Genetic Diversity and Population Structure of Normal Maize Germplasm Collected in South Sudan Revealed by SSR Markers
Abstract
:1. Introduction
2. Results
2.1. Polymorphism of SSR Loci for Maize Landrace Accessions
2.2. Genetic Diversity and Genetic Differentiation among Maize Landrace Populations
2.3. Cluster Analysis and Population Structure
3. Discussion
4. Materials and Methods
4.1. Plant Materials and DNA Extraction
4.2. SSR Analysis and DNA Electrophoresis
4.3. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shiferaw, B.; Prasanna, B.M.; Hellin, J.; Bänziger, M. Crops That Feed the World 6. Past Successes and Future Challenges to the Role Played by Maize in Global Food Security. Food Secur. 2011, 3, 307–327. [Google Scholar] [CrossRef] [Green Version]
- Vathana, Y.; Sa, K.J.; Lim, S.E.; Lee, J.K. Genetic Diversity and Association Analyses of Chinese Maize Inbred Lines Using SSR Markers. Plant Breed. Biotechnol. 2019, 7, 186–199. [Google Scholar] [CrossRef]
- Miracle, M.P. The Introduction and Spread of Maize in Africa. J. Afr. Hist. 1965, 6, 39–55. [Google Scholar] [CrossRef]
- Shepherd, D.N.; Martin, D.P.; Van Der Walt, E.; Dent, K.; Varsani, A.; Rybicki, E.P. Maize Streak Virus: An Old and Complex “emerging” Pathogen. Mol. Plant Pathol. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- African Development Bank. AfDB National Climate Change Profile; African Development Bank: Abidjan, Ivoorkust, 2018; pp. 1–28. [Google Scholar]
- Qin, Y.; Shi, G.; Sun, Y. Evaluation of Genetic Diversity in Pampus Argenteus Using SSR Markers. Genet. Mol. Res. 2013, 12, 5833–5841. [Google Scholar] [CrossRef]
- Aci, M.M.; Revilla, P.; Morsli, A.; Djemel, A.; Belalia, N.; Kadri, Y.; Khelifi-Saloui, M.; Ordás, B.; Khelifi, L. Genetic Diversity in Algerian Maize (Zea Mays L) Landraces Using SSR Markers. Maydica 2013, 58, 304–310. [Google Scholar]
- Security, F.; Republic, T.H.E.; South, O.F. Special Report–2020 FAO/WFP Crop and Food Security Assessment Mission (CFSAM) to the Republic of South Sudan; FAO: Rome, Italy, 2021; ISBN 9789251343470. [Google Scholar]
- Awata, L.A.O.; Tokwiny, S.A.; Kitara, I.; Zozimo, R.O.; Bennet, V.S.; Oyiki, C.O. Adaptation of Hybrid Maize to Greenbelt and Ironstone Plateau Agro-Ecologies of South Sudan. Plant Breed. Biotechnol. 2021, 9, 124–138. [Google Scholar] [CrossRef]
- PwC. Positioning Nigeria as Africa’s Leader in Maize Production for AfCFTA; PwC: London, UK, 2021. [Google Scholar]
- Kim, O.G.; Sa, K.J.; Lee, J.R.; Lee, J.K. Genetic Analysis of Maize Germplasm in the Korean Genebank and Association with Agronomic Traits and Simple Sequence Repeat Markers. Genes Genom. 2017, 39, 843–853. [Google Scholar] [CrossRef]
- European Union. South Sudan Rural development Programme South Sudan Crop Production. Farmer Ext. Guid. 2016, 15. Available online: https://europa.eu/capacity4dev/file/30356/download?token=pMPhuRFm (accessed on 1 October 2022).
