Identification of the OsCML4 Gene in Rice Related to Salt Stress Using QTL Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Linkage Map
2.2. Plant Materials and Mapping Population
2.3. Phenotypic Evaluation of Seedlings under Salt Stress
2.4. Phenotypic Data Collection
2.5. Statistical and QTL Analysis
2.6. Finding Candidate Genes through QTL Mapping
2.7. Gene Expression Levels of Candidate Genes Related to Salinity
3. Results
3.1. Phenotypic Variation
3.2. QTL Mapping of Traits Related to Salinity Stress
3.3. Screening Candidate Genes Related to Salinity Stress on the Basis of QTL Mapping
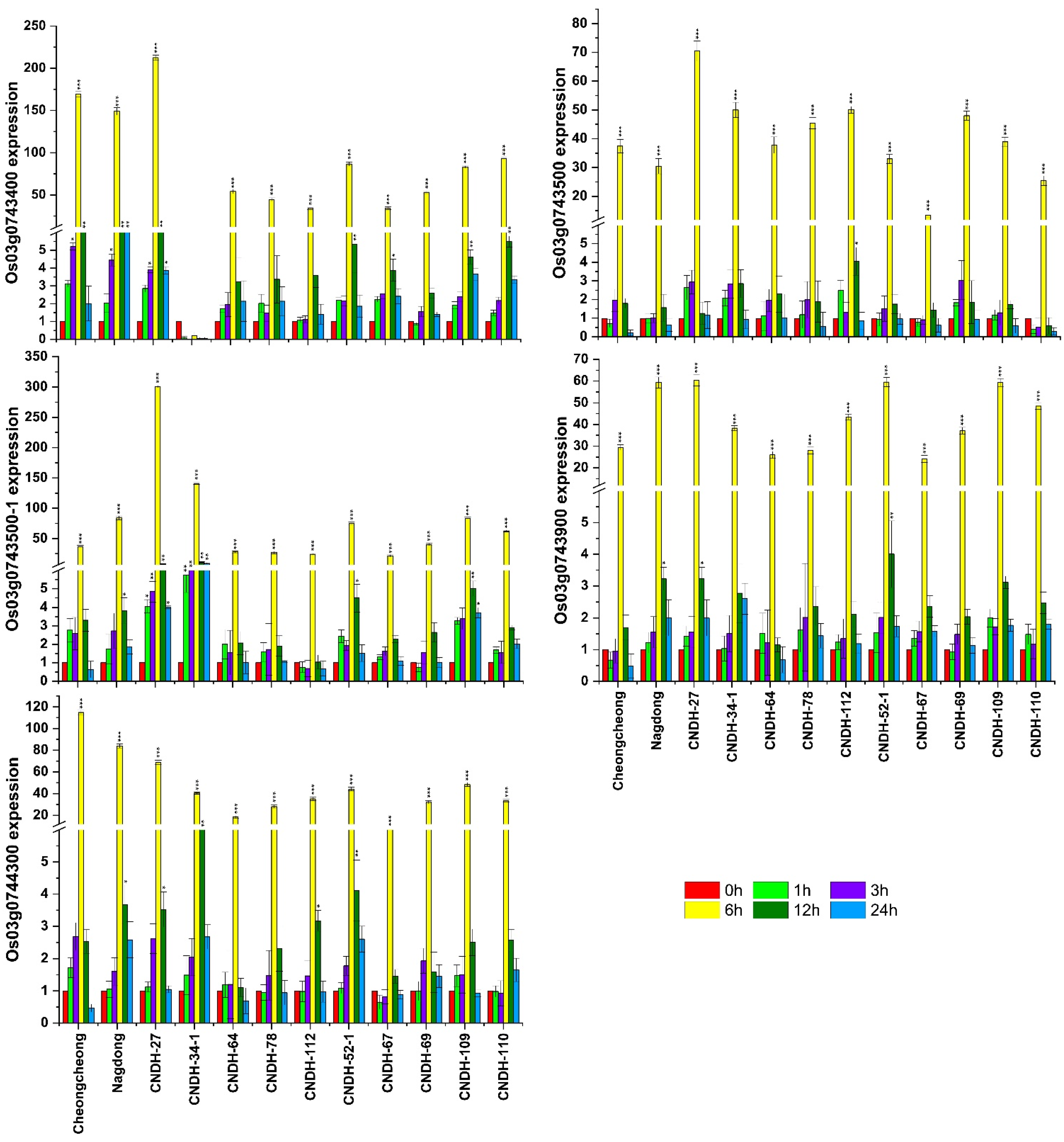
3.4. Relative Gene Expression of the Associated Genes in the Target Marker Region
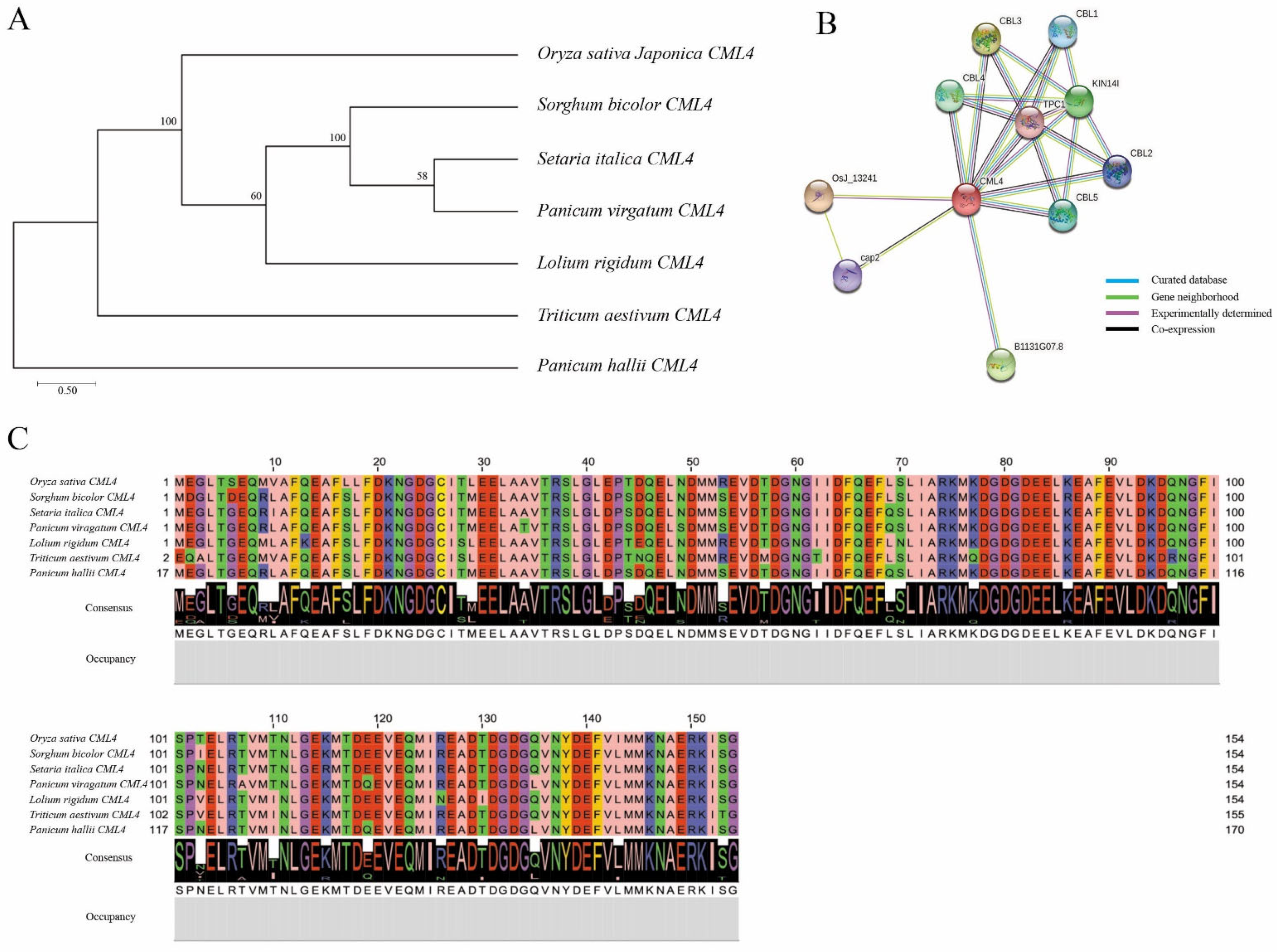
3.5. Phylogenetic Tree, Homologous Sequence, and Protein–Protein Interaction Analysis of OsCML4
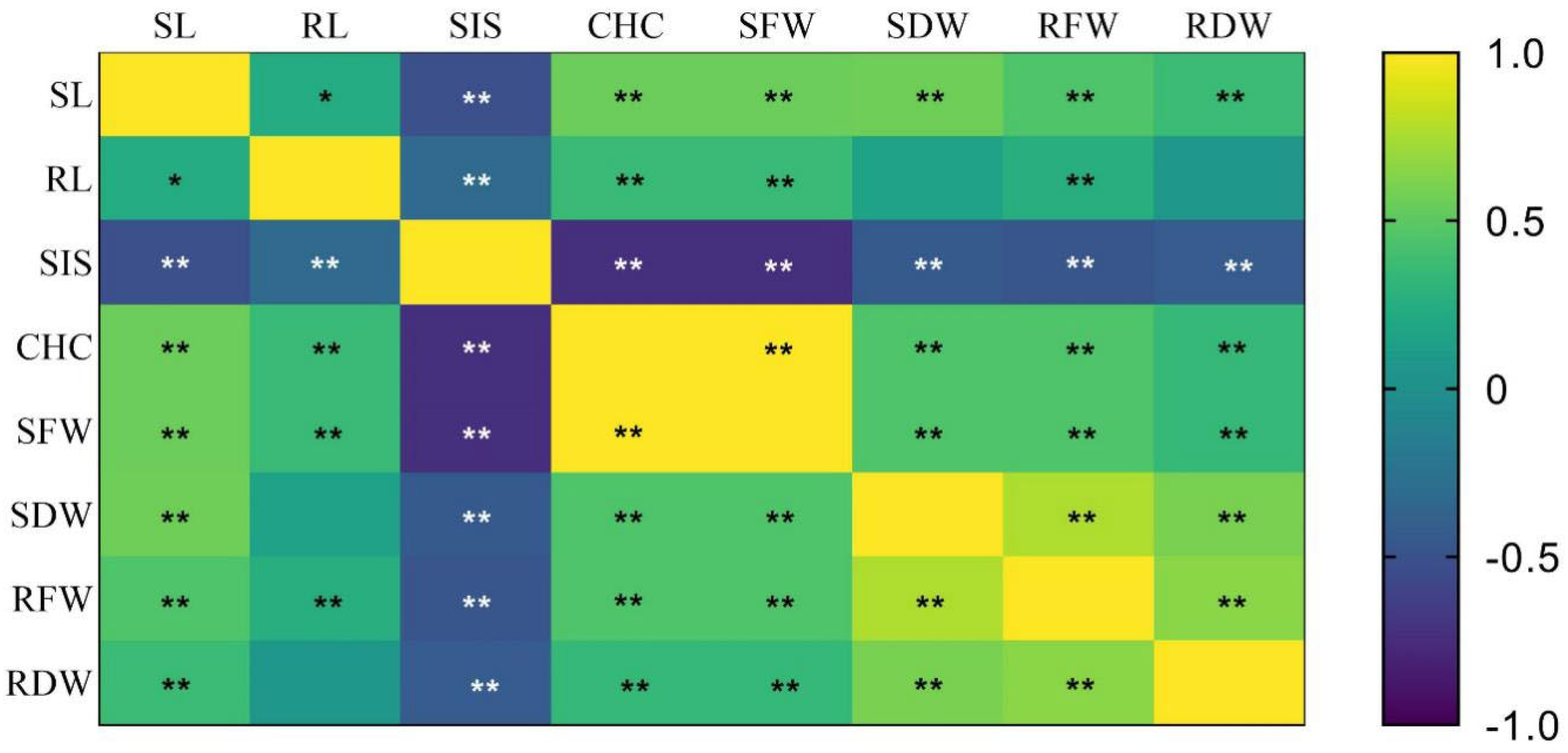
3.6. Correlation Analysis among the Different Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ammar, M.; Singh, R.; Singh, A.; Mohapatra, T.; Sharma, T.; Singh, N. Mapping QTLs for salinity tolerance at seedling stage in rice (Oryza sativa L.). In Proceedings of the African Crop Science Conference Proceedings, El-Minia, Egypt, 27–31 October 2007; pp. 617–620. [Google Scholar]
- Khan, M.A.; Hamayun, M.; Asaf, S.; Khan, M.; Yun, B.-W.; Kang, S.-M.; Lee, I.-J. Rhizospheric bacillus spp. Rescues Plant Growth under Salinity Stress via Regulating Gene Expression, Endogenous Hormones, and Antioxidant System of Oryza sativa L. Front. Plant Sci. 2021, 12, 1145. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Termaat, A. Whole-plant responses to salinity. Funct. Plant Biol. 1986, 13, 143–160. [Google Scholar] [CrossRef]
