Genome-Wide Analysis of UGT Genes in Petunia and Identification of PhUGT51 Involved in the Regulation of Salt Resistance
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Properties Analysis of PhUGT Family Members
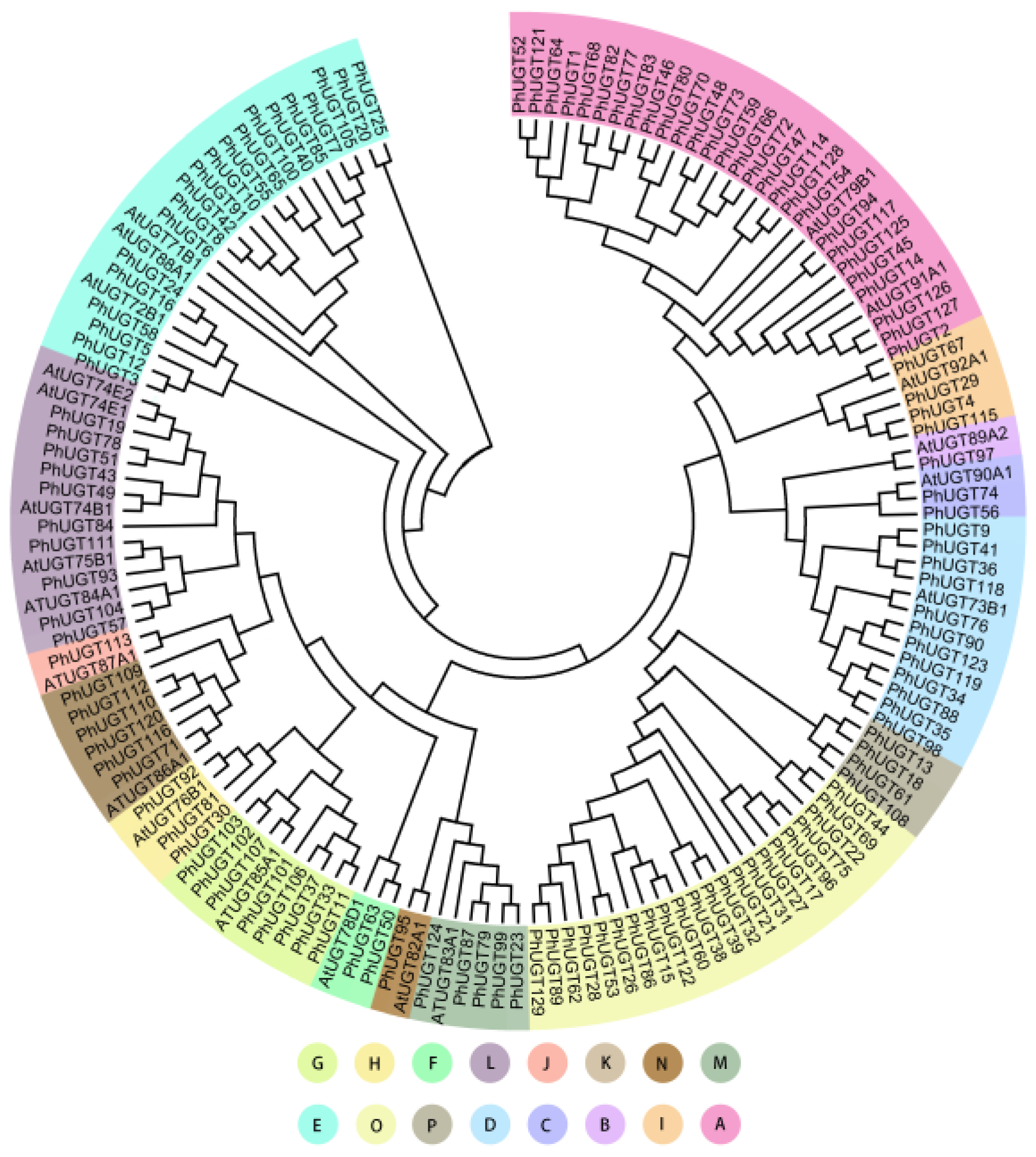
2.2. Phylogenetic Analysis of UGT Family in Petunia
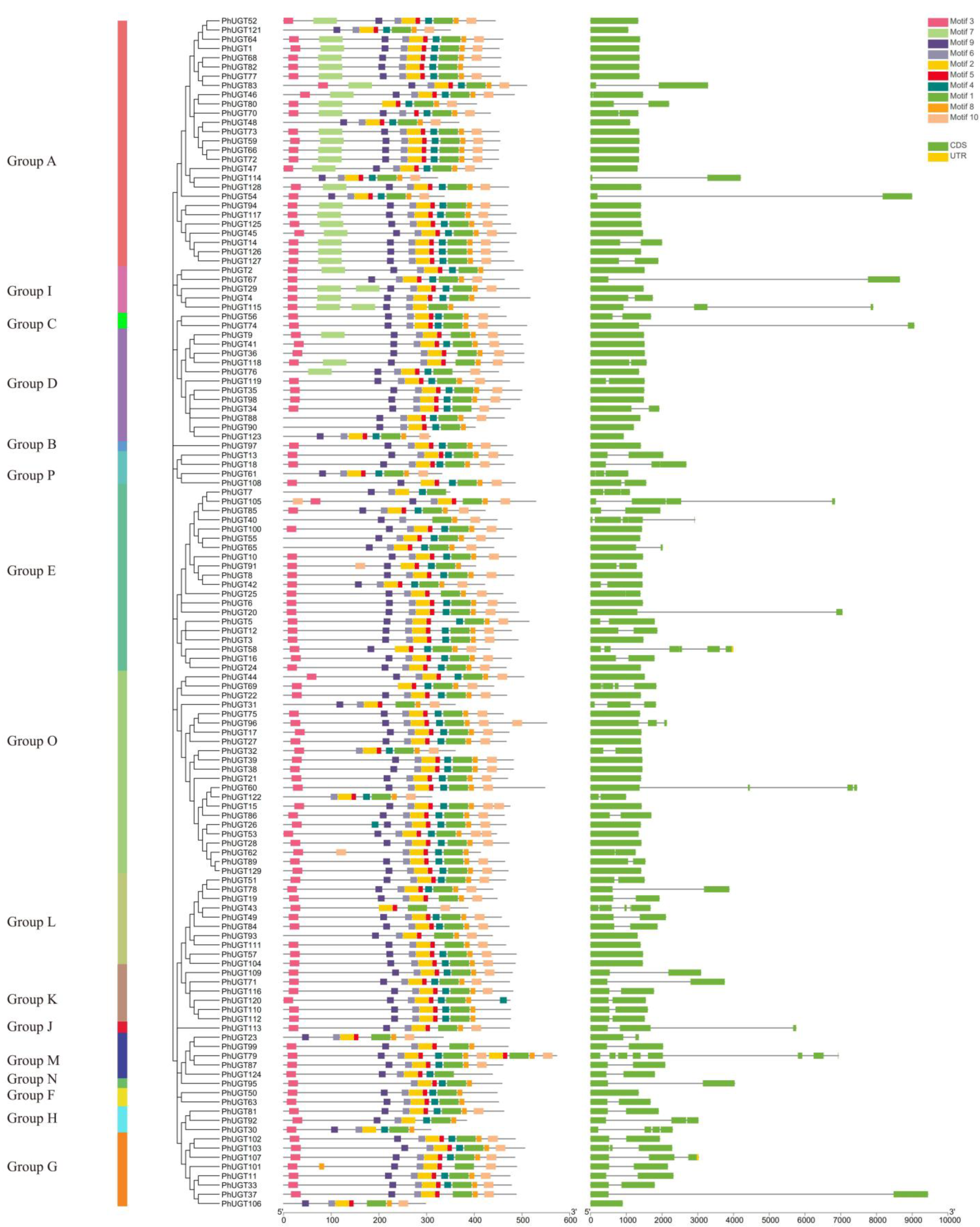
2.3. Analysis of the Petunia UGT Protein Motifs and Gene Structure
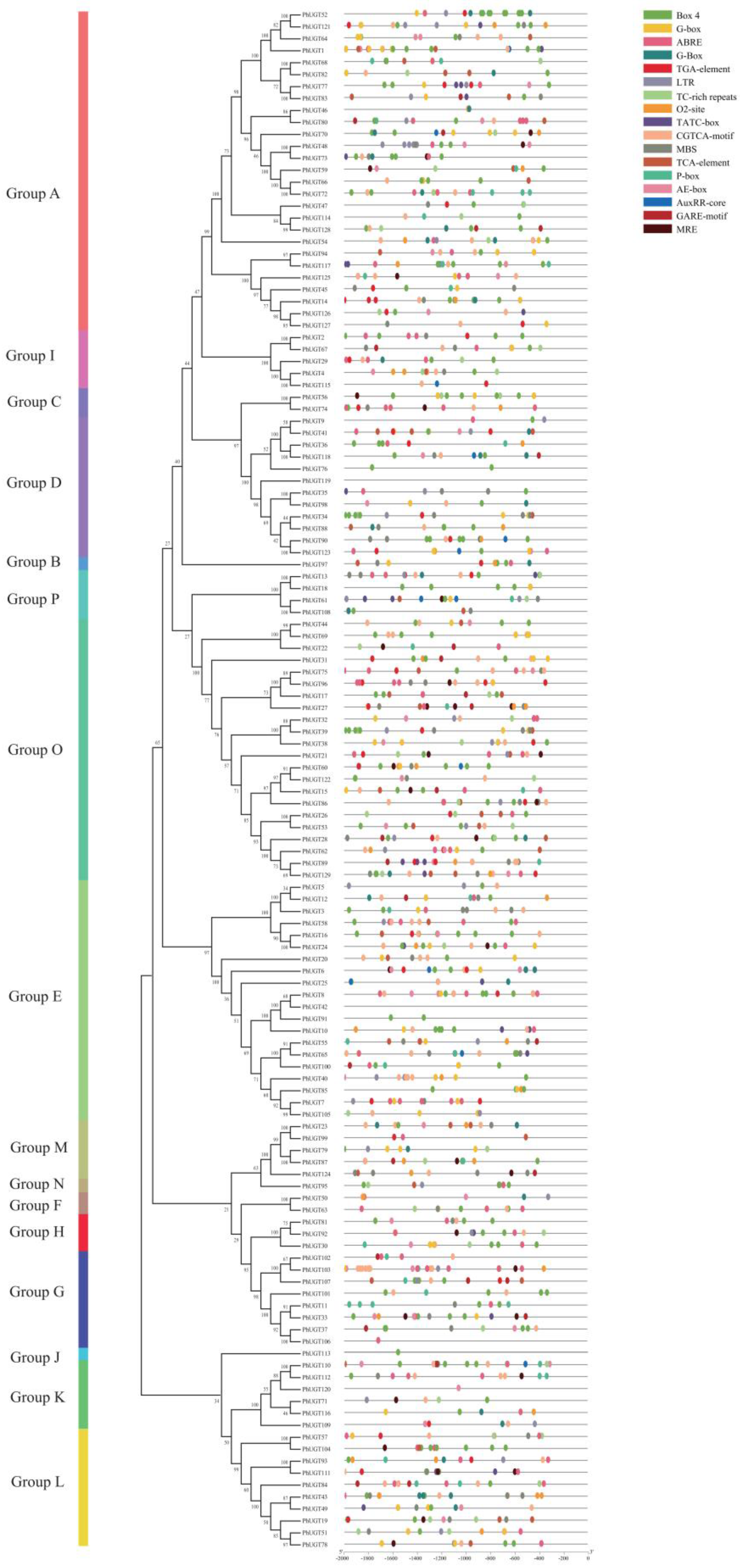
2.4. Analysis of Cis-Elements in the Promoters of PhUGT Genes
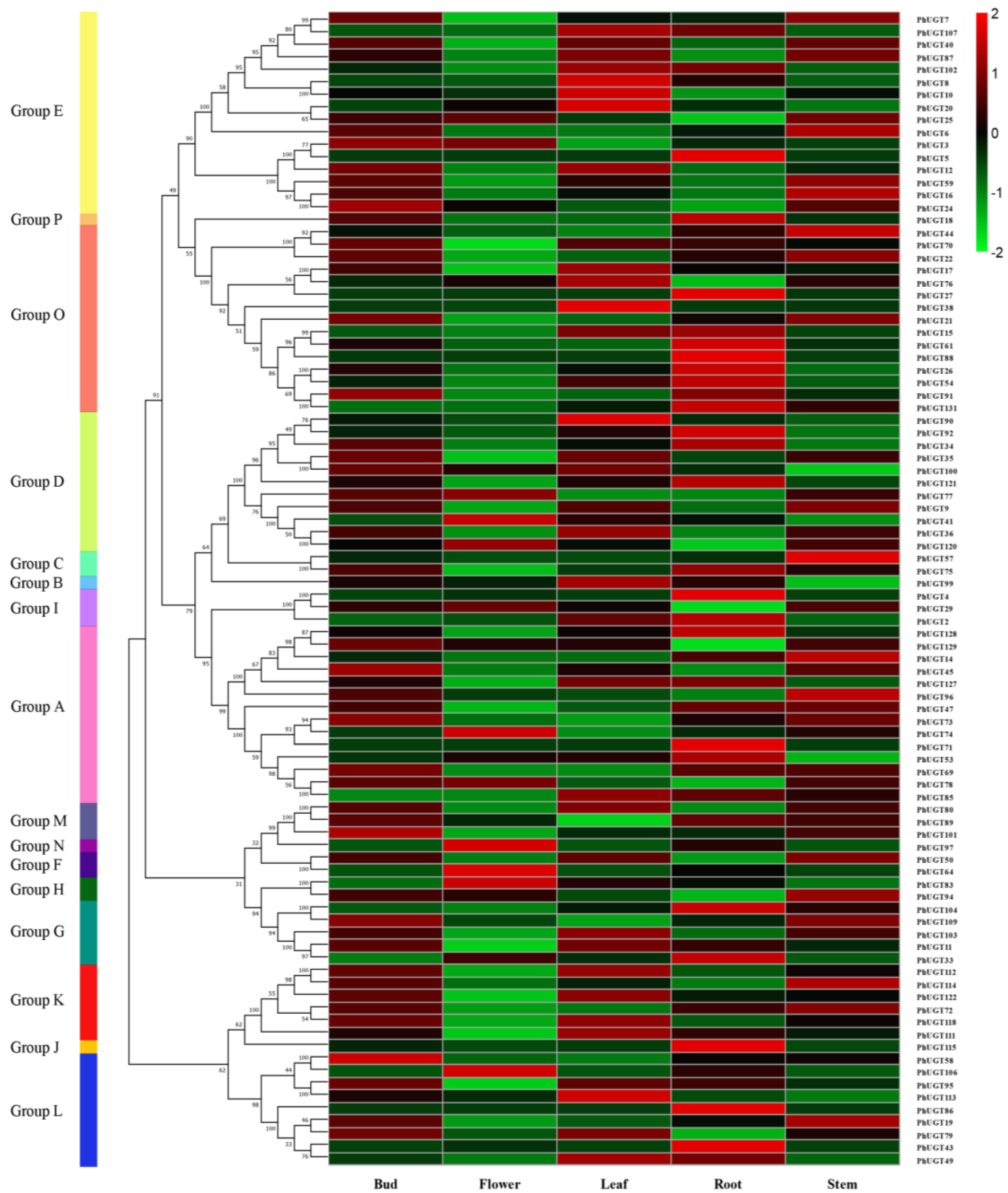
2.5. PhUGT Gene Expression Study in Five Tissues
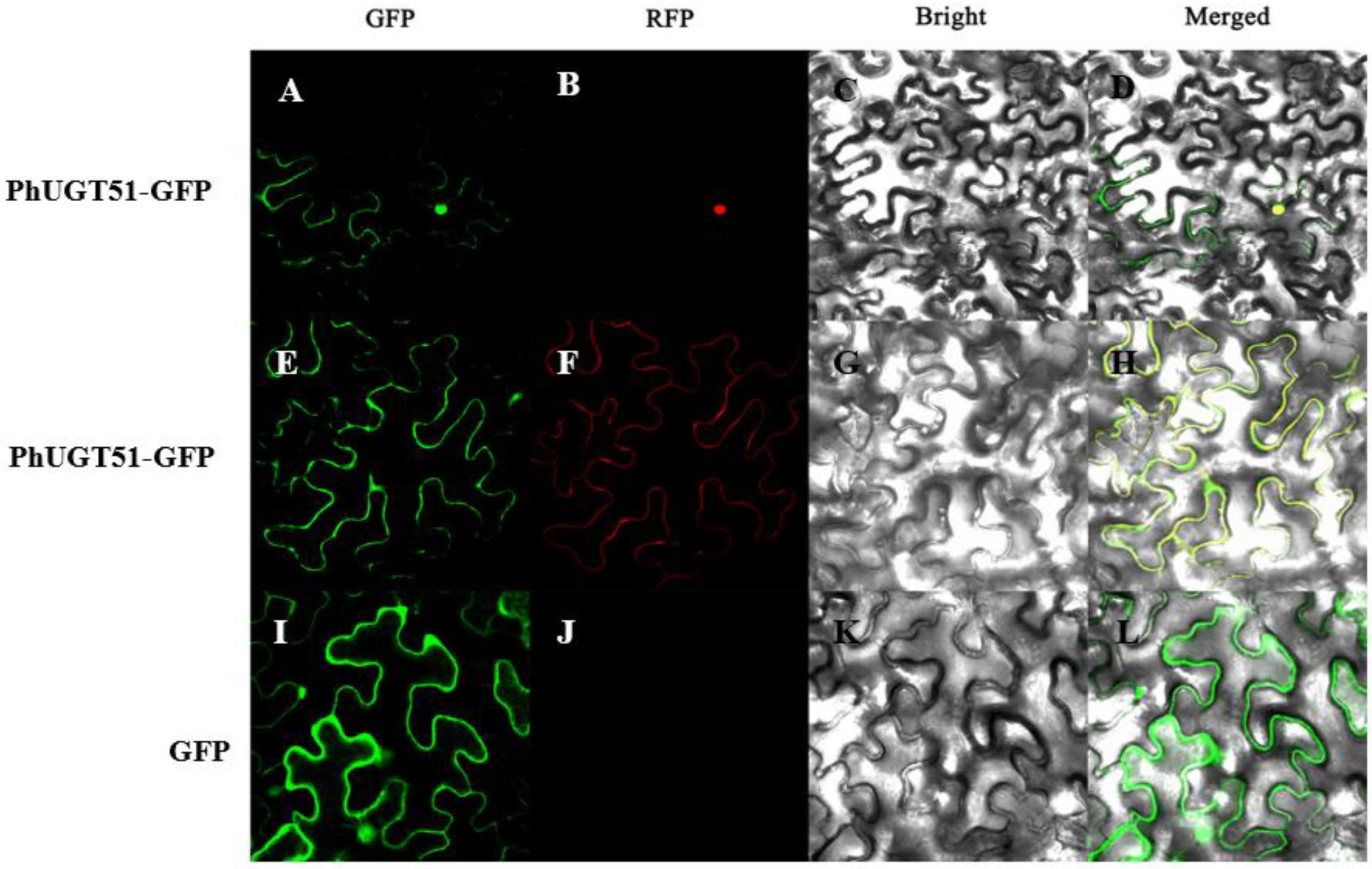
2.6. Subcellular Localization Analysis of PhUGT51
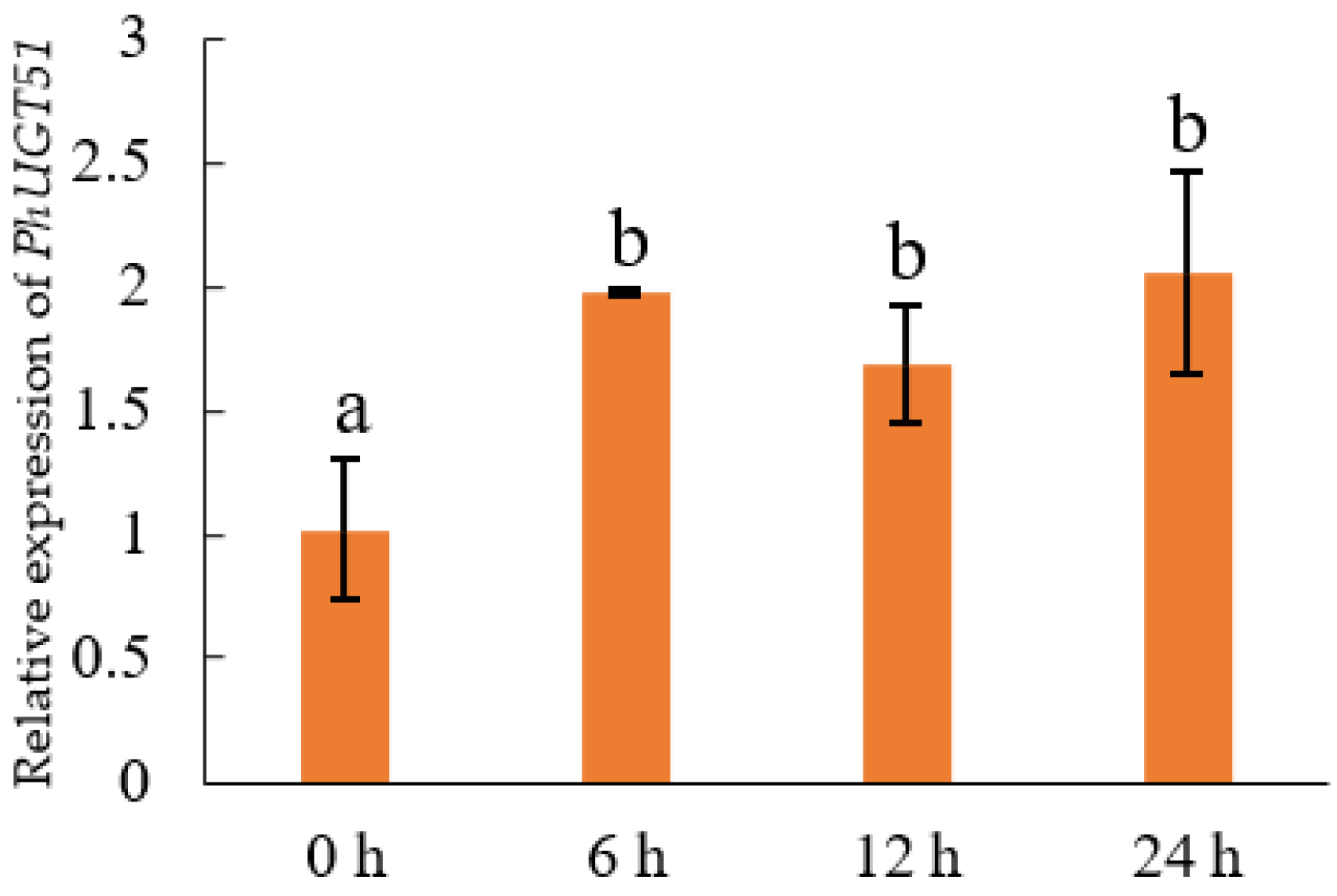
2.7. Expression Analysis of PhUGT51
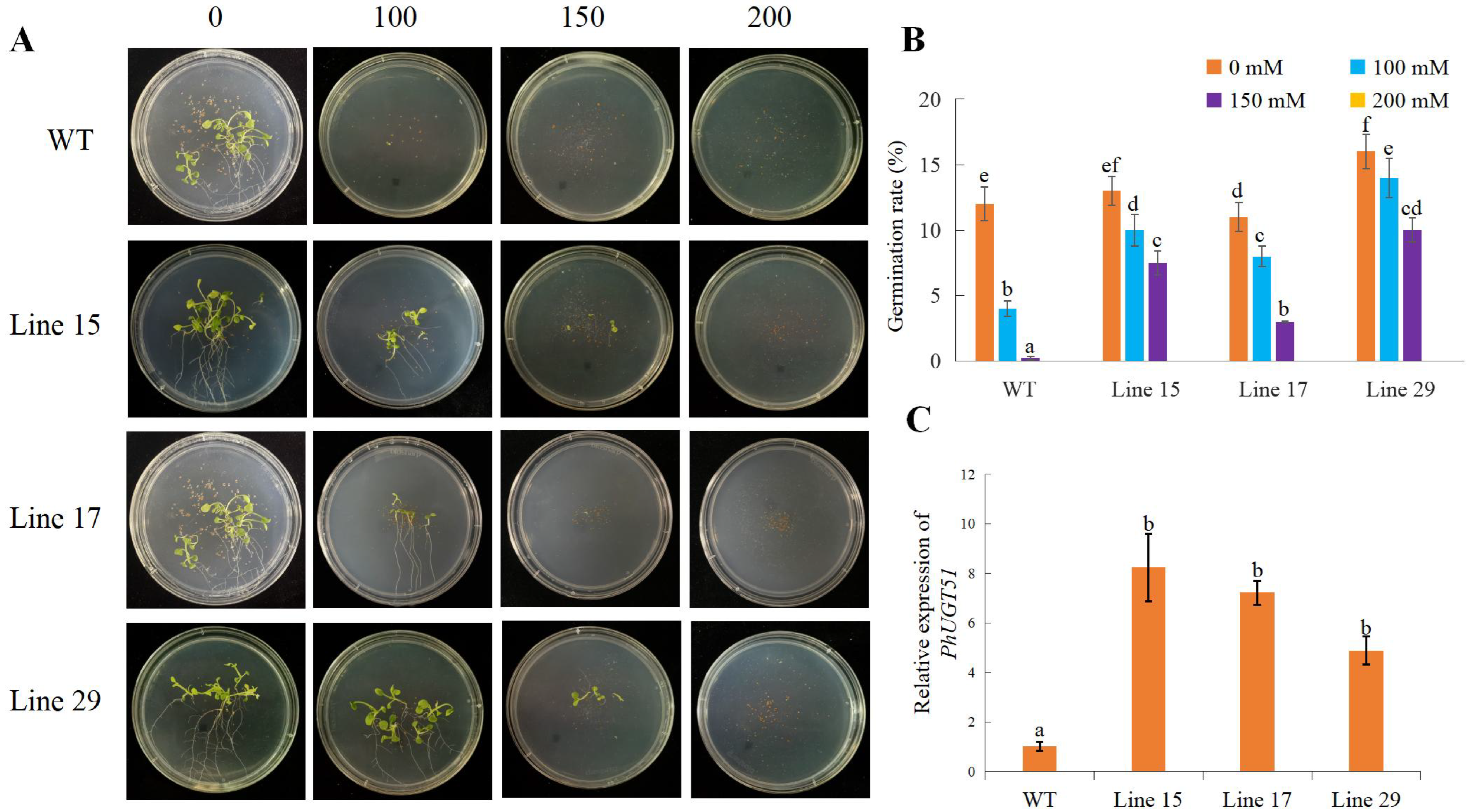
2.8. Phenotypes of Transgenic Petunia Plants Overexpressing PhUGT51
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Growth
5.2. Identification of UGT Genes in Petunia
5.3. Phylogenetic, Motif Distribution and Gene Structure Analysis
5.4. Expression Profile Analysis of Petunia UGT Genes
5.5. Analysis of the Cis-Regulatory Elements in PhUGT Genes
5.6. Subcellular Localization
