Uncovering the Genetic of Cadmium Accumulation in the Rice 3K Panel
Abstract
1. Introduction
2. Results
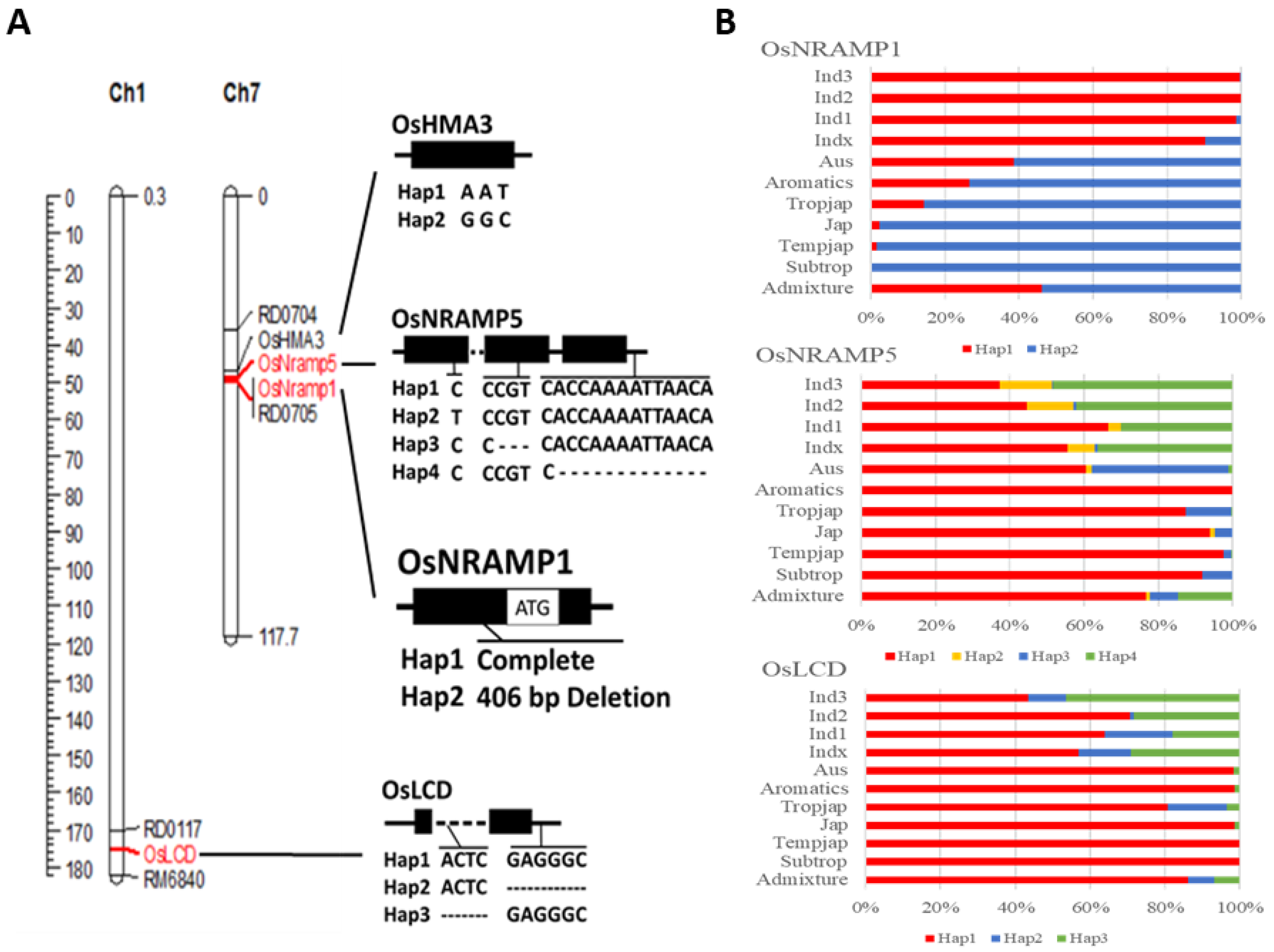
2.1. Genotype Organization
2.2. Correlation Analysis of Cd-Mobile Types and Cadmium Accumulation
2.3. Specific Marker Design and Genotype Analysis
3. Discussion
4. Materials and Methods
4.1. Gene Data Collection
4.2. Plant Materials
4.3. Hydroponic and Pot Experiments
4.4. Plant Collection, Digestion, and Analysis
4.5. Functional Marker Design and Genotyping
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krueger, W.S.; Wade, T.J. Elevated Blood Lead and Cadmium Levels Associated with Chronic Infections among Non-Smokers in a Cross-Sectional Analysis of NHANES Data. Environ. Health 2016, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.; Arumugam, P.; Lee, J.; Song, J.M. Impact of Environmental Pollutant Cadmium on the Establishment of a Cancer Stem Cell Population in Breast and Hepatic Cancer. ACS Omega 2017, 2, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, A. Cadmium Toxicity: Effects on Human Reproduction and Fertility. Rev. Environ. Health 2019, 34, 327–338. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Khalili, N.; Razi, S.; Keshavarz-Fathi, M.; Khalili, N.; Rezaei, N. Effects of Lead and Cadmium on the Immune System and Cancer Progression. J. Environ. Health Sci. Eng. 2020, 18, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Authority (EFSA), E.F.S. Cadmium in Food—Scientific Opinion of the Panel on Contaminants in the Food Chain. EFSA J. 2009, 7, 980. [Google Scholar] [CrossRef]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The Effects of Cadmium Toxicity. Int. J. Environ. Res. Public. Health 2020, 17, 3782. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Ishimaru, Y.; Igura, M.; Kuramata, M.; Abe, T.; Senoura, T.; Hase, Y.; Arao, T.; Nishizawa, N.K.; Nakanishi, H. Ion-Beam Irradiation, Gene Identification, and Marker-Assisted Breeding in the Development of Low-Cadmium Rice. Proc. Natl. Acad. Sci. USA 2012, 109, 19166–19171. [Google Scholar] [CrossRef]
- Honma, T.; Ohba, H.; Kaneko-Kadokura, A.; Makino, T.; Nakamura, K.; Katou, H. Optimal Soil Eh, PH, and Water Management for Simultaneously Minimizing Arsenic and Cadmium Concentrations in Rice Grains. Environ. Sci. Technol. 2016, 50, 4178–4185. [Google Scholar] [CrossRef]