- Asif, M.A.; Schilling, R.K.; Tilbrook, J.; Brien, C.; Dowling, K.; Rabie, H.; Short, L.; Trittermann, C.; Garcia, A.; Barrett-Lennard, E.G. Mapping of novel salt tolerance QTL in an Excalibur × Kukri doubled haploid wheat population. Theor. Appl. Genet. 2018, 131, 2179–2196. [Google Scholar] [CrossRef]
- Khan, M.A.; Asaf, S.; Khan, A.L.; Jan, R.; Kang, S.-M.; Kim, K.-M.; Lee, I.-J. Rhizobacteria AK1 remediates the toxic effects of salinity stress via regulation of endogenous phytohormones and gene expression in soybean. Biochem. J. 2019, 476, 2393–2409. [Google Scholar] [CrossRef]
- Kotuby-Amacher, J.; Koenig, R.; Kitchen, B. Salinity and Plant Tolerance; Electronic Publication AG-SO-03; Utah State University Extension: Logan, UT, USA, 2000. [Google Scholar]
- Hasegawa, M.; Sone, S.; Takashima, S.; Li, F.; Yang, Z.; Maruyama, Y.; Watanabe, T. Growth rate of small lung cancers detected on mass CT screening. Br. J. Radiol. 2000, 73, 1252–1259. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Blumwald, E. Developing salt-tolerant crop plants: Challenges and opportunities. Trends Plant Sci. 2005, 10, 615–620. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, Y.; Jahan, N.; Chen, G.; Ren, D.; Guo, L. Sensing of abiotic stress and ionic stress responses in plants. Int. J. Mol. Sci. 2018, 19, 3298. [Google Scholar] [CrossRef]
- Abbas, G.; Saqib, M.; Akhtar, J.; ul Haq, M.A. Interactive effects of salinity and iron deficiency on different rice genotypes. J. Plant Nutr. Soil Sci. 2015, 178, 306–311. [Google Scholar] [CrossRef]