- Prasanna, B.M. Diversity in Global Maize Germplasm: Characterization and Utilization. J. Biosci. 2012, 37, 843–855. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of Genetic Diversity Assessment in Crop Plants and Its Recent Advances: An Overview of Its Analytical Perspectives. Genet. Res. Int. 2015, 2015, 431487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauthier, P.; Gouesnard, B.; Dallard, J.; Redaelli, R.; Rebourg, C.; Charcosset, A.; Boyat, A. RFLP Diversity and Relationships among Traditional European Maize Populations. Theor. Appl. Genet. 2002, 105, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Belalia, N.; Lupini, A.; Djemel, A.; Morsli, A.; Mauceri, A.; Lotti, C.; Khelifi-Slaoui, M.; Khelifi, L.; Sunseri, F. Analysis of Genetic Diversity and Population Structure in Saharan Maize (Zea mays L.) Populations Using Phenotypic Traits and SSR Markers. Genet. Resour. Crop Evol. 2019, 66, 243–257. [Google Scholar] [CrossRef]
- Kamaluddin; Khan, M.A.; Kiran, U.; Ali, A.; Abdin, M.Z.; Zargar, M.Y.; Ahmad, S.; Sofi, P.A.; Gulzar, S. Molecular Markers and Marker-Assisted Selection in Crop Plants. In Plant Biotechnology: Principles and Applications; Springer: Singapore, 2017; pp. 295–328. [Google Scholar] [CrossRef]
- Rohini, M.R.; Sankaran, M.; Rajkumar, S.; Prakash, K.; Gaikwad, A.; Chaudhury, R.; Malik, S.K. Morphological Characterization and Analysis of Genetic Diversity and Population Structure in Citrus × Jambhiri Lush. Using SSR Markers. Genet. Resour. Crop Evol. 2020, 67, 1259–1275. [Google Scholar] [CrossRef]
- Dubreuil, P.; Rebourg, C.; Merlino, M.; Charcosset, A. Evaluation of a DNA Pooled-Sampling Strategy for Estimating the RFLP Diversity of Maize Populations. Plant Mol. Biol. Report. 1999, 17, 123–138. [Google Scholar] [CrossRef]
- Rebourg, C.; Gouesnard, B.; Charcosset, A. Large Scale Molecular Analysis of Traditional European Maize Populations. Relationships with Morphological Variation. Heredity 2001, 86, 574–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubreuil, P.; Warburton, M.; Chastanet, M.; Hoisington, D.; Charcosset, A. More on the Introduction of Temperate Maize into Europe: Large-Scale Bulk SSR Genotyping and New Historical Elements. Maydica 2006, 51, 281–291. [Google Scholar]
- Sharma, L.; Ramesh, B.M.; Ramesh, B. Phenotypic and microsatellite-based diversity and population genetic structure of maize landraces in India, especially from the North East Himalayan region. Genetica 2010, 138, 619–631. [Google Scholar] [CrossRef]
- Laborda, P.R.; Oliveira, K.M.; Garcia, A.A.F.; Paterniani, M.E.A.G.Z.; De Souza, A.P. Tropical Maize Germplasm: What Can We Say about Its Genetic Diversity in the Light of Molecular Markers? Theor. Appl. Genet. 2005, 111, 1288–1299. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, R.; Zhao, J.; Cai, Y.; Wang, F.; Cao, M.; Wang, R.; Shi, Y.; Song, Y.; Wang, T.; et al. Genetic Diversity of Two Important Groups of Maize Landraces with Same Name in China Revealed by M13 Tailed-Primer SSRs. Agric. Sci. China 2009, 8, 15–23. [Google Scholar] [CrossRef]
- Sa, K.J.; Choi, I.Y.; Park, K.C.; Lee, J.K. Genetic Diversity and Population Structure among Accessions of Perilla Frutescens (L.) Britton in East Asia Using New Developed Microsatellite Markers. Genes Genom. 2018, 40, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Lee, J.K.; Kim, N.S. Simple Sequence Repeat Polymorphisms (SSRPs) for Evaluation of Molecular Diversity and Germplasm Classification of Minor Crops. Molecules 2009, 14, 4546–4569. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.S.; Sa, K.J.; Park, H.; Hyun, D.Y.; Lee, S.; Rhee, J.H.; Lee, J.K. Genetic Variation of Native Perilla Germplasms Collected from South Korea Using Simple Sequence Repeat (Ssr) Markers and Morphological Characteristics. Plants 2021, 10, 1764. [Google Scholar] [CrossRef] [PubMed]
- Qi-Lun, Y.; Ping, F.; Ke-Cheng, K.; Guang-Tang, P. Genetic Diversity Based on SSR Markers in Maize (Zea mays L.) Landraces from Wuling Mountain Region in China. J. Genet. 2008, 87, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Oppong, A.; Bedoya, C.A.; Ewool, M.B.; Asante, M.D.; Thompson, R.N.; Adu-Dapaah, H.; Lamptey, J.N.L.; Ofori, K.; Offei, S.K.; Warburton, M.L. Bulk Genetic Characterization of Ghanaian Maize Landraces Using Microsatellite Markers. Maydica 2014, 59, 1–8. [Google Scholar]
- Cömertpay, G.; Baloch, F.S.; Kilian, B.; Ülger, A.C.; Özkan, H. Diversity Assessment of Turkish Maize Landraces Based on Fluorescent Labelled SSR Markers. Plant Mol. Biol. Rep. 2012, 30, 261–274. [Google Scholar] [CrossRef]