- Zhao, F.-J.; Wang, P. Arsenic and Cadmium Accumulation in Rice and Mitigation Strategies. Plant Soil 2020, 446, 1–21. [Google Scholar] [CrossRef]
- Uraguchi, S.; Fujiwara, T. Cadmium Transport and Tolerance in Rice: Perspectives for Reducing Grain Cadmium Accumulation. Rice 2012, 5, 5. [Google Scholar] [CrossRef]
- Uraguchi, S.; Fujiwara, T. Rice Breaks Ground for Cadmium-Free Cereals. Curr. Opin. Plant Biol. 2013, 16, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Luo, N.; Li, Y.W.; Cai, Q.Y.; Li, H.Y.; Mo, C.H.; Wong, M.H. Cadmium in Rice: Transport Mechanisms, Influencing Factors, and Minimizing Measures. Environ. Pollut. 2017, 224, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Ishimaru, Y.; Senoura, T.; Shimo, H.; Ishikawa, S.; Arao, T.; Nakanishi, H.; Nishizawa, N.K. The OsNRAMP1 Iron Transporter Is Involved in Cd Accumulation in Rice. J. Exp. Bot. 2011, 62, 4843–4850. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Yamaji, N.; Yokosho, K.; Ma, J.F. Nramp5 Is a Major Transporter Responsible for Manganese and Cadmium Uptake in Rice. Plant Cell 2012, 24, 2155–2167. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.-D.; Huang, S.; Konishi, N.; Wang, P.; Chen, J.; Huang, X.-Y.; Ma, J.F.; Zhao, F.-J. Overexpression of the Manganese/Cadmium Transporter OsNRAMP5 Reduces Cadmium Accumulation in Rice Grain. J. Exp. Bot. 2020, 71, 5705–5715. [Google Scholar] [CrossRef]
- Ueno, D.; Yamaji, N.; Kono, I.; Huang, C.F.; Ando, T.; Yano, M.; Ma, J.F. Gene Limiting Cadmium Accumulation in Rice. Proc. Natl. Acad. Sci. USA 2010, 107, 16500–16505. [Google Scholar] [CrossRef]
- Miyadate, H.; Adachi, S.; Hiraizumi, A.; Tezuka, K.; Nakazawa, N.; Kawamoto, T.; Katou, K.; Kodama, I.; Sakurai, K.; Takahashi, H.; et al. OsHMA3, a P1B-Type of ATPase Affects Root-to-Shoot Cadmium Translocation in Rice by Mediating Efflux into Vacuoles. New Phytol. 2011, 189, 190–199. [Google Scholar] [CrossRef]
- Takahashi, R.; Ishimaru, Y.; Shimo, H.; Ogo, Y.; Senoura, T.; Nishizawa, N.K.; Nakanishi, H. The OsHMA2 Transporter Is Involved in Root-to-Shoot Translocation of Zn and Cd in Rice. Plant Cell Environ. 2012, 35, 1948–1957. [Google Scholar] [CrossRef]
