Small RNA Differential Expression Analysis Reveals miRNAs Involved in Dormancy Progression in Sweet Cherry Floral Buds
Abstract
1. Introduction
2. Results
2.1. Identification and Annotation of miRNAs during Dormancy in Prunus avium L. var. Bing by Next-Generation Sequencing
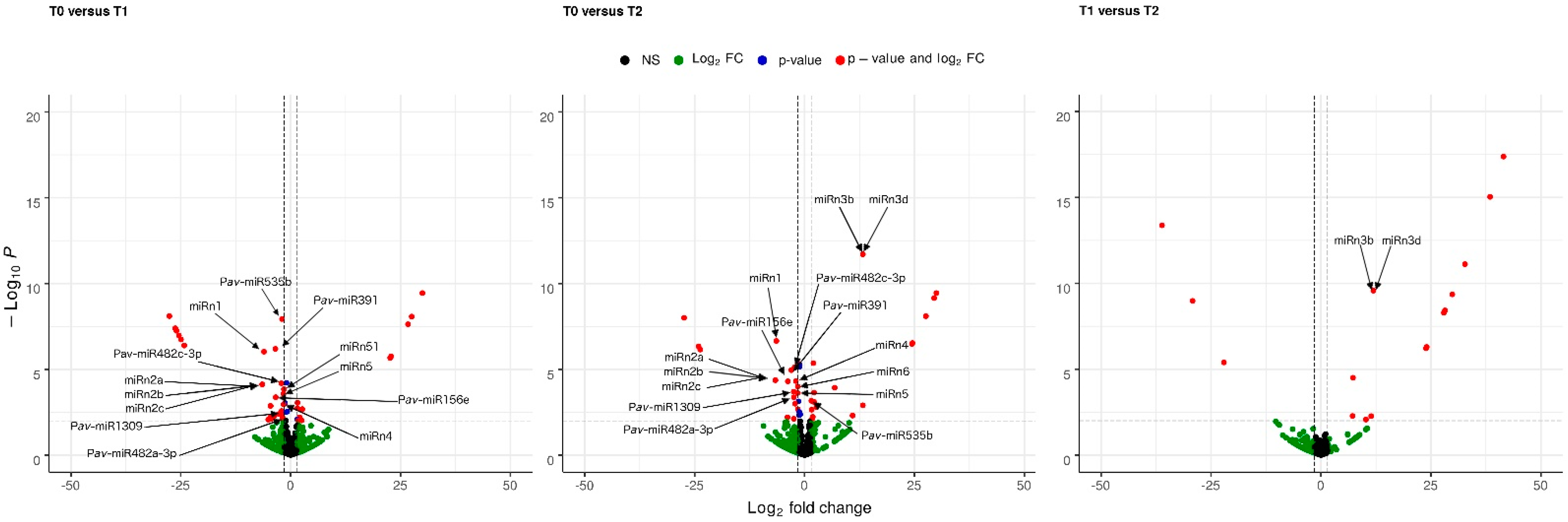
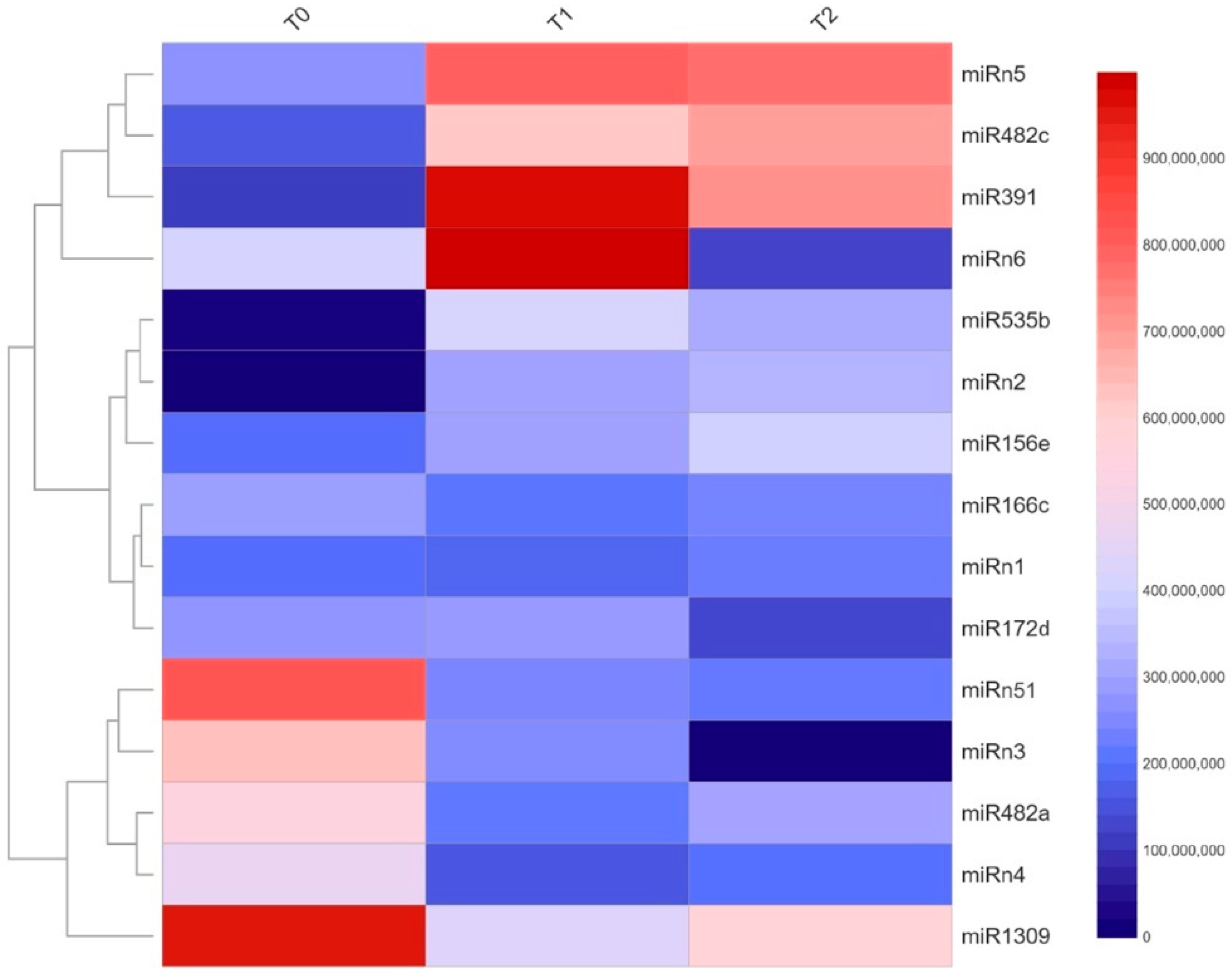
2.2. Expression Profiles of miRNAs during Dormancy in Prunus avium L. var. Bing
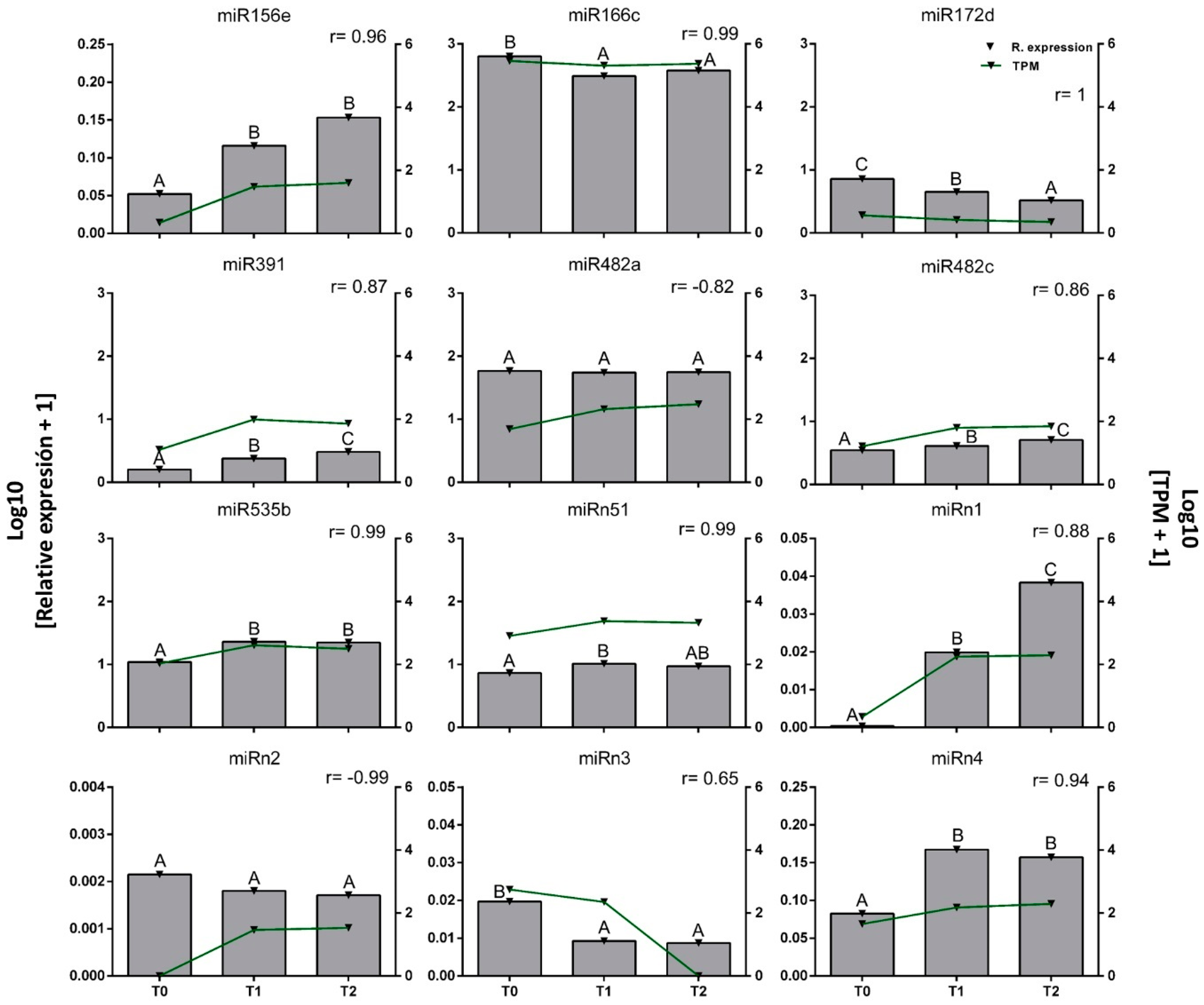
2.3. Effect of Environmental Fluctuations on the Levels of miR156e, miR172d, miR482c, miRn1, miRn2, and miRn3
2.4. Identification of the Target Genes of miRNAs in Prunus avium L. var. Bing
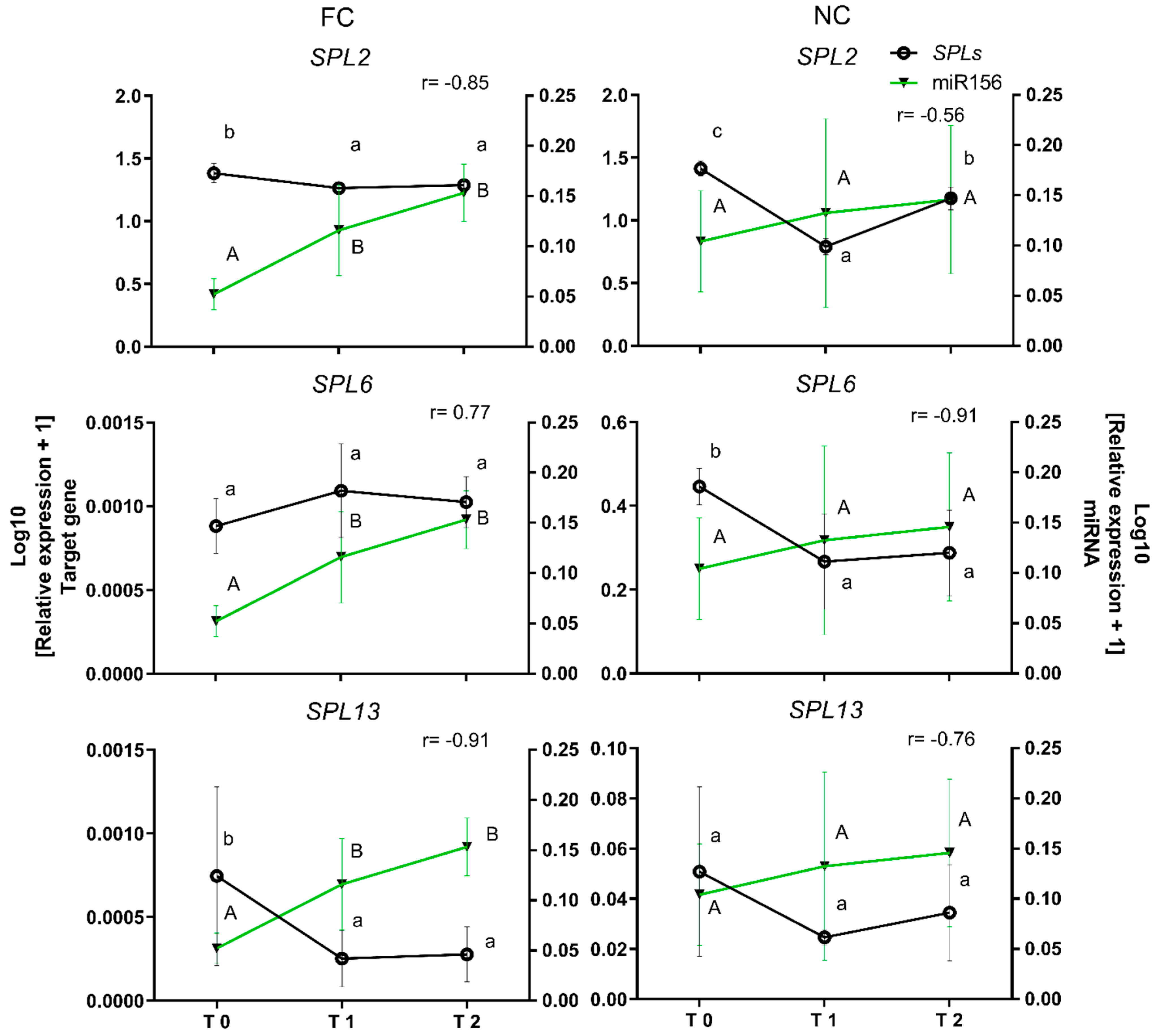
2.5. Functional Validation of miR156 and Its Target Genes SPL2, SPL6, and SPL13
3. Discussion
3.1. CR Determination of Prunus avium L. var. Bing
3.2. Identification of Dormancy Sensitive microRNAs in Prunus avium L. var. Bing by Massive Sequencing
3.3. Identification and Characterization of microRNAs and Target Genes during Dormancy in Prunus avium L. var. Bing
3.4. Effect of Environmental Fluctuations on the Expressions of miR156e, miR172d, miR482c, miRn1, miRn2, and miRn3 during Dormancy in Prunus avium L. var. Bing
3.5. Inverse Correlation between miR156e and the Target Genes SPL2, 6, and 13 under Field Chilling and Non-Stop Chilling Conditions during Dormancy in Prunus avium
3.6. Future Perspectives
3.7. Conclusions
4. Materials and Methods
4.1. Plant Material
4.2. Dormancy Analyses
4.3. Field Chilling (FC) Experiments and Temperature Data Collection
4.4. Chilling Requirement Determination under Forcing Conditions
4.5. Non-Stop Chilling (NC) Experiments