- Izadi, M.H.; Rabbani, J.; Emam, Y.; Pessarakli, M.; Tahmasebi, A. Effects of salinity stress on physiological performance of various wheat and barley cultivars. J. Plant Nutr. 2014, 37, 520–531. [Google Scholar] [CrossRef]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef]
- Jahan, N.; Zhang, Y.; Lv, Y.; Song, M.; Zhao, C.; Hu, H.; Cui, Y.; Wang, Z.; Yang, S.; Zhang, A.; et al. QTL analysis for rice salinity tolerance and fine mapping of a candidate locus qSL7 for shoot length under salt stress. Plant Growth Regul. 2020, 90, 307–319. [Google Scholar] [CrossRef]
- Gimhani, D.; Gregorio, G.B.; Kottearachchi, N.; Samarasinghe, W.L.G. SNP-based discovery of salinity-tolerant QTLs in a bi-parental population of rice (Oryza sativa). Mol. Genet. Genom. 2016, 291, 2081–2099. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Subudhi, P.K. Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a high-density GBS-based SNP linkage map. Rice 2016, 9, 52. [Google Scholar] [CrossRef]
- Gregorio, G.; Senadhira, D.; Mendoza, R.; Manigbas, N.; Roxas, J.; Guerta, C. Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 2002, 76, 91–101. [Google Scholar] [CrossRef]
- Thomson, M.J.; Zhao, K.; Wright, M.; McNally, K.L.; Rey, J.; Tung, C.-W.; Reynolds, A.; Scheffler, B.; Eizenga, G.; McClung, A.; et al. High-throughput single nucleotide polymorphism genotyping for breeding applications in rice using the BeadXpress platform. Mol. Breed. 2012, 29, 875–886. [Google Scholar] [CrossRef]
- Gregorio, G.B. Tagging Salinity Tolerance Genes in Rice Using Amplified Fragment Length Polymorphism (AFLP); Food and Agriculture Organization of the United Nations: Rome, Italy, 1997. [Google Scholar]