- Enoki, H.; Sato, H.; Koinuma, K. SSR Analysis of Genetic Diversity among Maize Inbred Lines Adapted to Cold Regions of Japan. Theor. Appl. Genet. 2002, 104, 1270–1277. [Google Scholar] [CrossRef]
- Thakur, N.; Prakash, J.; Thakur, K.; Sharma, J.K.; Kumari, R.; Rana, M.; Lata, S. Genetic Diversity and Structure of Maize Accessions of North Western Himalayas Based on Morphological and Molecular Markers. Proc. Natl. Acad. Sci. India Sect. B-Biol. Sci. 2017, 87, 1385–1398. [Google Scholar] [CrossRef]
- Lia, V.V.; Poggio, L.; Confalonieri, V.A. Microsatellite Variation in Maize Landraces from Northwestern Argentina: Genetic Diversity, Population Structure and Racial Affiliations. Theor. Appl. Genet. 2009, 119, 1053–1067. [Google Scholar] [CrossRef] [PubMed]
- Noldín, O.; Revilla, P.; Ordás, B. Genetic Diversity of the Floury Race of Maize Avati Morotî from the Guaraní Tribe in Paraguay. Span. J. Agric. Res. 2016, 14, e0707. [Google Scholar] [CrossRef] [Green Version]
- Reif, J.C.; Warburton, M.L.; Xia, X.C.; Hoisington, D.A.; Crossa, J.; Taba, S.; Muminović, J.; Bohn, M.; Frisch, M.; Melchinger, A.E. Grouping of Accessions of Mexican Races of Maize Revisited with SSR Markers. Theor. Appl. Genet. 2006, 113, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Eschholz, T.W.; Stamp, P.; Peter, R.; Leipner, J.; Hund, A. Genetic Structure and History of Swiss Maize (Zea mays L. Ssp. Mays) Landraces. Genet. Resour. Crop Evol. 2010, 57, 71–84. [Google Scholar] [CrossRef] [Green Version]
- Yao, Q.; Fang, P.; Kang, K.; Pan, G. Genetic diversity based on SSR markers in maize (Zea mays L) landraces from Wuling mountain region in China. J. Genet 2007, 87, 287–291. [Google Scholar]
- Njuguna, W. Development and Use of Molecular Tools in Fragaria. Ph.D. Thesis, Oregon State University, Corvallis, OR, USA, 2010. [Google Scholar]
- Hilmarsson, H.S.; Hytönen, T.; Isobe, S.; Göransson, M.; Toivainen, T.; Hallsson, J.H. Population Genetic Analysis of a Global Collection of Fragaria Vesca Using Microsatellite Markers. PLoS ONE 2017, 12, e0183384. [Google Scholar] [CrossRef]
- Chesser, R.K. Gene Diversity and Female Philopatry. Genetics 1991, 127, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Brock, J.; Dyer, J.M.; Kutchan, T.; Schachtman, D.; Augustin, M.; Ge, Y.; Fahlgren, N.; Abdel-Haleem, H. Genetic Diversity and Population Structure of a Camelina Sativa Spring Panel. Front. Plant Sci. 2019, 10, 184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pineda-Hidalgo, K.V.; Méndez-Marroquín, K.P.; Alvarez, E.V.; Chávez-Ontiveros, J.; Sánchez-Peña, P.; Garzón-Tiznado, J.A.; Vega-García, M.O.; López-Valenzuela, J.A. Microsatellite-Based Genetic Diversity among Accessions of Maize Landraces from Sinaloa in México. Hereditas 2013, 150, 53–59. [Google Scholar] [CrossRef]
- N’da, H.A.; Akanvou, L.; Pokou, N.D.; Akanza, K.P.; Kouakou, C.K.; Zoro, B.I.A. Genetic Diversity and Population Structure of Maize Landraces from Cte DIvoire. Afr. J. Biotechnol. 2016, 15, 2507–2516. [Google Scholar] [CrossRef] [Green Version]
- Wasala, S.K.; Prasanna, B.M. Microsatellite Marker-Based Diversity and Population Genetic Analysis of Selected Lowland and Mid-Altitude Maize Landrace Accessions of India. J. Plant Biochem. Biotechnol. 2013, 22, 392–400. [Google Scholar] [CrossRef]
- Louette, D.; Charrier, A.; Berthaud, J. In Situ Conservation of Maize in Mexico: Genetic Diversity and Maize Seed Management in a Traditional Community. Econ. Bot. 1997, 51, 20–38. [Google Scholar] [CrossRef]
- Singode, A.; Prasanna, B.M. Analysis of Genetic Diversity in the North Eastern Himalayan Maize Landraces Using Microsatellite Markers. J. Plant Biochem. Biotechnol. 2010, 19, 33–41. [Google Scholar] [CrossRef]
- Da Silva, T.A.; Cantagalli, L.B.; Saavedra, J.; Lopes, A.D.; Mangolin, C.A.; da Silva Machado, M.D.F.P.; Scapima, C.A. Population Structure and Genetic Diversity of Brazilian Popcorn Germplasm Inferred by Microsatellite Markers. Electron. J. Biotechnol. 2015, 18, 181–187. [Google Scholar] [CrossRef] [Green Version]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA mini-preparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Yeh, F.C.; Yang, R.C.; Boyle, T. POPGENE 1.32: A Free Program for the Analysis of Genetic Variation Among and Within Populations Using Co-Dominant and Dominant Markers; Uni Alberta: Edmonton, AB, Canada, 2000. [Google Scholar]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Rohlf, F. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System; Version Exter Software: Setauket, NY, USA, 2000. [Google Scholar]
- Pritchard, J.K.; Wen, W.; Falush, D. Documentation for Structure Software: Version 2.3. 2003. Available online: http://pritc.h.bsd.uchicago.edu/structure.html (accessed on 1 November 2021).