4.6. RNA Isolation
4.7. Genome-Wide Identification of Cherry miRNAs and Their Expression
4.8. Prediction of Target Genes
4.9. Experimental Determination of microRNAs and Target Genes
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fadón, E.; Herrero, M.; Rodrigo, J. Flower development in sweet cherry framed in the BBCH scale. Sci. Hortic. 2015, 192, 141–147. [Google Scholar] [CrossRef]
- Lang, G.A. Dormancy: A new universal terminology. HortScience 1987, 22, 817–820. [Google Scholar]
- Van der Schoot, C.; Rinne, P.L. Dormancy cycling at the shoot apical meristem: Transitioning between self-organization and self-arrest. Plant Sci. 2011, 180, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Campoy, J.A.; Ruiz, D.; Egea, J. Dormancy in temperate fruit trees in a global warming context: A review. Sci. Hortic. 2011, 130, 357–372. [Google Scholar] [CrossRef]
- Benmoussa, H.; Ghrab, M.; Ben Mimoun, M.; Luedeling, E. Chilling and heat requirements for local and foreign almond (Prunus dulcis Mill.) cultivars in a warm Mediterranean location based on 30 years of phenology records. Agric. For. Meteorol. 2017, 239, 34–46. [Google Scholar] [CrossRef]
- Ghrab, M.; Ben Mimoun, M.; Masmoudi, M.M.; Ben Mechlia, N. The behaviour of peach cultivars under warm climatic conditions in the Mediterranean area. Int. J. Environ. Stud. 2014, 71, 3–14. [Google Scholar] [CrossRef]
- Miranda, C.; Santesteban, G.; Royo, B.; De Pamplona, U. Variability in the relationship between frost temperature and injury level for some cultivated Prunus species. Hortscience 2005, 40, 357–361. [Google Scholar] [CrossRef]
