidOut DNA Removal Kit (Thermo Scientific™, Waltham, MA, USA). Reverse transcriptase reactions were performed with the ImProm-II reverse transcription system (Promega, Madison, WI, USA). According to the manufacturer’s instructions, the obtained cDNA was amplified by qPCR using gene-specific primers (Table S1) and Luna® Universal qPCR Master Mix (NEB, Hertfordshire, UK). The PCR mixtures were prepared in a total volume of 10 µL and contained 1 µL of template cDNA, 5 µL of Luna® Universal qPCR Master Mix (2×), and forward and reverse primer in a final concentration of 0.3 µM. qPCR temperature profile consisted of a 150-sec denaturation step at 95 °C followed by 45 cycles consisting of a 20-sec denaturation step at 95 °C and a 40-sec annealing extension step at 60 °C.
4.5. Phytohormone Levels Analysis
Phytohormone levels were measured 24 h after solution treatment (5 mM BABA, 5 mM BABA with 5 mM Asp, 5 mM BABA with 5 mM Gln, 5 mM Asp, 5 mM Gln, pH = 7). A total of 50 mg of leaf powder with 20 ng of internal standard (o-anisic acid) were extracted twice into 1 mL of cold 10% methanol with 3 min of sonication and 30 min of shaking at 4 °C and 700 rpm. Samples were centrifuged for 10 min at 4 °C and 15,000× g, the supernatants were applied to SPE columns (HLB, Merck, Darmstadt, Germany) washed with 2 mL of water and 1 mL of water, and samples were applied and washed with 1 mL of 10% MeOH and eluted with 2 mL of methanol. The samples were dried, thoroughly resuspended in 40 µL of 0.04% formic acid and 15% acetonitrile, and filtered through 0.22 µm filters. Phytohormone levels were measured by LC–MS/MS (6545, Agilent, Santa Clara, CA, USA). Phytohormones were separated by reverse-phase column (Poroshell 120 SB-C18 2.1 × 100 mm2, 2.7 µm, Agilent, Santa Clara, CA, USA) and gradient solution technique. A total of 0.04 % formic acid (A) and acetonitrile (B) were used as mobile phases with the following gradient conditions: 15% B from 0 to 5 min, 15 to 45% B from 5 to 15 min, 45 to 47% B from 15 to 22 min and finally, 100% from 22 to 32 min, at a flow rate of 0.4 mL/min and injection volume of 10 µL. Conditions of electrospray ion source were as follows: acquisition mode 100–1700 m/z, gas flow 8 L/min, gas temperature 240 °C, capillary voltage 3000 V, fragmentor voltage 150 V. Positive ion polarity was used for the detection of methyl jasmonate and jasmonoyl–isoleucine and negative for the o-anisic, jasmonic and salicylic acid. The analytes were qualified by comparison of retention times and mass spectra with standards and quantified using molecular ion peak areas. MassHunter Quantitative Analysis software was used for drawing calibration curves.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11212908/s1, Table S1: Sequences of used primers.