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peakall, R.; Smouse, P.E. GenALEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research-an Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Pagnotta, M. Comparison among Methods and Statistical Software Packages to Analyze Germplasm Genetic Diversity b Means of Codominant Markersy. J-Multidiscip. Sci. J. 2018, 1, 197–215. [Google Scholar] [CrossRef]
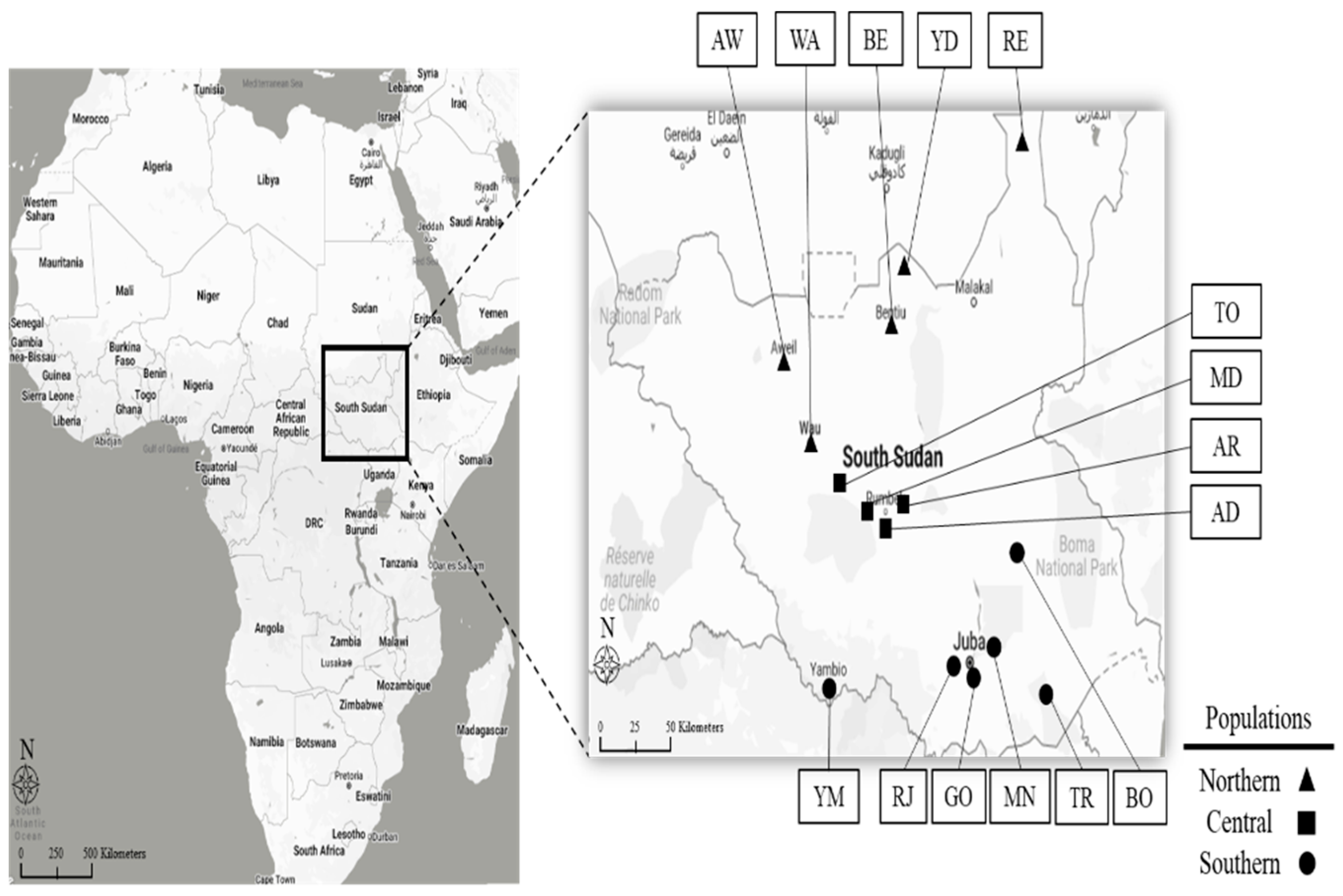
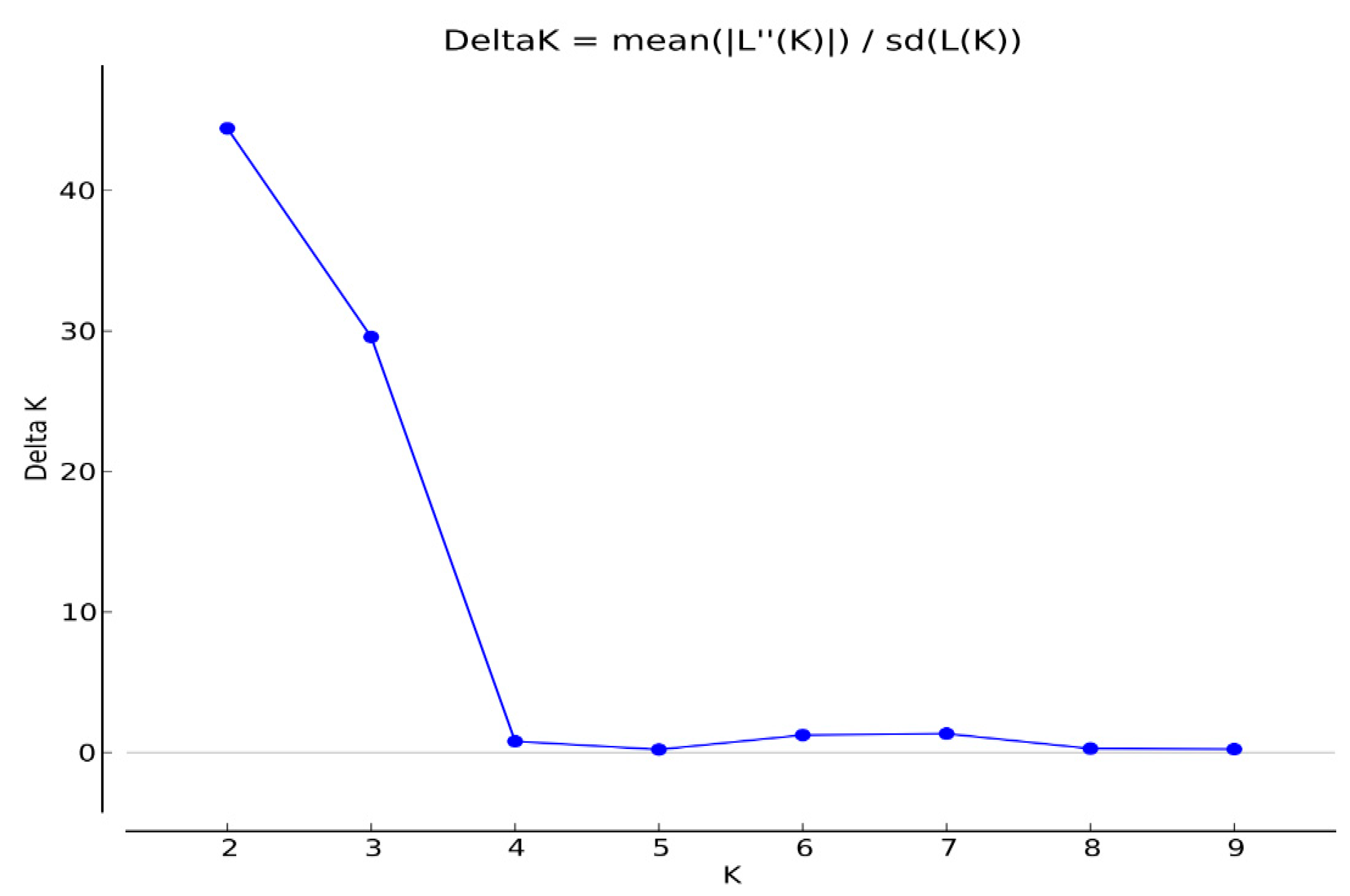
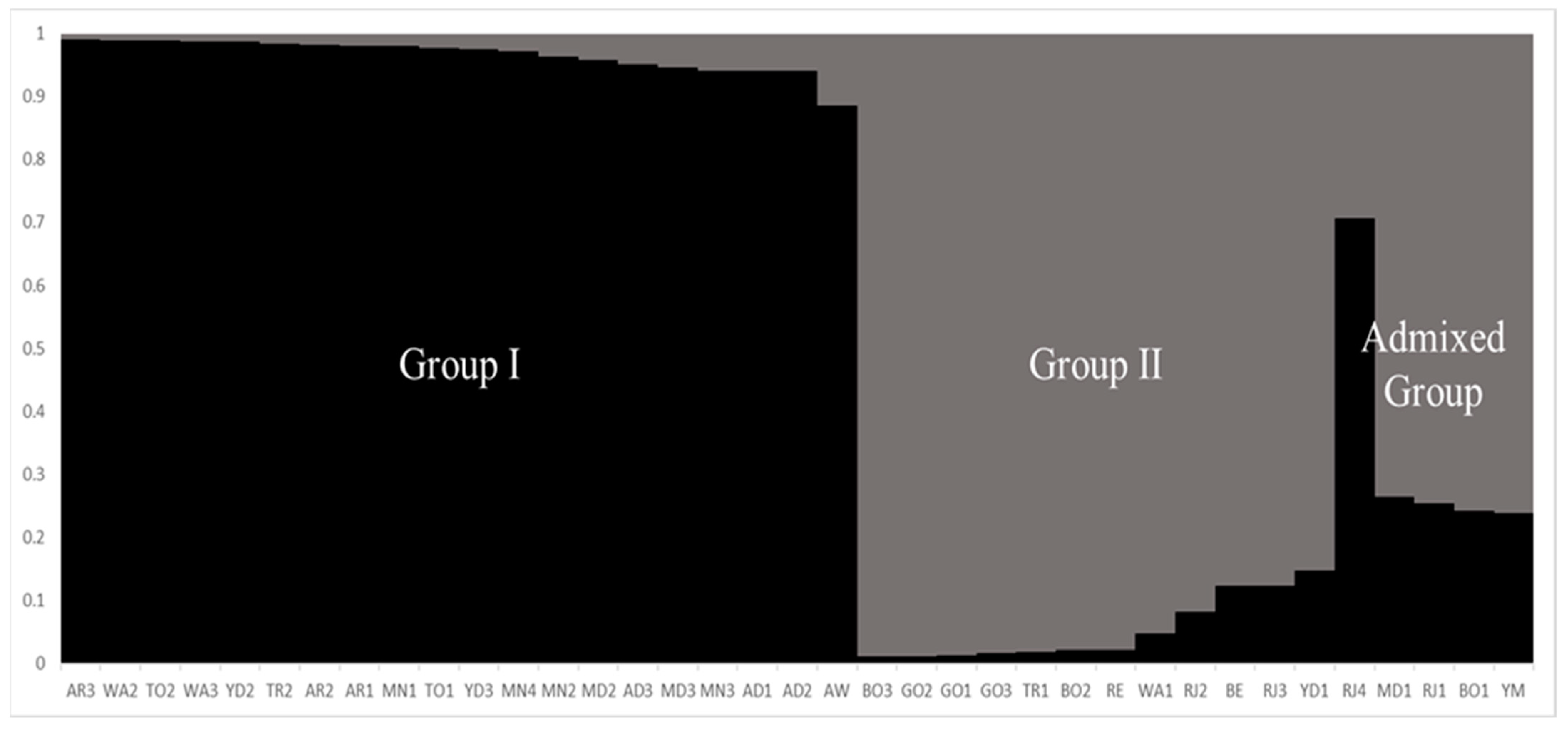

| Population | Reference Name (Abbr) | Field-Plot Name | Field-Plot Location | No. Genotype Sampled | Biological Status | Geographical Coordinates | |
|---|---|---|---|---|---|---|---|
| Latitude | Longitude | ||||||
| Southern populations | BO | Bor | Bor, Gonglei State | 3 | Landrace | 6°12′47.8″ | 31°33′56.0″ |
| RJ | Rejaf | Central Equatoria State | 4 | Landrace | 4°45′07.9″ | 31°34′18.8″ | |
| MN | Mangalla | Central Equatoria State | 4 | Landrace | 5°10′48.0″ | 31°46′04.8″ | |
| GO | Gondokor | Central Equatoria State | 3 | Landrace | 4°54′00.0″ | 31°40′00.1″ | |
| TR | Torit | Torit, Magwi, Eastern Equatoria | 2 | Landrace | 4°24′36.7″ | 32°34′26.4″ | |
| YM | Yambio | Western Equatoria State | 1 | Landrace | 4°34′39.4″ | 28°23′55.7″ | |
| Central populations | AR | Aber | Rumbek, Lake State | 3 | Landrace | 6°48′25.9″ | 29°40′44.0″ |
| MD | Madhok | Rumbek, Lake State | 3 | Landrace | 6°42′22.7″ | 29°40′46.6″ | |
| AD | Adull | Rumbek, Lake State | 3 | Landrace | 6°37′00.1″ | 29°57′00.0″ | |
| TO | Tonj | Tonj, Kuajok, warrap State | 2 | Landrace | 8°18′14.4″ | 27°59′36.2″ | |