- Yamaji, N.; Xia, J.; Mitani-Ueno, N.; Yokosho, K.; Feng Ma, J. Preferential Delivery of Zinc to Developing Tissues in Rice Is Mediated by P-Type Heavy Metal ATPase OsHMA2. Plant Physiol. 2013, 162, 927–939. [Google Scholar] [CrossRef]
- Uraguchi, S.; Kamiya, T.; Sakamoto, T.; Kasai, K.; Sato, Y.; Nagamura, Y.; Yoshida, A.; Kyozuka, J.; Ishikawa, S.; Fujiwara, T. Low-Affinity Cation Transporter (OsLCT1) Regulates Cadmium Transport into Rice Grains. Proc. Natl. Acad. Sci. USA 2011, 108, 20959–20964. [Google Scholar] [CrossRef]
- Shimo, H.; Ishimaru, Y.; An, G.; Yamakawa, T.; Nakanishi, H.; Nishizawa, N.K. Low Cadmium (LCD), a Novel Gene Related to Cadmium Tolerance and Accumulation in Rice. J. Exp. Bot. 2011, 62, 5727–5734. [Google Scholar] [CrossRef] [PubMed]
- Norton, G.J.; Deacon, C.M.; Xiong, L.; Huang, S.; Meharg, A.A.; Price, A.H. Genetic Mapping of the Rice Ionome in Leaves and Grain: Identification of QTLs for 17 Elements Including Arsenic, Cadmium, Iron and Selenium. Plant Soil 2010, 329, 139–153. [Google Scholar] [CrossRef]
- Ishikawa, S.; Abe, T.; Kuramata, M.; Yamaguchi, M.; Ando, T.; Yamamoto, T.; Yano, M. A Major Quantitative Trait Locus for Increasing Cadmium-Specific Concentration in Rice Grain Is Located on the Short Arm of Chromosome 7. J. Exp. Bot. 2010, 61, 923–934. [Google Scholar] [CrossRef]
- Tezuka, K.; Miyadate, H.; Katou, K.; Kodama, I.; Matsumoto, S.; Kawamoto, T.; Masaki, S.; Satoh, H.; Yamaguchi, M.; Sakurai, K.; et al. A Single Recessive Gene Controls Cadmium Translocation in the Cadmium Hyperaccumulating Rice Cultivar Cho-Ko-Koku. Theor. Appl. Genet. 2010, 120, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Taguchi-Shiobara, F.; Kojima, Y.; Ebitani, T.; Kuramata, M.; Yamamoto, T.; Yano, M.; Ishikawa, S. Detection of a QTL for Accumulating Cd in Rice That Enables Efficient Cd Phytoextraction from Soil. Breed. Sci. 2011, 61, 43–51. [Google Scholar] [CrossRef]