- Bizimana, J.B.; Luzi-Kihupi, A.; Murori, R.W.; Singh, R.K. Identification of quantitative trait loci for salinity tolerance in rice (Oryza sativa L.) using IR29/Hasawi mapping population. J. Genet. 2017, 96, 571–582. [Google Scholar] [CrossRef]
- Rahman, M.A.; Thomson, M.J.; Shah-E-Alam, M.; de Ocampo, M.; Egdane, J.; Ismail, A.M. Exploring novel genetic sources of salinity tolerance in rice through molecular and physiological characterization. Ann. Bot. 2016, 117, 1083–1097. [Google Scholar] [CrossRef]
- Yang, T.; Poovaiah, B.W. Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 2003, 8, 505–512. [Google Scholar] [CrossRef]
- Yin, X.; Huang, L.; Wang, M.; Cui, Y.; Xia, X. OsDSR-1, a calmodulin-like gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Mol. Breed. 2017, 37, 75. [Google Scholar] [CrossRef]
- Snedden, W.A.; Fromm, H. Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 2001, 151, 35–66. [Google Scholar] [CrossRef]
- McCormack, E.; Tsai, Y.-C.; Braam, J. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005, 10, 383–389. [Google Scholar] [CrossRef]
- Chinpongpanich, A.; Limruengroj, K.; Phean-o-pas, S.; Limpaseni, T.; Buaboocha, T. Expression analysis of calmodulin and calmodulin-like genes from rice, Oryza sativa L. BMC Res. Notes 2012, 5, 625. [Google Scholar] [CrossRef]
- Boonburapong, B.; Buaboocha, T. Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 2007, 7, 4. [Google Scholar] [CrossRef][Green Version]
- Vanderbeld, B.; Snedden, W.A. Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39. Plant Mol. Biol. 2007, 64, 683–697. [Google Scholar] [CrossRef]
- Kim, E.-G.; Yun, S.; Park, J.-R.; Kim, K.-M. Identification of F3H, Major Secondary Metabolite-Related Gene That Confers Resistance against Whitebacked Planthopper through QTL Mapping in Rice. Plants 2021, 10, 81. [Google Scholar] [CrossRef]