5.7. Overexpression of PhUGT51 and Phenotype Analysis in Petunia
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shahid, M.A.; Sarkhosh, A.; Khan, N.; Balal, R.M.; Ali, S.; Rossi, L.; Gómez, C.; Mattson, N.; Nasim, W.; Garcia-Sanchez, F. Insights into the Physiological and Biochemical Impacts of Salt Stress on Plant Growth and Development. Agronomy 2020, 10, 938. [Google Scholar] [CrossRef]
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant Salt Stress: Adaptive Responses, Tolerance Mechanism and Bioengineering for Salt Tolerance. Bot. Rev. 2016, 82, 371–406. [Google Scholar] [CrossRef]
- Wilson, A.E.; Tian, L. Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 2019, 100, 1273–1288. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.; Isayenkova, J.; Lim, E.-K.; Poppenberger, B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 2005, 8, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; Jakubowska, A. Udp-Glycosyltransferases of Plant Hormones. Adv. Cell Biol. 2014, 4, 43–60. [Google Scholar] [CrossRef]
- Rahimi, S.; Kim, J.; Mijakovic, I.; Jung, K.-H.; Choi, G.; Kim, S.-C.; Kim, Y.-J. Triterpenoid-biosynthetic UDP-glycosyltransferases from plants. Biotechnol. Adv. 2019, 37, 107394. [Google Scholar] [CrossRef]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef] [PubMed]