Author Contributions
Conceptualisation, A.J. and J.L.; methodology, K.D., J.L. and M.Z.; formal analysis, A.J., K.D. and M.Z.; investigation, A.J.; writing—original draft preparation, A.J., K.D. and M.Z.; writing—review and editing, J.L.; supervision, M.Z.; project administration, J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Institutional Research Fund of Masaryk University, MUNI/A/1492/2021.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vijayakumari, K.; Jisha, K.C.; Puthur, J.T. GABA/BABA Priming: A Means for Enhancing Abiotic Stress Tolerance Potential of Plants with Less Energy Investments on Defence Cache. Acta Physiol. Plant. 2016, 38, 230. [Google Scholar] [CrossRef]
- Baccelli, I.; Mauch-Mani, B. Beta-Aminobutyric Acid Priming of Plant Defense: The Role of ABA and Other Hormones. Plant Mol. Biol. 2016, 91, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Vaknin, M.; Mauch-Mani, B. BABA-Induced Resistance: Milestones along a 55-Year Journey. Phytoparasitica 2016, 44, 513–538. [Google Scholar] [CrossRef]
- Thevenet, D.; Pastor, V.; Baccelli, I.; Balmer, A.; Vallat, A.; Neier, R.; Glauser, G.; Mauch-Mani, B. The Priming Molecule β-Aminobutyric Acid Is Naturally Present in Plants and Is Induced by Stress. New Phytol. 2017, 213, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Baccelli, I.; Glauser, G.; Mauch-Mani, B. The Accumulation of β-Aminobutyric Acid Is Controlled by the Plant’s Immune System. Planta 2017, 246, 791–796. [Google Scholar] [CrossRef]
- Schwarzenbacher, R.E.; Wardell, G.; Stassen, J.; Guest, E.; Zhang, P.; Luna, E.; Ton, J. The IBI1 Receptor of β-Aminobutyric Acid Interacts with VOZ Transcription Factors to Regulate Abscisic Acid Signaling and Callose-Associated Defense. Mol. Plant 2020, 13, 1455–1469. [Google Scholar] [CrossRef]
- Wu, C.-C.; Singh, P.; Chen, M.-C.; Zimmerli, L. L-Glutamine Inhibits Beta-Aminobutyric Acid-Induced Stress Resistance and Priming in Arabidopsis. J. Exp. Bot. 2010, 61, 995–1002. [Google Scholar] [CrossRef]
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 14 September 2022).
- Kimura, S.; Sinha, N. Tomato (Solanum lycopersicum): A Model Fruit-Bearing Crop. Cold Spring Harb. Protoc. 2008, 2008, pdb.emo105. [Google Scholar] [CrossRef]
- Bengtsson, T.; Weighill, D.; Proux-Wéra, E.; Levander, F.; Resjö, S.; Burra, D.D.; Moushib, L.I.; Hedley, P.E.; Liljeroth, E.; Jacobson, D.; et al. Proteomics and Transcriptomics of the BABA-Induced Resistance Response in Potato Using a Novel Functional Annotation Approach. BMC Genom. 2014, 15, 315. [Google Scholar] [CrossRef]
- Satková, P.; Starý, T.; Plešková, V.; Zapletalová, M.; Kašparovský, T.; Činčalová-Kubienová, L.; Luhová, L.; Mieslerová, B.; Mikulík, J.; Lochman, J.; et al. Diverse Responses of Wild and Cultivated Tomato to BABA, Oligandrin and Oidium Neolycopersici Infection. Ann. Bot. 2017, 119, 829–840. [Google Scholar] [CrossRef][Green Version]
- Zimmerli, L.; Jakab, G.; Metraux, J.P.; Mauch-Mani, B. Potentiation of Pathogen-Specific Defense Mechanisms in Arabidopsis by Beta -Aminobutyric Acid. Proc. Natl. Acad. Sci. USA 2000, 97, 12920–12925. [Google Scholar] [CrossRef] [PubMed]