- Ueno, D.; Koyama, E.; Kono, I.; Ando, T.; Yano, M.; Ma, J.F. Identification of a Novel Major Quantitative Trait Locus Controlling Distribution of Cd Between Roots and Shoots in Rice. Plant Cell Physiol. 2009, 50, 2223–2233. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Liang, S.; Qiao, K.; Wang, F.; Zhang, Y.; Chai, T. Co-Expression of Multiple Heavy Metal Transporters Changes the Translocation, Accumulation, and Potential Oxidative Stress of Cd and Zn in Rice (Oryza Sativa). J. Hazard. Mater. 2019, 380, 120853. [Google Scholar] [CrossRef]
- Tang, L.; Mao, B.; Li, Y.; Lv, Q.; Zhang, L.; Chen, C.; He, H.; Wang, W.; Zeng, X.; Shao, Y.; et al. Knockout of OsNramp5 Using the CRISPR/Cas9 System Produces Low Cd-Accumulating Indica Rice without Compromising Yield. Sci. Rep. 2017, 7, 14438. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, Y.; Huang, C. Reduction in Cadmium Accumulation in Japonica Rice Grains by CRISPR/Cas9-Mediated Editing of OsNRAMP5. J. Integr. Agric. 2019, 18, 688–697. [Google Scholar] [CrossRef]
- Wang, K.; Yan, T.; Xu, S.; Yan, X.; Zhou, Q.; Zhao, X.; Li, Y.; Wu, Z.; Qin, P.; Fu, C.; et al. Validating a Segment on Chromosome 7 of Japonica for Establishing Low-Cadmium Accumulating Indica Rice Variety. Sci. Rep. 2021, 11, 6053. [Google Scholar] [CrossRef]
- Wu, P.-J.; Lin, Y.-W.; Li, C.-P.; Syu, C.-H.; Jwo, W.-S.; Yen, H.-M.; Guo, H.-Y.; Lai, M.-H.; Wu, D.-H. A Novel Indica Rice Breeding Line Selection with Low Cadmium Accumulation Level by Functional Markers Assisted Backcross Strategy. J. Taiwan Agric. Res. 2017, 66, 248–260. [Google Scholar] [CrossRef]
- Yan, J.; Wang, P.; Wang, P.; Yang, M.; Lian, X.; Tang, Z.; Huang, C.-F.; Salt, D.E.; Zhao, F.J. A Loss-of-Function Allele of OsHMA3 Associated with High Cadmium Accumulation in Shoots and Grain of Japonica Rice Cultivars. Plant Cell Environ. 2016, 39, 1941–1954. [Google Scholar] [CrossRef]
- Alexandrov, N.; Tai, S.; Wang, W.; Mansueto, L.; Palis, K.; Fuentes, R.R.; Ulat, V.J.; Chebotarov, D.; Zhang, G.; Li, Z.; et al. SNP-Seek Database of SNPs Derived from 3000 Rice Genomes. Nucleic Acids Res. 2015, 43, D1023–D1027. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.A.; Shin, R.; Eide, D.J.; Schachtman, D.P. Differential Metal Selectivity and Gene Expression of Two Zinc Transporters from Rice. Plant Physiol. 