- Dooner, H.K.; Nelson, O.E. Controlling element-induced alterations in UDPglucose: Flavonoid glucosyltransferase, the enzyme specified by the bronze locus in maize. Proc. Natl. Acad. Sci. USA 1977, 74, 5623–5627. [Google Scholar] [CrossRef]
- Kim, M.J.; Zheng, J.; Liao, M.H.; Jang, I.C. Overexpression of SrUGT76G1 in Stevia alters major steviol glycosides composition towards improved quality. Plant Biotechnol. J. 2019, 17, 1037–1047. [Google Scholar] [CrossRef]
- Langenbach, C.; Campe, R.; Schaffrath, U.; Goellner, K.; Conrath, U. UDP-glucosyltransferase UGT84A2/BRT1 is required for Arabidopsis nonhost resistance to the Asian soybean rust pathogen Phakopsora pachyrhizi. New Phytol. 2013, 198, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.; Garg, A.; Misra, R.C.; Chanotiya, C.S.; Ghosh, S. UGT86C11 is a novel plant UDP-glycosyltransferase involved in labdane diterpene biosynthesis. J. Biol. Chem. 2021, 297, 101045. [Google Scholar] [CrossRef]
- Sun, Y.; Ji, K.; Liang, B.; Du, Y.; Jiang, L.; Wang, J.; Kai, W.; Zhang, Y.; Zhai, X.; Chen, P.; et al. Suppressing ABA uridine diphosphate glucosyltransferase (SlUGT75C1) alters fruit ripening and the stress response in tomato. Plant J. 2017, 91, 574–589. [Google Scholar] [CrossRef]