- Fan, S.; Bielenberg, D.G.; Zhebentyayeva, T.N.; Reighard, G.; Okie, W.; Holland, D.; Abbott, A. Mapping quantitative trait loci associated with chilling requirement, heat requirement and bloom date in peach (Prunus persica). New Phytol. 2010, 185, 917–930. [Google Scholar] [CrossRef]
- Sánchez-Pérez, R.; Dicenta, F.; Martínez-Gómez, P. Inheritance of chilling and heat requirements for flowering in almond and QTL analysis. Tree Genet. Genomes 2012, 8, 379–389. [Google Scholar] [CrossRef]
- Castède, S.; Campoy, J.A.; García, J.Q.; Le Dantec, L.; Lafargue, M.; Barreneche, T.; Wenden, B.; Dirlewanger, E. Genetic determinism of phenological traits highly affected by climate change in Prunus avium: Flowering date dissected into chilling and heat requirements. New Phytol. 2014, 202, 703–715. [Google Scholar] [CrossRef]
- Lloret, A.; Badenes, M.L.; Ríos, G. Modulation of dormancy and growth responses in reproductive buds of temperate trees. Front. Plant Sci. 2018, 9, 1368. [Google Scholar] [CrossRef]
- Lloret, A.; Quesada-Traver, C.; Conejero, A.; Arbona, V.; Gómez-Mena, C.; Sánchez-Navarro, J.A.; Zuriaga, E.; Leída, C.; Badenes, M.L.; Ríos, G. Regulatory circuits involving bud dormancy factor PpeDAM6. Hortic. Res. 2021, 8, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Bennett, D.; Dardick, C.; Zhebentyayeva, T.; Abbott, A.G.; Liu, Z.; Staton, M.E. Genome-Wide Changes of Regulatory Non-Coding RNAs Reveal Pollen Development Initiated at Ecodormancy in Peach. Front. Mol. Biosci. 2021, 8, 61288. [Google Scholar] [CrossRef]
- Rothkegel, K.; Sánchez, E.; Montes, C.; Greve, M.; Tapia, S.; Bravo, S.; Prieto, H.; Almeida, A.M. DNA methylation and small interference RNAs participate in the regulation of MADS-box genes involved in dormancy in sweet cherry (Prunus avium L.). Tree Physiol. 2017, 37, 1739–1751. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed]