| Northern populations | WA | Wau | Wau, Raja, western bahr ghazhal State | 3 | Landrace | 7°42′33.1″ | 27°59′00.6″ |
| AW | Aweil | Northern Bahr el Ghazal State | 1 | Landrace | 8°46′01.6″ | 27°23′59.3″ | |
| YD | Yida | Ruweng Administrative Area | 3 | Landrace | 10°06′13.0″ | 30°05′25.1″ | |
| RE | Renk | Renk, Upper Nile State | 1 | Landrace | 11°44′34.8″ | 32°48′16.1″ | |
| BE | Bentiu | Bentiu, Unity State | 1 | Landrace | 9°13′53.4″ | 29°48′01.8″ | |
| Total | 37 | ||||||
| SSR Loci Name | Chr. | Allele Size | Sample Size | Allele No | Na | Ne | I | Ho | He | MAF | GD | PIC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bnlg1605 | 9 | 100–150 | 74 | 8 | 5 | 1.744 | 0.883 | 0.351 | 0.432 | 0.595 | 0.614 | 0.590 |
| umc1024 | 1 | 102–150 | 70 | 6 | 3 | 1.895 | 0.785 | 0.086 | 0.479 | 0.595 | 0.580 | 0.532 |
| umc1066 | 7 | 110–150 | 72 | 6 | 3 | 1.882 | 0.762 | 0.389 | 0.475 | 0.459 | 0.652 | 0.590 |
| umc1082 | 8 | 150–200 | 66 | 9 | 4 | 3.571 | 1.326 | 0.485 | 0.731 | 0.243 | 0.859 | 0.844 |
| umc1101 | 4 | 110–150 | 38 | 6 | 3 | 2.431 | 0.972 | 0.105 | 0.605 | 0.486 | 0.673 | 0.627 |
| umc1108 | 8 | 100–130 | 70 | 13 | 4 | 3.695 | 1.342 | 0.914 | 0.740 | 0.378 | 0.808 | 0.792 |
| umc1130 | 8 | 100–130 | 74 | 3 | 2 | 1.810 | 0.640 | 0.351 | 0.454 | 0.486 | 0.614 | 0.536 |
| umc1175 | 7 | 60–80 | 66 | 7 | 4 | 3.081 | 1.224 | 0.273 | 0.686 | 0.324 | 0.790 | 0.762 |
| umc1227 | 5 | 60–110 | 56 | 8 | 4 | 3.045 | 1.218 | 0.214 | 0.684 | 0.243 | 0.824 | 0.800 |
| umc1303 | 3 | 90–120 | 72 | 5 | 3 | 1.861 | 0.704 | 0.417 | 0.469 | 0.432 | 0.650 | 0.583 |