- Wilson, A.E.; Wu, S.; Tian, L. PgUGT95B2 preferentially metabolizes flavones/flavonols and has evolved independently from flavone/flavonol UGTs identified in Arabidopsis thaliana. Phytochemistry 2019, 157, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kwon, C.S.; Woo, J.-Y.; Lee, G.-J.; Kim, Y.J.; Paek, K.-H. Suppression of UDP-glycosyltransferase-coding Arabidopsis thaliana UGT74E2 Gene Expression Leads to Increased Resistance to Psuedomonas syringae pv. tomato DC3000 Infection. Plant Pathol. J. 2011, 27, 170–182. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Van Aken, O.; Morreel, K.; Vandenbroucke, K.; van de Cotte, B.; De Clercq, I.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W.; et al. Perturbation of Indole-3-Butyric Acid Homeostasis by the UDP-Glucosyltransferase UGT74E2 Modulates Arabidopsis Architecture and Water Stress Tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef] [PubMed]
- Annotation, P.R. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2008, 36 (Suppl. S1), D1028–D1033. [Google Scholar]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463. [Google Scholar]
- Mamoon Rehman, H.; Amjad Nawaz, M.; Bao, L.; Hussain Shah, Z.; Lee, J.M.; Ahmad, M.Q.; Chung, G.; Yang, S.H. Genome-wide analysis of Family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. J. Plant Physiol. 2016, 206, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Liu, X.; Xu, K.; Zhang, B. Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (Citrus grandis) fruit. BMC Plant Biol. 2020, 20, 459. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Wei, G.; Zhou, H.; Gu, C.; Vimolmangkang, S.; Liao, L.; Han, Y. Unraveling the Mechanism Underlying the Glycosylation and Methylation of Anthocyanins in Peach. Plant Physiol. 2014, 166, 1044–1058. [Google Scholar] [CrossRef]
- Dong, L.; Liu, T.; Gao, D.; Li, J.; Qian, J. Isolation of the Histone Lysine Methyltransferase Gene PhSDG8 and Characterization of Function in Shoot Branching. J. Am. Soc. Hortic. Sci. 2020, 145, 299–307. [Google Scholar] [CrossRef]
- Banks, J.A.; Nishiyama, T.; Hasebe, M.; Bowman, J.L.; Gribskov, M.; de Pamphilis, C.; Albert, V.A.; Aono, N.; Aoyama, T.; Ambrose, B.A.; et al. The Selaginella Genome Identifies Genetic Changes Associated with the Evolution of Vascular Plants. Science 2011, 332, 960–963. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Gen. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Ji, X.; Jiao, Z.; Luo, Y.; Zhang, G.-Q.; Tao, S.; Lei, Z.; Zhang, J.; Wang, Y.; Liu, Z.-J.; et al. Functional analysis of a novel C-glycosyltransferase in the orchid Dendrobium catenatum. Hortic. Res. 2020, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Kadoo, N.Y.; Gupta, V.S. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genom. 2012, 13, 175. [Google Scholar] [CrossRef]
- Li, Y.; Liu, F.; Li, P.; Wang, T.; Zheng, C.; Hou, B. An Arabidopsis Cytokinin-Modifying Glycosyltransferase UGT76C2 Improves Drought and Salt Tolerance in Rice. Front. Plant Sci. 2020, 11, 560696. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, C.; Wang, Y.; Zhou, W.; Chen, J.; Yan, X.; Li, Z.; Huang, S.; Li, M.; Tang, Y.; et al. Overexpression of the glucosyltransferase gene BoaUGT74B1 enhances the accumulation of indole glucosinolates in Chinese kale. Sci. Hortic. 2021, 288, 110302. [Google Scholar] [CrossRef]
- Cheng, X.; Muhammad, A.; Li, G.; Zhang, J.; Cheng, J.; Qiu, J.; Jiang, T.; Jin, Q.; Cai, Y.; Lin, Y. Family-1 UDP glycosyltransferases in pear (Pyrus bretschneideri): Molecular identification, phylogenomic characterization and expression profiling during stone cell formation. Mol. Biol. Rep. 2019, 46, 2153–2175. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef]
- Zhang, K.; Sun, Y.; Li, M.; Long, R. CrUGT87A1, a UDP-sugar glycosyltransferases (UGTs) gene from Carex rigescens, increases salt tolerance by accumulating flavonoids for antioxidation in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 159, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, P.; Zhang, L.; Shu, J.; Court, M.H.; Sun, Z.; Jiang, L.; Zheng, C.; Shu, H.; Ji, L.; et al. Genome-wide analysis of the apple family 1 glycosyltransferases identified a flavonoid-modifying UGT, MdUGT83L3, which is targeted by MdMYB88 and contributes to stress adaptation. Plant Sci. 2022, 321, 111314. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Yang, T.; Qian, J.; Deng, X.; Dong, L. Genome-Wide Analysis and Exploration of WRKY Transcription Factor Family Involved in the Regulation of Shoot Branching in Petunia. Genes 2022, 13, 855. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Guo, Y.-L.; Yu, Y.; Yang, Z.; Qin, X.-T.; Ma, J.; Han, Y.; Yang, X.; Li, M.