2003, 133, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Takahashi, R.; Bashir, K.; Shimo, H.; Senoura, T.; Sugimoto, K.; Ono, K.; Yano, M.; Ishikawa, S.; Arao, T.; et al. Characterizing the Role of Rice NRAMP5 in Manganese, Iron and Cadmium Transport. Sci. Rep. 2012, 2, 286. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, Y.; Zhang, L.; Hu, J.; Zhang, X.; Lu, K.; Dong, H.; Wang, D.; Zhao, F.-J.; Huang, C.-F.; et al. OsNRAMP5 Contributes to Manganese Translocation and Distribution in Rice Shoots. J. Exp. Bot. 2014, 65, 4849–4861. [Google Scholar] [CrossRef]
- Takahashi, R.; Ishimaru, Y.; Shimo, H.; Bashir, K.; Senoura, T.; Sugimoto, K.; Ono, K.; Suzui, N.; Kawachi, N.; Ishii, S.; et al. From Laboratory to Field: OsNRAMP5-Knockdown Rice Is a Promising Candidate for Cd Phytoremediation in Paddy Fields. PLoS ONE 2014, 9, e98816. [Google Scholar] [CrossRef]
- Songmei, L.; Jie, J.; Yang, L.; Jun, M.; Shouling, X.; Yuanyuan, T.; Youfa, L.; Qingyao, S.; Jianzhong, H. Characterization and Evaluation of OsLCT1 and OsNramp5 Mutants Generated Through CRISPR/Cas9-Mediated Mutagenesis for Breeding Low Cd Rice. Rice Sci. 2019, 26, 88–97. [Google Scholar] [CrossRef]
- Sun, L.; Xu, X.; Jiang, Y.; Zhu, Q.; Yang, F.; Zhou, J.; Yang, Y.; Huang, Z.; Li, A.; Chen, L.; et al. Genetic Diversity, Rather than Cultivar Type, Determines Relative Grain Cd Accumulation in Hybrid Rice. Front. Plant Sci. 2016, 7, 1407. [Google Scholar] [CrossRef]
- Arao, T.; Ae, N. Genotypic Variations in Cadmium Levels of Rice Grain. Soil Sci. Plant Nutr. 2003, 49, 473–479. [Google Scholar] [CrossRef]
- Wang, T.; Li, Y.; Fu, Y.; Xie, H.; Song, S.; Qiu, M.; Wen, J.; Chen, M.; Chen, G.; Tian, Y.; et al. Mutation at Different Sites of Metal Transporter Gene OsNramp5 Affects Cd Accumulation and Related Agronomic Traits in Rice (Oryza Sativa L.). Front. Plant Sci. 2019, 10, 1081. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Ae, N.; Yano, M. Chromosomal Regions with Quantitative Trait Loci Controlling Cadmium Concentration in Brown Rice (Oryza Sativa). New Phytol. 