- Park, M.Y.; Wu, G.; Gonzalez-Sulser, A.; Vaucheret, H.; Poethig, R.S. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 3691–3696. [Google Scholar] [CrossRef]
- Rubio-Somoza, I.; Weigel, D. MicroRNA networks and developmental plasticity in plants. Trends Plant Sci. 2011, 16, 258–264. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and Their Regulatory Roles in Plant-Environment Interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Barakat, A.; Sriram, A.; Park, J.; Zhebentyayeva, T.; Main, D.; Abbott, A. Genome wide identification of chilling responsive microRNAs in Prunus persica. BMC Genom. 2012, 13, 481. [Google Scholar] [CrossRef]
- Niu, Q.; Li, J.; Cai, D.; Qian, M.; Jia, H.; Bai, S.; Hussain, S.; Liu, G.; Teng, Y.; Zheng, X. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 2016, 67, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Saito, T.; Ito, A.; Tuan, P.A.; Xu, Y.; Teng, Y.; Moriguchi, T. Small RNA and PARE sequencing in flower bud reveal the involvement of sRNAs in endodormancy release of Japanese pear (Pyrus pyrifolia ‘Kosui’). BMC Genom. 2016, 17, 230. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Ghazanfari, F.; Fadaei, A.; Ahmadi, L.; Shiran, B.; Rabei, M.; Fallahi, H. The Small-RNA Profiles of Almond (Prunus dulcis Mill.) Reproductive Tissues in Response to Cold Stress. PLoS ONE 2016, 11, e0156519. [Google Scholar]