- Zhao, D.-D.; Park, J.-R.; Jang, Y.-H.; Kim, E.-G.; Du, X.-X.; Farooq, M.; Yun, B.-J.; Kim, K.-M. Identification of One Major QTL and a Novel Gene OsIAA17q5 Associated with Tiller Number in Rice Using QTL Analysis. Plants 2022, 11, 538. [Google Scholar] [CrossRef]
- Zeng, Z.-B. Precision mapping of quantitative trait loci. Genetics 1994, 136, 1457–1468. [Google Scholar] [CrossRef]
- McCough, S.R.; Doerge, R.W. QTL mapping in rice. Trends Genet. 1995, 11, 482–487. [Google Scholar] [CrossRef]
- Krishnamurthy, S.L.; Pundir, P.; Warraich, A.S.; Rathor, S.; Lokeshkumar, B.M.; Singh, N.K.; Sharma, P.C. Introgressed Saltol QTL Lines Improves the Salinity Tolerance in Rice at Seedling Stage. Frontiers 2020, 11, 833. [Google Scholar] [CrossRef] [PubMed]
- Sabouri, H.; Rezai, A.M.; Moumeni, A.; Kavousi, A.; Katouzi, M.; Sabouri, A. QTLs mapping of physiological traits related to salt tolerance in young rice seedlings. Biol. Plant. 2009, 53, 657–662. [Google Scholar] [CrossRef]
- Yiliang, Q.; Hui, W.; Manyuan, C. Detection of Salt-Tolerant QTL Using BC2F3 Yield Selected Introgression Lines of Rice? Oryza sativa L.? Food and Agriculture Organization of the United Nations: Rome, Italy, 2009. [Google Scholar]
- Liang, J.-L.; Qu, Y.-P.; Yang, C.-G.; Ma, X.-D.; Cao, G.-L.; Zhao, Z.-W.; Zhang, S.-Y.; Zhang, T.; Han, L.-Z. Identification of QTLs associated with salt or alkaline tolerance at the seedling stage in rice under salt or alkaline stress. Euphytica 2015, 201, 441–452. [Google Scholar] [CrossRef]
- Haitao, C.; Hua, J.; Meixian, Y. QTL-Mapping Comparison of Tolerance to Alkali at Germination Period and Early Seeding Stage between Two Different Double Haploid Populations in Rice; Food and Agriculture Organization of the United Nations: Rome, Italy, 2008. [Google Scholar]