2005, 168, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, T.; Shindoh, K.; Hirotsu, N.; Ishimaru, K. Evidence for Separate Translocation Pathways in Determining Cadmium Accumulation in Grain and Aerial Plant Parts in Rice. BMC Plant Biol. 2009, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Sheng, Z.; Li, Q.; Chen, W.; Wei, X.; Xie, L.; Jiao, G.; Shao, G.; Wang, J.; Tang, S.; et al. Identification of QTLs Associated with Cadmium Concentration in Rice Grains. J. Integr. Agric. 2018, 17, 1563–1573. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, W.; Zhang, S.; Yang, T.; Liu, Q.; Dong, J.; Fu, H.; Mao, X.; Liu, B. Genome-Wide Association Study and Candidate Gene Analysis of Rice Cadmium Accumulation in Grain in a Diverse Rice Collection. Rice 2018, 11, 61. [Google Scholar] [CrossRef]
- Hao, X.; Wu, C.; Wang, R.; Tian, L.; Song, T.; Tan, H.; Peng, Y.; Zeng, M.; Chen, L.; Liang, M.; et al. Association between Sequence Variants in Cadmium-Related Genes and the Cadmium Accumulation Trait in Thermo-Sensitive Genic Male Sterile Rice. Breed. Sci. 2019, 69, 18191. [Google Scholar] [CrossRef]
- Nakanishi, H.; Ogawa, I.; Ishimaru, Y.; Mori, S.; Nishizawa, N.K. Iron Deficiency Enhances Cadmium Uptake and Translocation Mediated by the Fe 2+ Transporters OsIRT1 and OsIRT2 in Rice. Soil Sci. Plant Nutr. 2006, 52, 464–469. [Google Scholar] [CrossRef]
- Satoh-Nagasawa, N.; Mori, M.; Nakazawa, N.; Kawamoto, T.; Nagato, Y.; Sakurai, K.; Takahashi, H.; Watanabe, A.; Akagi, H. Mutations in Rice (Oryza Sativa) Heavy Metal ATPase 2 (OsHMA2) Restrict the Translocation of Zinc and Cadmium. Plant Cell Physiol. 2012, 53, 213–224. [Google Scholar] [CrossRef]
- Oda, K.; Otani, M.; Uraguchi, S.; Akihiro, T.; Fujiwara, T. Rice ABCG43 Is Cd Inducible and Confers Cd Tolerance on Yeast. Biosci. Biotechnol. Biochem. 2011, 75, 1211–1213. [Google Scholar] [CrossRef]
- Mclean, E.O. Soil PH and Lime Requirement. In Methods of Soil Analysis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1983; pp. 199–224. ISBN 978-0-89118-977-0. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic Matter. In Methods of Soil Analysis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1983; pp. 539–579. ISBN 978-0-89118-977-0. [Google Scholar]
- Chapman, H.D. Cation-Exchange Capacity. In Methods of Soil Analysis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1965; pp. 891–901. ISBN 978-0-89118-204-7. [Google Scholar]