| umc1315 | 4 | 90–120 | 74 | 4 | 3 | 1.938 | 0.809 | 0.568 | 0.491 | 0.432 | 0.614 | 0.534 |
| umc1316 | 8 | 95–130 | 72 | 11 | 4 | 3.011 | 1.205 | 0.389 | 0.677 | 0.324 | 0.831 | 0.815 |
| umc1339 | 2 | 110–150 | 52 | 9 | 4 | 3.108 | 1.220 | 0.192 | 0.692 | 0.297 | 0.801 | 0.774 |
| umc1380 | 7 | 130–150 | 70 | 6 | 3 | 1.724 | 0.724 | 0.143 | 0.426 | 0.622 | 0.574 | 0.543 |
| umc1454 | 4 | 130–160 | 68 | 7 | 3 | 1.959 | 0.852 | 0.412 | 0.497 | 0.459 | 0.723 | 0.693 |
| umc1466 | 2 | 150–200 | 60 | 5 | 3 | 1.744 | 0.765 | 0.067 | 0.434 | 0.568 | 0.621 | 0.582 |
| umc1607 | 7 | 100–140 | 70 | 11 | 4 | 2.162 | 1.014 | 0.314 | 0.545 | 0.324 | 0.830 | 0.813 |
| umc1718 | 4 | 150–200 | 70 | 4 | 2 | 1.994 | 0.692 | 0.257 | 0.506 | 0.378 | 0.690 | 0.628 |
| umc1872 | 4 | 100–130 | 74 | 7 | 4 | 3.009 | 1.208 | 0.432 | 0.677 | 0.297 | 0.811 | 0.786 |
| umc2075 | 2 | 140–200 | 68 | 4 | 2 | 1.745 | 0.618 | 0.265 | 0.433 | 0.514 | 0.644 | 0.591 |
| umc2135 | 8 | 150–200 | 52 | 7 | 4 | 2.136 | 0.887 | 0.231 | 0.542 | 0.351 | 0.754 | 0.717 |
| umc2275 | 4 | 130–200 | 74 | 11 | 5 | 3.423 | 1.410 | 0.865 | 0.718 | 0.270 | 0.840 | 0.822 |
| umc2286 | 2 | 70–110 | 56 | 6 | 3 | 2.085 | 0.892 | 0.214 | 0.530 | 0.405 | 0.741 | 0.704 |
| umc2329 | 10 | 150–200 | 66 | 10 | 4 | 2.447 | 1.036 | 0.182 | 0.601 | 0.297 | 0.830 | 0.810 |
| umc2334 | 7 | 50–100 | 74 | 10 | 4 | 3.695 | 1.341 | 0.595 | 0.739 | 0.135 | 0.887 | 0.876 |
| umc2378 | 3 | 90–130 | 62 | 12 | 6 | 4.178 | 1.576 | 0.548 | 0.773 | 0.216 | 0.874 | 0.861 |
| umc2540 | 7 | 140–200 | 64 | 5 | 3 | 2.293 | 0.931 | 0.219 | 0.573 | 0.405 | 0.717 | 0.671 |