- Slaughter, A.; Daniel, X.; Flors, V.; Luna, E.; Hohn, B.; Mauch-Mani, B. Descendants of Primed Arabidopsis Plants Exhibit Resistance to Biotic Stress. Plant Physiol. 2012, 158, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; van Hulten, M.; Zhang, Y.; Berkowitz, O.; López, A.; Pétriacq, P.; Sellwood, M.A.; Chen, B.; Burrell, M.; van de Meene, A.; et al. Plant Perception of β-Aminobutyric Acid Is Mediated by an Aspartyl-TRNA Synthetase. Nat. Chem. Biol. 2014, 10, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Vega-Mas, I.; Rossi, M.T.; Gupta, K.J.; González-Murua, C.; Ratcliffe, R.G.; Estavillo, J.M.; González-Moro, M.B. Tomato Roots Exhibit in Vivo Glutamate Dehydrogenase Aminating Capacity in Response to Excess Ammonium Supply. J. Plant Physiol. 2019, 239, 83–91. [Google Scholar] [CrossRef]
- Vega-Mas, I.; Cukier, C.; Coleto, I.; González-Murua, C.; Limami, A.M.; González-Moro, M.B.; Marino, D. Isotopic Labelling Reveals the Efficient Adaptation of Wheat Root TCA Cycle Flux Modes to Match Carbon Demand under Ammonium Nutrition. Sci. Rep. 2019, 9, 8925. [Google Scholar] [CrossRef]
- Ton, J.; Mauch-Mani, B. β-Amino-Butyric Acid-Induced Resistance against Necrotrophic Pathogens Is Based on ABA-Dependent Priming for Callose. Plant J. 2004, 38, 119–130. [Google Scholar] [CrossRef]
- Zimmerli, L.; Métraux, J.-P.; Mauch-Mani, B. β-Aminobutyric Acid-Induced Protection of Arabidopsis against the Necrotrophic Fungus Botrytis Cinerea. Plant Physiol. 2001, 126, 517–523. [Google Scholar] [CrossRef]
- Hamiduzzaman, M.M.; Jakab, G.; Barnavon, L.; Neuhaus, J.-M.; Mauch-Mani, B. Beta-Aminobutyric Acid-Induced Resistance against Downy Mildew in Grapevine Acts through the Potentiation of Callose Formation and Jasmonic Acid Signaling. Mol. Plant-Microbe Interact. 2005, 18, 819–829. [Google Scholar] [CrossRef]
- Ton, J.; Jakab, G.; Toquin, V.; Flors, V.; Iavicoli, A.; Maeder, M.N.; Métraux, J.-P.; Mauch-Mani, B. Dissecting the Beta-Aminobutyric Acid-Induced Priming Phenomenon in Arabidopsis. Plant Cell 2005, 17, 987–999. [Google Scholar] [CrossRef]
- Skottke, K.R.; Yoon, G.M.; Kieber, J.J.; DeLong, A. Protein Phosphatase 2A Controls Ethylene Biosynthesis by Differentially Regulating the Turnover of ACC Synthase Isoforms. PLoS Genet. 2011, 7, e1001370. [Google Scholar] [CrossRef]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense Priming: An Adaptive Part of Induced Resistance. Annu. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef] [PubMed]
- Piękna-Grochala, J.; Kępczyńska, E. Induction of Resistance against Pathogens by β-Aminobutyric Acid. Acta Physiol. Plant. 2013, 35, 1735–1748. [Google Scholar] [CrossRef]
- Scott, J.W.; Harbaugh, B.K. Micro-Tom: A Miniature Dwarf Tomato; Circular (University of Florida, Agricultural Expriment Station); Agricultural Experiment Station, Institute of Food and Agricultural Sciences, University of Florida: Gainesville, FL, USA, 1989. [Google Scholar]
- Xin, X.-F.; He, S.Y. Pseudomonas Syringae Pv. Tomato DC3000: A Model Pathogen for Probing Disease Susceptibility and Hormone Signaling in Plants. Annu. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar] [CrossRef] [PubMed]
- Martí, E.; Gisbert, C.; Bishop, G.J.; Dixon, M.S.; García-Martínez, J.L. Genetic and Physiological Characterization of Tomato cv. Micro-Tom. J. Exp. Bot. 2006, 57, 2037–2047. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Shimizu, A.; Arie, T.; Rosmalawati, S.; Fukushima, S.; Kikuchi, M.; Hikichi, Y.; Kanda, A.; Takahashi, A.; Kiba, A.; et al. Catalog of Micro-Tom Tomato Responses to Common Fungal, Bacterial, and Viral Pathogens. J. Gen. Plant Pathol. 2005, 71, 8–22. [Google Scholar] [CrossRef]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling Mechanisms in Pattern-Triggered Immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.M.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.-A.; Pieterse, C.M.J.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting Ready for Battle. MPMI 2006, 19, 1062–1071. [Google Scholar] [CrossRef]