- Nelson, J.L.; Boawn, L.C.; Viets, F.G.J. Method for Assessing Zinc Status of Soils Using Acid-Extractable Zinc and “Titratable Alkalinity” Values. Soil Sci. 1959, 88, 275–283. [Google Scholar] [CrossRef]
- Lin, Y.-W.; Liu, T.-S.; Guo, H.-Y.; Chiang, C.-M.; Tang, H.-J.; Chen, H.-T.; Chen, J.-H. Relationships between Cd Concentrations in Different Vegetables and Those in Arable Soils, and Food Safety Evaluation of Vegetables in Taiwan. Soil Sci. Plant Nutr. 2015, 61, 983–998. [Google Scholar] [CrossRef]
- Meharg, A.A.; Rahman, M.M. Arsenic Contamination of Bangladesh Paddy Field Soils: Implications for Rice Contribution to Arsenic Consumption. Environ. Sci. Technol. 2003, 37, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999, 41, 95–98. [Google Scholar]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- De Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. R Package Version 2014, 1, 1–4. [Google Scholar]
- Allaire, J. RStudio: Integrated Development Environment for R. Boston MA 2012, 770, 165–171. [Google Scholar]
- Wickham, H. Ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
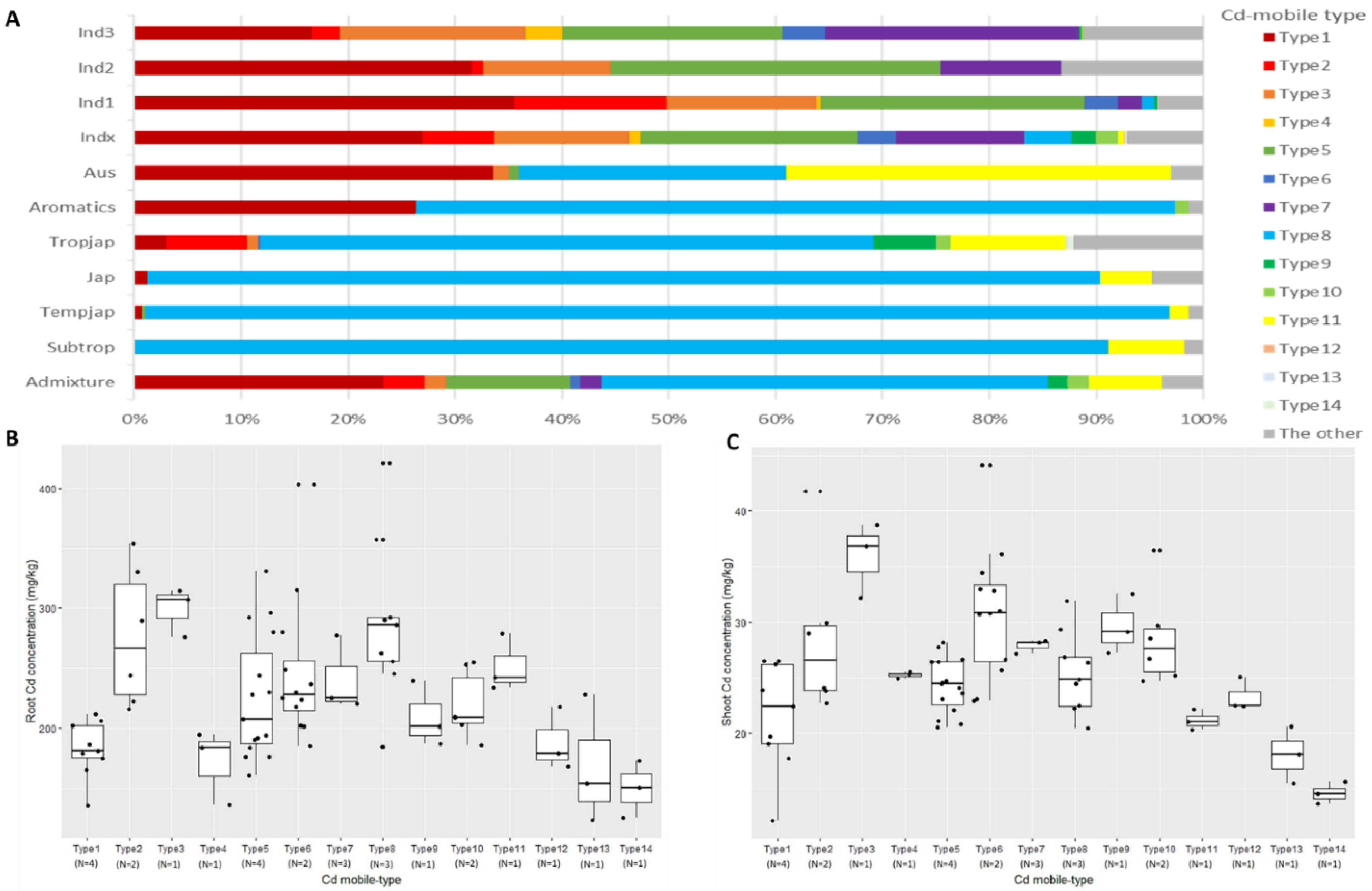

| Cd Mobile-Type | NR1 | NR5 | LCD | Cd Concentration (mg kg−1) | Variety | ||
|---|---|---|---|---|---|---|---|
| Root 1 | Shoot | Shoot/Root Ratio | |||||
| Type1 | Hap1 | Hap1 | Hap1 | 195.19 bc | 22.64 cd | 11.60% abc | Habataki, Taichung Sen 10, Kurulutudu, Maranhao Branco |
| Type2 | Hap1 | Hap1 | Hap2 | 275.91 a | 28.55 b | 10.35% bc | Taichung Sen 17, Chang Le San Shu Zao |
| Type3 | Hap1 | Hap1 | Hap3 | 299.13 a | 35.91 a | 12.01% abc | Hsinchu Ai Chio Chieng |
| Type4 | Hap1 | Hap2 | Hap2 | 171.31 de | 25.26 cd | 14.75% a | Hsi’-T’ao Yuan Ching Yu |
| Type5 | Hap1 | Hap4 | Hap1 | 223.47 cde | 23.91 ef | 10.70% abc | Taichung Sen Waxy 2, Ncs840, Psbrc50, IR 19661-364-1-2-3 |
| Type6 | Hap1 | Hap4 | Hap2 | 278.80 a | 35.19 a | 12.62% ab | B 6136-3-TB-0-1-5, B 6136 E-3-TB-0-1-5 |
| Type7 | Hap1 | Hap4 | Hap3 | 224.41 bc | 27.08 bc | 12.07% abc | Ncs771A, Arc14868, IR 80310-12-B-1-3-B |
| Type8 | Hap2 | Hap1 | Hap1 | 288.23 a | 25.42 cd | 8.82% c | Nipponbare, Tainan 11, Taitung 30 |
| Type9 | Hap2 | Hap1 | Hap2 | 209.07 bcde | 29.63 b | 14.18% ab | Jin Jun Dao |