- Wang, S.; Cao, M.; Ma, X.; Chen, W.; Zhao, J.; Sun, C.; Tan, L.; Liu, F. Integrated RNA Sequencing and QTL Mapping to Identify Candidate Genes from Oryza rufipogon Associated with Salt Tolerance at the Seedling Stage. Frontiers 2017, 8, 1427. [Google Scholar] [CrossRef]
- Singh, V.K.; Jain, M. Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Frontiers 2015, 6, 918. [Google Scholar] [CrossRef]
- Jung, H.; Lee, D.-K.; Choi, Y.D.; Kim, J.-K. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 2015, 236, 304–312. [Google Scholar] [CrossRef]
- Liscum, E.; Reed, J.W. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002, 49, 387–400. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 47–59. [Google Scholar] [CrossRef]
- Kalluri, U.C.; DiFazio, S.P.; Brunner, A.M.; Tuskan, G.A. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 2007, 7, 59. [Google Scholar] [CrossRef]
- Li, G.; Ye, Y.; Ren, X.; Qi, M.; Zhao, H.; Zhou, Q.; Chen, X.; Wang, J.; Yuan, C.; Wang, F. The rice Aux/IAA transcription factor gene OsIAA18 enhances salt and osmotic tolerance in Arabidopsis. Biol. Plant. 2020, 64, 454–464. [Google Scholar] [CrossRef]
- Cho, K.M.; Nguyen, H.T.K.; Kim, S.Y.; Shin, J.S.; Cho, D.H.; Hong, S.B.; Shin, J.S.; Ok, S.H. CML 10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytol. Found. 2016, 209, 664–678. [Google Scholar] [CrossRef]