- Garighan, J.; Dvorak, E.; Estevan, J.; Loridon, K.; Huettel, B.; Sarah, G.; Farrera, I.; Leclercq, J.; Grynberg, P.; Coiti Togawa, R.; et al. The Identification of Small RNAs Differentially Expressed in Apple Buds Reveals a Potential Role of the Mir159-MYB Regulatory Module during Dormancy. Plants 2021, 10, 2665. [Google Scholar] [CrossRef] [PubMed]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3' UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Hileman, L.C. Functional Evolution in the Plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) Gene Family. Front. Plant Sci. 2013, 4, 80. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Mathieu, J.; Yant, L.J.; Mürdter, F.; Küttner, F.; Schmid, M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009, 7, e1000148. [Google Scholar] [CrossRef]
- Jung, J.H.; Seo, P.J.; Kang, S.K.; Park, C.M. miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol. Biol. 2011, 76, 35–45. [Google Scholar] [CrossRef]
- Jung, J.H.; Seo, P.J.; Ahn, J.H.; Park, C.M. Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. J. Biol. Chem. 2012, 287, 16007–16016. [Google Scholar] [CrossRef]
- Cho, H.J.; Kim, J.J.; Lee, J.H.; Kim, W.; Jung, J.-H.; Park, C.-M.; Ahn, J.H. SHORT VEGETATIVE PHASE (SVP) protein negatively regulates miR172 transcription via direct binding to the pri-miR172a promoter in Arabidopsis. FEBS Lett. 2012, 586, 2332–2337. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, W.; Sun, L.; Zhao, F.; Huang, B.; Yang, W.; Tao, Y.; Wang, J.; Yuan, Z.; Fan, G.; et al. The genome of Prunus mume. Nat. Commun. 2012, 3, 1318. [Google Scholar] [CrossRef]
- Fogle, H.W.; Snyder, J.C.; Baker, H.; Cameron, H.R.; Cochran, L.C.; Schomer, H.A.; Yang, H.Y. Sweet Cherries: Production, Marketing, and Processing; USDA Agri. Handbook 442: Washington, DC, USA, 1973.