-Y. Over-expressing PMADS20-SRDX Repressor Leads to the Formation of Ectopic Trichome and Stoma on Petals and Pistils in Petunia. Acta Hortic. Sin. 2014, 41, 509–520. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, L.; Tang, Z.; Yang, T.; Hao, F.; Deng, X. Genome-Wide Analysis of UGT Genes in Petunia and Identification of PhUGT51 Involved in the Regulation of Salt Resistance. Plants 2022, 11, 2434. https://doi.org/10.3390/plants11182434
Dong L, Tang Z, Yang T, Hao F, Deng X. Genome-Wide Analysis of UGT Genes in Petunia and Identification of PhUGT51 Involved in the Regulation of Salt Resistance. Plants. 2022; 11(18):2434. https://doi.org/10.3390/plants11182434
Chicago/Turabian StyleDong, Lili, Ziyan Tang, Tianyin Yang, Fuling Hao, and Xinyi Deng. 2022. "Genome-Wide Analysis of UGT Genes in Petunia and Identification of PhUGT51 Involved in the Regulation of Salt Resistance" Plants 11, no. 18: 2434. https://doi.org/10.3390/plants11182434
APA StyleDong, L., Tang, Z., Yang, T., Hao, F., & Deng, X. (2022). Genome-Wide Analysis of UGT Genes in Petunia and Identification of PhUGT51 Involved in the Regulation of Salt Resistance. Plants, 11(18), 2434. https://doi.org/10.3390/plants11182434