- Delk, N.A.; Johnson, K.A.; Chowdhury, N.I.; Braam, J. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 2005, 139, 240–253. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, W.; Liu, L.; Su, Y.; Li, Y.; Jia, W.; Jiao, B.; Wang, J.; Yang, F.; Dong, F.; et al. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in wheat (Triticum aestivum L.). Plant Signal. Behav. 2022, 17, 2013646. [Google Scholar] [CrossRef]
- Gu, Z.; Ma, B.; Jiang, Y.; Chen, Z.; Su, X.; Zhang, H. Expression analysis of the calcineurin B-like gene family in rice (Oryza sativa L.) under environmental stresses. Gene 2008, 415, 1–12. [Google Scholar] [CrossRef]

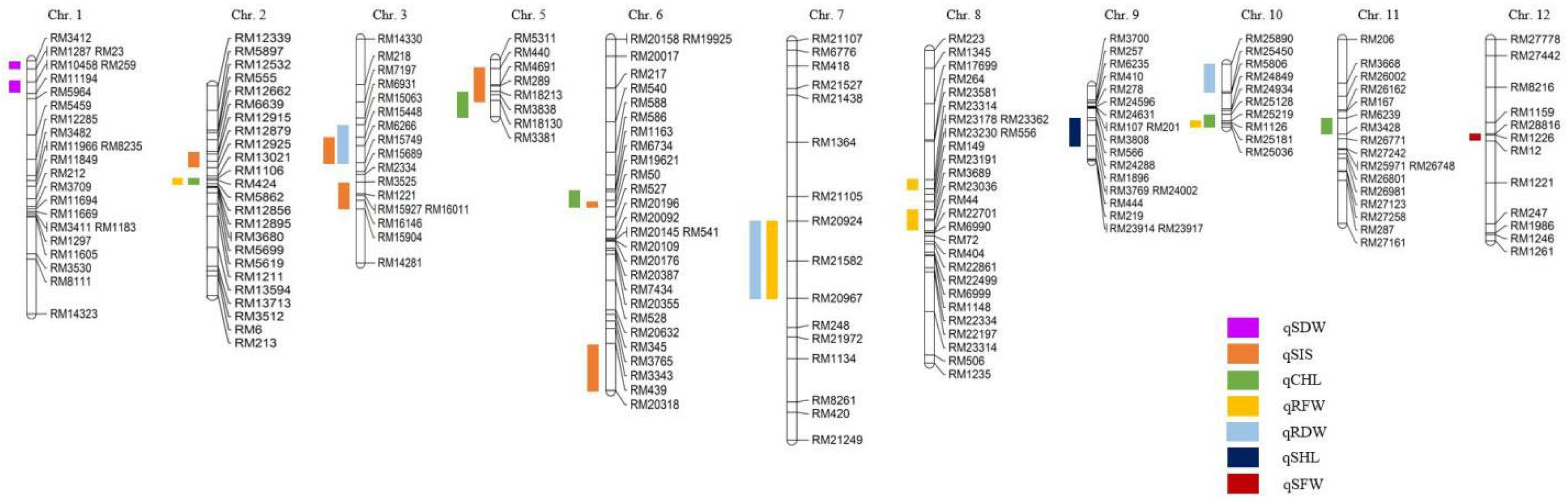


| Characteristics | QTLs | Chr | Interval Markers z | LOD | Additive effect y | R2 x | Increasing effects w |
|---|---|---|---|---|---|---|---|
| Shoot length | qSHL-9 | 9 | RM24288-RM3769 | 2.42 | −2.61 | 0.25 | Nagdong |
| SIS | qSIS-2 | 2 | RM6639-RM12879 | 2.76 | 0.66 | 0.22 | Cheongcheong |
| qSIS-3a | 3 | RM15448-RM15749 | 2.77 | 0.65 | 0.24 | Cheongcheong | |
| qSIS-3b | 3 | RM3525-RM15904 | 6 | −0.98 | 0.31 | Nagdong | |
| qSIS-5 | 5 | RM440-RM18130 | 2.5 | −0.53 | 0.27 | Nagdong | |
| qSIS-6a | 6 | RM19621-RM50 | 2.13 | 0.48 | 0.26 | Cheongcheong | |
| qSIS-6b | 6 | RM439-RM20318 | 2.44 | 0.73 | 0.34 | Cheongcheong | |
| Chlorophyll content | qCHL-2 | 2 | RM1106-RM5862 | 2.36 | −2.89 | 0.23 | Nagdong |
| qCHL-5 | 5 | RM18213-RM3381 | 2.45 | 2.64 | 0.2 | Cheongcheong | |
| qCHL-6 | 6 | RM1163-RM50 | 2.17 | −2.75 | 0.21 | Nagdong | |
| qCHL-10 | 10 | RM25128-RM25036 | 2.3 | 2.64 | 0.18 | Cheongcheong | |
| qCHL-11 | 11 | RM6239-RM3428 | 2.29 | −2.63 | 0.23 | Nagdong | |
| Shoot fresh weight | qSFW-12 | 12 | RM28816-RM12 | 2.3 | 29.98 | 0.22 | Cheongcheong |
| Shoot dry weight | qSDW-1a | 1 | RM3412-RM1287 | 4.09 | −15.14 | 0.25 | Nagdong |
| qSDW-1b | 1 | RM10458-RM11194 | 4.46 | 15.49 | 0.25 | Cheongcheong | |