| Type10 | Hap2 | Hap1 | Hap3 | 219.09 bcd | 28.54 b | 13.03% ab | Lobang (white), Balibud |
| Type11 | Hap2 | Hap3 | Hap1 | 251.53 ab | 21.16 ef | 8.41% c | Uprh166 |
| Type12 | Hap2 | Hap2 | Hap3 | 188.28 cde | 23.32 de | 12.39% abc | Ai Jiao Zi |
| Type13 | Hap2 | Hap3 | Hap2 | 168.37 de | 18.09 fg | 10.75% abc | Landeo |
| Type14 | Hap2 | Hap3 | Hap3 | 149.88 e | 14.64 g | 9.77% bc | Asu |
| Cd in Soil (mg kg−1) | Cd-Mobile Type | NR1 | NR5 | LCD | Cd Concentration (mg kg−1) | Cd Concentration Ratio | Variety | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Root 1 | Shoot | Brown Rice | Shoot/Root | Brown Rice/Root | Brown Rice/Shoot | ||||||||||||
| 3.76 | Type8 | Hap2 | Hap1 | Hap1 | 13.29 | b | 1.24 | ab | 0.04 | b | 9.33% | a | 0.30% | b | 3.23% | a | TN11 |
| Type8 | Hap2 | Hap1 | Hap1 | 7.01 | bc | 0.68 | c | 0.07 | b | 9.70% | a | 1.00% | a | 10.29% | a | TCS10-OsNRAMP1 | |
| Type11 | Hap2 | Hap3 | Hap1 | 22.54 | a | 1.33 | a | 0.16 | a | 5.90% | a | 0.71% | ab | 12.03% | a | Uprh166 | |
| Type13 | Hap2 | Hap3 | Hap2 | 8.96 | bc | 0.88 | bc | 0.06 | b | 9.82% | a | 0.67% | ab | 6.82% | a | Landeo | |
| Type14 | Hap2 | Hap3 | Hap3 | 4.98 | c | 0.68 | c | 0.03 | b | 13.65% | a | 0.60% | ab | 4.41% | a | Asu | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syu, C.-H.; Nieh, T.-I.; Hsieh, M.-T.; Lo, Y.-C.; Du, P.-R.; Lin, Y.-W.; Wu, D.-H. Uncovering the Genetic of Cadmium Accumulation in the Rice 3K Panel. Plants 2022, 11, 2813. https://doi.org/10.3390/plants11212813
Syu C-H, Nieh T-I, Hsieh M-T, Lo Y-C, Du P-R, Lin Y-W, Wu D-H. Uncovering the Genetic of Cadmium Accumulation in the Rice 3K Panel. Plants. 2022; 11(21):2813. https://doi.org/10.3390/plants11212813
Chicago/Turabian StyleSyu, Chien-Hui, Ting-Iun Nieh, Meng-Ting Hsieh, Yu-Ching Lo, Pei-Rong Du, Yu-Wen Lin, and Dong-Hong Wu. 2022. "Uncovering the Genetic of Cadmium Accumulation in the Rice 3K Panel" Plants 11, no. 21: 2813. https://doi.org/10.3390/plants11212813
APA StyleSyu, C.-H., Nieh, T.-I., Hsieh, M.-T., Lo, Y.-C., Du, P.-R., Lin, Y.-W., & Wu, D.-H. (2022). Uncovering the Genetic of Cadmium Accumulation in the Rice 3K Panel. Plants, 11(21), 2813. https://doi.org/10.3390/plants11212813