| Max | 74 | 13 | 6 | 4.178 | 1.576 | 0.914 | 0.773 | 0.622 | 0.887 | 0.876 | ||
| Min | 38 | 3 | 2 | 1.724 | 0.618 | 0.067 | 0.426 | 0.135 | 0.574 | 0.532 | ||
| Mean | 66.1 | 7.4 | 3.6 | 2.506 | 1.001 | 0.351 | 0.578 | 0.390 | 0.735 | 0.699 | ||
| Total | 1784 | 200 |
| Geographic Population | Sample Size | Na | Ne | I | Ho | He | F |
|---|---|---|---|---|---|---|---|
| AD | 5 | 2.039 | 1.778 | 0.583 | 0.263 | 0.468 | 0.438 |
| AR | 5 | 2.148 | 1.925 | 0.601 | 0.395 | 0.457 | 0.135 |
| AW | 2 | 1.240 | 1.240 | 0.166 | 0.240 | 0.240 | 0.000 |
| BE | 2 | 1.192 | 1.192 | 0.133 | 0.192 | 0.192 | 0.000 |
| BO | 5 | 2.148 | 1.790 | 0.593 | 0.444 | 0.474 | 0.063 |
| GO | 5 | 1.852 | 1.610 | 0.476 | 0.259 | 0.383 | 0.322 |
| MD | 6 | 2.185 | 1.893 | 0.620 | 0.364 | 0.473 | 0.230 |
| MN | 7 | 2.407 | 2.058 | 0.736 | 0.463 | 0.562 | 0.176 |
| RE | 2 | 1.160 | 1.160 | 0.111 | 0.160 | 0.160 | 0.000 |
| RJ | 7 | 2.077 | 1.730 | 0.524 | 0.292 | 0.373 | 0.219 |
| TO | 4 | 1.800 | 1.677 | 0.495 | 0.400 | 0.467 | 0.143 |
| TR | 4 | 1.889 | 1.770 | 0.525 | 0.296 | 0.488 | 0.392 |
| WA | 6 | 2.185 | 1.846 | 0.612 | 0.377 | 0.475 | 0.208 |
| YD | 6 | 2.407 | 2.066 | 0.726 | 0.457 | 0.553 | 0.174 |
| YM | 2 | 1.429 | 1.429 | 0.297 | 0.429 | 0.429 | 0.000 |
| Max | 7 | 2.407 | 2.066 | 0.736 | 0.463 | 0.562 | 0.438 |
| Min | 2 | 1.160 | 1.160 | 0.111 | 0.160 | 0.160 | 0.000 |
| Mean | 4.5 | 1.877 | 1.678 | 0.480 | 0.335 | 0.413 | 0.167 |
| Total | 68 |
| Population | No. Populations | Sample Size | Na | Ne | I | Ho | He | F |
|---|---|---|---|---|---|---|---|---|