| Root fresh weight | qRFW-2 | 2 | RM1106-RM5862 | 2.36 | −2.84 | 0.22 | Nagdong |
| qRFW-7 | 7 | RM20924-RM20967 | 4.36 | 31.51 | 0.28 | Cheongcheong | |
| qRFW-8a | 8 | RM23314-RM23178 | 2.14 | −26.57 | 0.31 | Nagdong | |
| qRFW-8b | 8 | RM23191-RM44 | 2.51 | 27.32 | 0.3 | Cheongcheong | |
| qRFW-10 | 10 | RM25219-RM25036 | 2.22 | 2.75 | 0.24 | Cheongcheong | |
| Root dry weight | qRDW-3 | 3 | RM15063-RM15749 | 3.52 | −3.83 | 0.28 | Nagdong |
| qRDW-7 | 7 | RM20924-RM20967 | 3.44 | 3.36 | 0.25 | Cheongcheong | |
| qRDW-10 | 10 | RM25890-RM5806 | 3.07 | 4.42 | 0.31 | Cheongcheong |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asif, S.; Kim, E.-G.; Jang, Y.-H.; Jan, R.; Kim, N.; Asaf, S.; Lubna; Farooq, M.; Kim, K.-M. Identification of the OsCML4 Gene in Rice Related to Salt Stress Using QTL Analysis. Plants 2022, 11, 2467. https://doi.org/10.3390/plants11192467
Asif S, Kim E-G, Jang Y-H, Jan R, Kim N, Asaf S, Lubna, Farooq M, Kim K-M. Identification of the OsCML4 Gene in Rice Related to Salt Stress Using QTL Analysis. Plants. 2022; 11(19):2467. https://doi.org/10.3390/plants11192467
Chicago/Turabian StyleAsif, Saleem, Eun-Gyeong Kim, Yoon-Hee Jang, Rahmatullah Jan, Nari Kim, Sajjad Asaf, Lubna, Muhammad Farooq, and Kyung-Min Kim. 2022. "Identification of the OsCML4 Gene in Rice Related to Salt Stress Using QTL Analysis" Plants 11, no. 19: 2467. https://doi.org/10.3390/plants11192467
APA StyleAsif, S., Kim, E.-G., Jang, Y.-H., Jan, R., Kim, N., Asaf, S., Lubna, Farooq, M., & Kim, K.-M. (2022). Identification of the OsCML4 Gene in Rice Related to Salt Stress Using QTL Analysis. Plants, 11(19), 2467. https://doi.org/10.3390/plants11192467