- Vimont, N.; Fouché, M.; Campoy, J.A.; Tong, M.; Arkoun, M.; Yvin, J.-C.; Wigge, P.A.; Dirlewanger, E.; Cortijo, S.; Wenden, B. From bud formation to flowering: Transcriptomic state defines the cherry developmental phases of sweet cherry bud dormancy. BMC Genom. 2019, 20, 974. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Chen, P.Y.; Zhong, S.; Dardick, C.; Callahan, A.; An, Y.-Q.; van Knocker, S.; Yang, Y.; Zhong, G.-Y.; Abbott, A.; et al. Thermal-responsive genetic and epigenetic regulation of DAM cluster controlling dormancy and chilling requirement in peach floral buds. Hortic. Res. 2020, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Liu, Y.; Zhu, A.; Wu, X.; Ye, J.; Yu, K.; Guo, W.; Deng, X. Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genom. 2010, 11, 246. [Google Scholar] [CrossRef]
- Zhu, H.; Xia, R.; Zhao, B.; An, Y.Q.; Dardick, C.D.; Callahan, A.M.; Liu, Z. Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 2012, 12, 149. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef]
- Bergonzi, S.; Albani, M.C.; Ver Loren van Themaat, E.; Nordström, K.J.V.; Wang, R.; Schneeberger, K.; Moerland, P.D.; Coupland, G. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 2013, 340, 1094–1097. [Google Scholar] [CrossRef]
- Esposito, S.; Aversano, R.; Bradeen, J.M.; Di Matteo, A.; Villano, C.; Carputo, D. Deep-sequencing of Solanum commersonii small RNA libraries reveals riboregulators involved in cold stress response. Plant Biol. 2020, 22, 133–142. [Google Scholar] [CrossRef]
- Zhou, M.; Tang, W. MicroRNA156 amplifies transcription factor-associated cold stress tolerance in plant cells. Mol. Genet. Genom. 2019, 294, 379–393. [Google Scholar] [CrossRef]
- Floyd, S.K.; Bowman, J.L. Gene regulation: Ancient microRNA target sequences in plants. Nature 2004, 428, 485–486. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Fukuda, H. HD-Zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol. 2003, 44, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jung, J.H.; Reyes, J.L.; Kim, Y.-S.; Kim, S.-Y.; Chung, K.-S.; Kim, J.A.; Lee, M.; Lee, Y.; Kim, V.N.; et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005, 42, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Gattolin, S.; Cirilli, M.; Pacheco, I.; Ciacciulli, A.; Linge, C.S.; Mauroux, J.-B.; Lambert, P.; Cammarata, E.; Bassi, D.; Pascal, T.; et al. Deletion of the miR172 target site in a TOE-type gene is a strong candidate variant for dominant double-flower trait in Rosaceae. Plant J. 2018, 96, 358–371. [Google Scholar] [CrossRef]
- Cirilli, M.; Rossini, L.; Chiozzotto, R.; Baccichet, I.; Florio, F.E.; Mazzaglia, A.; Turco, S.; Bassi, D.; Gattolin, S. Less is more: Natural variation disrupting a miR172 gene at the di locus underlies the recessive double-flower trait in peach (P. persica L. Batsch). BMC Plant Biol. 2022, 22, 318. [Google Scholar] [CrossRef]
- Wang, F.; Jing, Y.; Wang, Z.; Mao, T.; Šamaj, J.; Yuan, M.; Ren, H. Arabidopsis profilin isoforms, PRF1 and PRF2 show distinctive binding activities and subcellular distributions. J. Integr. Plant Biol. 2009, 51, 113–121. [Google Scholar] [CrossRef]
- Fei, Y.; Luo, C.; Tang, W. Differential expression of microRNAs during root formation in Taxus chinensis var. mairei cultivars. Open Life Sci. 2019, 14, 97–109. [Google Scholar] [CrossRef]
- Attri, K.; Zhang, Z.; Singh, A.; Sharrock, R.A.; Xie, Z. Rapid sequence and functional diversification of a miRNA superfamily targeting calcium signaling components in seed plants. New Phytol. 2022, 235, 1082–1095. [Google Scholar] [CrossRef]
- Shivaprasad, P.V.; Chen, H.M.; Patel, K.; Bond, D.M.; Santos, B.A.; Baulcombe, D.C. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef]
- Zhang, Y.; Waseem, M.; Zeng, Z.; Xu, J.; Chen, C.; Liu, Y.; Zhai, J.; Xia, R. MicroRNA482/2118, a miRNA superfamily essential for both disease resistance and plant development. New Phytol. 2021, 233, 2047–2057. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.; Lavagi-Craddock, I.; Bodaghi, S.; Vidalakis, G. Next-Generation Sequencing Identification and Characterization of MicroRNAs in Dwarfed Citrus Trees Infected With Citrus Dwarfing Viroid in High-Density Plantings. Front. Microbiol. 2021, 12, 646273. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Shen, Y.; Li, H.; Yanga, J.; Cai, X.; Zheng, G.; Zhu, Y.; Jia, B.; Sun, X. The multiple roles of OsmiR535 in modulating plant height, panicle branching and grain shape. Plant Sci. 2019, 283, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhang, Y.; Tang, R.; Qu, H.; Duan, X.; Jiang, Y. Banana sRNAome and degradome identify microRNAs functioning in differential responses to temperature stress. BMC Genom. 2019, 20, 33. [Google Scholar] [CrossRef]