- Dutton, C.; Hõrak, H.; Hepworth, C.; Mitchell, A.; Ton, J.; Hunt, L.; Gray, J.E. Bacterial Infection Systemically Suppresses Stomatal Density. Plant Cell Environ. 2019, 42, 2411–2421. [Google Scholar] [CrossRef]
- Kadotani, N.; Akagi, A.; Takatsuji, H.; Miwa, T.; Igarashi, D. Exogenous Proteinogenic Amino Acids Induce Systemic Resistance in Rice. BMC Plant Biol. 2016, 16, 60. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological Implications of Arginine Metabolism in Plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Maximova, S.N.; Jensen, P.J.; Lehman, B.L.; Ngugi, H.K.; McNellis, T.W. FIBRILLIN4 Is Required for Plastoglobule Development and Stress Resistance in Apple and Arabidopsis. Plant Physiol. 2010, 154, 1281–1293. [Google Scholar] [CrossRef] [PubMed]
- Heldt, H.-W.; Piechulla, B. Plant Biochemistry, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2011; ISBN 978-0-12-384986-1. [Google Scholar]
- Hartmann, M.; Zeier, J. L-Lysine Metabolism to N-Hydroxypipecolic Acid: An Integral Immune-Activating Pathway in Plants. Plant J. 2018, 96, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Jakab, G.; Cottier, V.; Toquin, V.; Rigoli, G.; Zimmerli, L.; Métraux, J.-P.; Mauch-Mani, B. β-Aminobutyric Acid-Induced Resistance in Plants. Eur. J. Plant Pathol. 2001, 107, 29–37. [Google Scholar] [CrossRef]
- Wang, H.; Liu, G.; Li, C.; Powell, A.L.T.; Reid, M.S.; Zhang, Z.; Jiang, C.-Z. Defence Responses Regulated by Jasmonate and Delayed Senescence Caused by Ethylene Receptor Mutation Contribute to the Tolerance of Petunia to Botrytis Cinerea. Mol. Plant Pathol. 2013, 14, 453–469. [Google Scholar] [CrossRef]
- Boller, T. Ethylene in Pathogenesis and Disease Resistance. In The Plant Hormone Ethylene; CRC Press: Boca Raton, FL, USA, 1991; ISBN 978-1-351-07576-3. [Google Scholar]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for Enhanced Defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef]
- Worrall, D.; Holroyd, G.H.; Moore, J.P.; Glowacz, M.; Croft, P.; Taylor, J.E.; Paul, N.D.; Roberts, M.R. Treating Seeds with Activators of Plant Defence Generates Long-Lasting Priming of Resistance to Pests and Pathogens. New Phytol. 2012, 193, 770–778. [Google Scholar] [CrossRef]
- Zimmerli, L.; Hou, B.-H.; Tsai, C.-H.; Jakab, G.; Mauch-Mani, B.; Somerville, S. The Xenobiotic Beta-Aminobutyric Acid Enhances Arabidopsis Thermotolerance. Plant J. 2008, 53, 144–156. [Google Scholar] [CrossRef]
- Yan, Z.; Reddy, M.S.; Ryu, C.-M.; McInroy, J.A.; Wilson, M.; Kloepper, J.W. Induced Systemic Protection against Tomato Late Blight Elicited by Plant Growth-Promoting Rhizobacteria. Phytopathology 2002, 92, 1329–1333. [Google Scholar] [CrossRef]
- Li, X.; Sun, Z.; Shao, S.; Zhang, S.; Ahammed, G.J.; Zhang, G.; Jiang, Y.; Zhou, J.; Xia, X.; Zhou, Y.; et al. Tomato–Pseudomonas Syringae Interactions under Elevated CO2 Concentration: The Role of Stomata. J. Exp. Bot. 2015, 66, 307–316. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Gómez-Alonso, S.; Hermosín-Gutiérrez, I.; García-Romero, E. Simultaneous HPLC Analysis of Biogenic Amines, Amino Acids, and Ammonium Ion as Aminoenone Derivatives in Wine and Beer Samples. J. Agric. Food Chem. 2007, 55, 608–613. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).