| Southern populations | 6 | 30 | 3.296 | 2.293 | 0.913 | 0.353 | 0.553 | 0.361 |
| Central populations | 4 | 20 | 3.074 | 2.364 | 0.903 | 0.355 | 0.563 | 0.369 |
| Northern populations | 5 | 17 | 3.000 | 2.310 | 0.880 | 0.336 | 0.552 | 0.392 |
| Mean | 22.3 | 3.123 | 2.322 | 0.899 | 0.348 | 0.556 | 0.374 | |
| Total | 67 |
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among Regions | 2 | 63.780 | 31.890 | 1.282 | 7% |
| Within Regions | 34 | 567.193 | 16.682 | 16.682 | 93% |
| Total | 36 | 630.973 | 17.964 | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathiang, E.A.; Sa, K.J.; Park, H.; Kim, Y.J.; Lee, J.K. Genetic Diversity and Population Structure of Normal Maize Germplasm Collected in South Sudan Revealed by SSR Markers. Plants 2022, 11, 2787. https://doi.org/10.3390/plants11202787
Mathiang EA, Sa KJ, Park H, Kim YJ, Lee JK. Genetic Diversity and Population Structure of Normal Maize Germplasm Collected in South Sudan Revealed by SSR Markers. Plants. 2022; 11(20):2787. https://doi.org/10.3390/plants11202787
Chicago/Turabian StyleMathiang, Emmanuel Andrea, Kyu Jin Sa, Hyeon Park, Yeon Joon Kim, and Ju Kyong Lee. 2022. "Genetic Diversity and Population Structure of Normal Maize Germplasm Collected in South Sudan Revealed by SSR Markers" Plants 11, no. 20: 2787. https://doi.org/10.3390/plants11202787
APA StyleMathiang, E. A., Sa, K. J., Park, H., Kim, Y. J., & Lee, J. K. (2022). Genetic Diversity and Population Structure of Normal Maize Germplasm Collected in South Sudan Revealed by SSR Markers. Plants, 11(20), 2787. https://doi.org/10.3390/plants11202787