- Morin, R.D.; Aksay, G.; Dolgosheina, E.; Ebhardt, H.A.; Magrini, V.; Mardis, E.R.; Sahinalp, S.C.; Unrau, P.J. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 2008, 18, 571–584. [Google Scholar] [CrossRef]
- Garcia, M.E.; Lynch, T.; Peeters, J.; Snowden, C.; Finkelstein, R. A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 2008, 67, 643–658. [Google Scholar] [CrossRef]
- Bassett, C.L.; Nickerson, M.L.; Farrell, R.E., Jr.; Artlip, T.S.; El Ghaouth, A.; Wilson, C.L.; Wisniewski, M.E. Characterization of an S-locus receptor protein kinase-like gene from peach. Tree Physiol. 2005, 25, 403–411. [Google Scholar] [CrossRef][Green Version]
- Egelund, J.; Obel, N.; Ulvskov, P.; Geshi, N.; Pauly, M.; Bacic, A.; Petersen, B.L. Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol. Biol. 2007, 64, 439–451. [Google Scholar] [CrossRef]
- Hyun, Y.; Richter, R.; Coupland, G. Competence to Flower: Age-Controlled Sensitivity to Environmental Cues. Plant Physiol. 2017, 173, 36–46. [Google Scholar] [CrossRef]
- Howe, G.T.; Horvath, D.P.; Dharmawardhana, P.; Priest, H.D.; Mockler, T.C.; Strauss, S.H. Extensive Transcriptome Changes During Natural Onset and Release of Vegetative Bud Dormancy in Populus. Front. Plant. Sci. 2015, 6, 989. [Google Scholar] [CrossRef]
- Mezzetti, B.; Arpaia, S.; Baraldi, E.; Dietz-Pfeilstetter, A.; Smagghe, G.; Ventura, V.; Sweet, J.B. Editorial: Advances and Challenges of RNAi Based Technologies for Plants—Volume 2. Front. Plant. Sci. 2022, 13, 930851. [Google Scholar] [CrossRef] [PubMed]
- Aranzana, M.J.; Decroocq, V.; Dirlewanger, E.; Eduardo, I.; Gao, Z.S.; Gasic, K.; Iezzoni, A.; Jung, S.; Peace, C.; Prieto, H.; et al. Prunus genetics and applications after de novo genome sequencing: Achievements and prospects. Hortic. Res. 2019, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, J.H. Chilling requirements of peach varieties. J. Am. Soc. Hortic. Sci. 1950, 56, 122–128. [Google Scholar]
- Davis, M.P.; van Dongen, S.; Abreu-Goodger, C.; Bartonicek, N.; Enright, A.J. Kraken: A set of tools for quality control and analysis of high-throughput sequence data. Methods 2013, 63, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–4670. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Bateman, A.; Marshall, M.; Khanna, A.; Eddy, S.R. Rfam: An RNA family database. Nucleic Acids. Res. 2003, 31, 439–441. [Google Scholar] [CrossRef]
- Cáceres-Molina, J.; Rothkegel, K.; Sánchez, E.; Carrasco-Valenzuela, T.; Meneses, C.; Prieto, H.; Almeida, A.M. A draft genome of Prunus avium ‘Karina’ as a tool for genomic studies. Acta Hortic. 2019, 1235, 85–92. [Google Scholar] [CrossRef]
- Lei, J.; Sun, Y. miR-PREFeR: An accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 2014, 30, 2837–2839. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids. Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Yan, G.; Zhou, Y.; Zhang, K. Over-expression of the PaAP1 gene from sweet cherry (Prunus avium L.) causes early flowering in Arabidopsis thaliana. J. Plant. Physiol. 2013, 170, 315–320. [Google Scholar] [CrossRef]


| FC 1 | NC 2 | ||
|---|---|---|---|
| Chilling Hours | Observed Bud Burst | Chilling Hours | Observed Bud Burst |
| 0 (T0) | 0% | 0 (T0) | 0% |
| 254 | 0% | 218 | 0% |
| 356 | 0% | 386 | 0% |
| 545 | 0% | 530 | 0% |
| 693 | 0% | 698 | 0% |
| 853 (T1) | 14% | 890 (T1) | 36% |
| 909 (T2) | 58% | 914 (T2) | 58% |
| 989 | 83% | 1250 | 87% |
| Name | Best Match miRBase | Mature Sequence (5’-3’) | miRNA Levels | Target(s) (psRNAtarget) | GO-Molecular Function | GO-Biological Process | Function |
|---|---|---|---|---|---|---|---|
| miR156e | ppe-miR156e | UGACAGAAGAGAGUGAGCAC | Increased | Squamosa promoter-binding-like protein | DNA binding; DNA binding transcription factor activity; metal ion binding | Defense response to bacterium; regulation of gene expression; regulation of transcription, DNA-templated; anther development (SPL13) | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5’-TNCGTACAA-3’. |
| miR166c | ppe-miR166c | UCGGACCAGGCUUCAUUCCCC | Decreased | Homeobox-leucine zipper protein ATHB-15 | DNA binding; lipid binding | Cell differentiation; regulation of transcription, DNA-templated | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulate procambial and vascular tissue formation or maintenance and vascular development in inflorescence stems. |
| miR172d | ppe-miR172d | GGAAUCUUGAUGAUGCUGCAG | Decreased | AP2-like ethylene-responsive transcription factor | DNA binding; DNA binding transcription factor activity | Ethylene-activated signaling pathway; multicellular organism development; transcription, DNA templated | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (by similarity). May negatively regulate the transition to flowering time and confers flowering time delay. |
| miR391 | mdm-miR391 | UACGCAGGAGAGAUGGCGCUG | Increased | Profilin | Actin monomer binding | Actin polymerization or depolymerization; inflorescence development; lateral root development; leaf development; sequestering of actin monomers; unidimensional cell growth | Binds to actin and affects the structure of the cytoskeleton. At high concentrations, profilin prevents the polymerization of actin, whereas it enhances it at low concentrations. By binding to PIP2, it inhibits the formation of IP3 and DG (by similarity). |
| miR482a | ppe-miR482a-3p | UUUCCGAAACCUCCCAUUCCAA | unchanged | Disease resistance protein At4g27190-like | ADP binding; ATP binding | Defense response; signal transduction | Disease resistance protein |
| miR482c | ppe-miR482c-3p | UUGCCAACCCCGCCCAUUCCAA | Increased | Putative disease resistance RPP13-like protein 1 | ATP binding | Plant-type hypersensitive responser; signal transduction | Potential disease resistance protein |
| miR535b | ppe-miR535d | UUGACGACGAGAGAGAGCACG | Increased | Periodic tryptophan protein 2 homolog | RNA binding; snoRNA binding | Maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA); ribosomal small subunit assembly; rRNA processing | Not in plants |
| miR1309 | pta-miR1309 | UUGAUGGACCAUUUGAAUGAA | not validated | WD repeat-containing protein 7 | Hematopoietic progenitor cell differentiation | / | Not present in plants |
| miRn51 | / | UUUGGCGCGUUGCUGUGGAUU | Increased | Metal transporter Nramp5-like | / | Metal ion transmembrane transporter activity | / |
| miRn1 | / | CAUAGGAUGCUUAGGAAACUU | Increased | Putative receptor protein kinase | ATP binding; protein serine/threonine kinase activity | Recognition of pollen, self-incompatibility | Probable receptor. Interaction with a ligand in the extracellular domain triggers the protein kinase activity of the cytoplasmic domain. |
| miRn2 | / | UAGUCAAUUAAUGAGGAUUAGU | unchanged | n.a | / | / | / |
| miRn3 | / | UUUUCUGAAGCAUUUGGCAUC | Decreased | Ninja-family protein [AFP3] | / | Signal transduction; protein binding | Acts as a negative regulator of abscisic acid (ABA) response and stress responses. |
| miRn4 | / | UUCUUGGAGGCAUGAAGCACC | Increased | Arabinosyltransferase RRA3-like | Transferase, transfering glycosyl groups | Cell wall biogenesis; cell wall organization; root hair cell development | Plays a role in the arabinosylation of cell wall components. Involved in the arabinosylation of extensin proteins in root hair cells. Extensins are structural glycoproteins present in cell walls, and their arabinosylation is important for root hair cell development and root hair tip growth. |
| miRn5 | / | UGGCAUCGAGGACGAACAGCU | not validated | psbP domain-containing protein 7, chloroplastic | Calcium ion binding | Photosynthesis | / |
| miRn6 | / | UUACAAAGUAUCUUAUGGGUCU | not validated | Pentatricopeptide repeat-containing protein At1g32415, mitochondrial | Endonuclease activity; RNA binding | RNA modification | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soto, E.; Sanchez, E.; Nuñez, C.; Montes, C.; Rothkegel, K.; Andrade, P.; Prieto, H.; Almeida, A.M. Small RNA Differential Expression Analysis Reveals miRNAs Involved in Dormancy Progression in Sweet Cherry Floral Buds. Plants 2022, 11, 2396. https://doi.org/10.3390/plants11182396
Soto E, Sanchez E, Nuñez C, Montes C, Rothkegel K, Andrade P, Prieto H, Almeida AM. Small RNA Differential Expression Analysis Reveals miRNAs Involved in Dormancy Progression in Sweet Cherry Floral Buds. Plants. 2022; 11(18):2396. https://doi.org/10.3390/plants11182396
Chicago/Turabian StyleSoto, Esteban, Evelyn Sanchez, Carlos Nuñez, Christian Montes, Karin Rothkegel, Paola Andrade, Humberto Prieto, and Andrea Miyasaka Almeida. 2022. "Small RNA Differential Expression Analysis Reveals miRNAs Involved in Dormancy Progression in Sweet Cherry Floral Buds" Plants 11, no. 18: 2396. https://doi.org/10.3390/plants11182396
APA StyleSoto, E., Sanchez, E., Nuñez, C., Montes, C., Rothkegel, K., Andrade, P., Prieto, H., & Almeida, A. M. (2022). Small RNA Differential Expression Analysis Reveals miRNAs Involved in Dormancy Progression in Sweet Cherry Floral Buds. Plants, 11(18), 2396. https://doi.org/10.3390/plants11182396






