Advancement in the Breeding, Biotechnological and Genomic Tools towards Development of Durable Genetic Resistance against the Rice Blast Disease
Abstract
1. Introduction
2. Economic Impacts of Rice Blast
3. Pathogenesis of the Causal Fungus Magnoporthe oryzae
3.1. Attachment and Germination
3.2. Germ Tube Elongation and Recognition of Host
3.3. Appressorium Formation and Maturation
3.4. Penetration Peg Formation and Invasion
3.5. Invasion and Defense Suppression
4. Pathogenomics of Magnaporthe oryzae
5. Genetics of Blast Disease Resistance in Rice
6. Disease Screening Protocols for Blast Resistance in Rice
6.1. Field Screening Technique
6.2. Screening Techniques under Greenhouse/Polyhouse/Controlled Conditions
6.3. Molecular-Marker-Based Screening of Rice Genotypes for Blast Resistance
7. Strategies to Develop Durable Resistance in Rice Genotypes against the Blast Disease
7.1. Conventional Breeding Strategies for Developing Resistance against Rice Blast
7.2. Mutation Breeding for Blast Resistance in Rice
7.3. Molecular-Marker-Based Approaches for Resistance to Rice Blast Disease
| S. No. | Name of Blast-Resistant Genes | Name of Source Genotype | Reporting Year | Chromosome Number | Genomic Position (Mb) | Name of Linked Molecular Marker | Name of Country | References |
|---|---|---|---|---|---|---|---|---|
| 1 * | Pb2 | Jiangnanwan | 2022 | 11 | 1.47 | SNP | China | Yu et al. [15] |
| 2 | Pi67 | Tetep | 2019 | 12 | 12.09 | SSR | India | Joshi et al. [186] |
| 3 | Pi57(t) | IL-E1454 | 2017 | 12 | 10.8 | SSR, STS | Myanmar | Dong et al. [187] |
| 4 | Pi65(t) | Gangyu 129 | 2016 | 11 | 28.22 | SNP, InDel | – | Zheng et al. [114] |
| 5 | Pi-jnw1 | Jiangnanwan | 2016 | 11 | 27.36 | SSR, InDel | – | Wang et al. [188] |
| 6 | Pi66(t) | AS20-1 | 2016 | 3 | 26.78 | SSR | Australia | Liang et al. [189] |
| 7 | Pita3(t) | IRBLta2-Re | 2015 | 12 | 9.89 | SSR | – | Chen et al. [190] |
| 8 * | Pik-e | Xiangzao 143 | 2015 | 11 | 28 | SSR, InDel | China | Chen et al. [190] |
| 9 | Pi-h2(t) | HR4 | 2015 | 1 | 7.9 | SSR | India | Xiao et al. [191] |
| 10 | Pih3(t) | HR4 | 2015 | 12 | 12.95 | SSR | India | Xiao et al. [191] |
| 11 | Pi-h1(t) | HR4 | 2015 | 11 | 28.11 | SSR, InDel | India | Xiao et al. [191] |
| 12 * | Pi64 | Yangmaogu | 2015 | 1 | 32.31 | SSR, InDel | Japan | Ma et al. [192] |
| 13 | Pitb | Zixuan | 2013 | 12 | 9.37 | SSR, InDel | – | Sun et al. [193] |
| 14 | Pi61(t) | 93-11 | 2013 | 12 | 9.98 | InDel, SSR | China | Lei et al. [194] |
| 15 | Pi60(t) | 93-11 | 2013 | 11 | 6.62 | SSR, InDel | China | Lei et al. [194] |
| 16 | Pi58(t) | Haoru | 2013 | 12 | 10.42 | SSR | Myanmar | Koide et al. [195] |
| 17 | Pi51(t) | D69 | 2012 | 6 | 10.38 | InDel, SSR | – | Xiao et al. [196] |
| 18 * | Pi50(t) | EBZ, EBZ × LTH F2 and (EBZ × LTH) × LTH, BC1F2 | 2012 | 6 | 10.41 | SSR, CAPS | – | Zhu et al. [197] and Jiang et al. [198] |
| 19 | Pi-hk1 | Heikezijing | 2012 | 11 | 27.66 | SSR | – | Wu et al. [199] and Liu et al. [200] |
| 20 | Pihk2 | Heikezijing | 2012 | 9 | 10.17 | SSR, InDel | – | He et al. [201] |
| 21 | Pias(t) | Asominori | 2012 | 4 | 31.26 | SSR, CAPS | China | Endo et al. [202] |
| 22 | Pi51(t) | Tianjingyeshengdao | 2012 | 12 | 11.95 | SSR, SFP | China | Wang et al. [203] |
| 23 | pi55(t) | Yuejingsimiao 2 | 2012 | 8 | 25.58 | SSR, STS | China | He et al. [201] |
| 24 | Pi46(t) | H4 | 2011 | 11 | 27.74 | SSR, InDel | – | Xiao et al. [204] |
| 25 | Pi-48 | Xiangzi 3150 | 2011 | 12 | 11.95 | SSR | China | Huang et al. [205] |
| 26 * | Pi-a | Aichi Asahi | 2011 | 11 | 6.49 | SSR, InDel | Japan | Zeng et al. [206] |
| 27 | Pi-45(t) | Moroberekan | 2011 | 4 | 31.49 | SSR | Japan | Kim et al. [148] |
| 28 | Pi-42(t) | DHR9 | 2010 | 12 | 10.62 | RAPD, SSR, STS | India | Kumar et al. [207] |
| 29 | Pi43(t) | Zhe733 | 2009 | 11 | 27.67 | SSR | – | Lee et al. [208] |
| 30 | Pi-41 | 93-11 | 2009 | 12 | 16.74 | SSR, STS | China | Yang et al. [209] |
| 31 * | Pid3 | Digu | 2009 | 6 | 13.05 | STS | China | Shang et al. [210] |
| 32 * | Pik-p | K60, HR22 | 2009 | 11 | 28.05 | SSR, CAPS | China | Wang et al. [211] |
| 33 | Pi2-2 | Jefferson | 2008 | 6 | 10.2 | SSR | – | Jiang et al. [198] and Ballini et al. [179] |
| 34 * | Pikahei- 1(t) | Kahei | 2008 | 4 | 31.67 | SSR, SNP | – | Xu et al. [212] |
| 35 * | Pik-h | IRBLkh-K3, HP2216, and Tetep | 2008 | 11 | 24.99 | SNP | India | Xu et al. [213] |
| 36 | Pir2-3(t) | IR64 | 2008 | 2 | – | SSR | Indonesia | Dwinita et al. [214] |
| 37 | Pirf2-1(t) | O. rufipogon | 2008 | 2 | – | SSR | Indonesia | Dwinita et al. [214] |
| 38 | Pi39(t) | Chubu 111, Q15 | 2007 | 4, 12 | – | SSR | China | Liu et al. [215] |
| 39 | Pi-39(t) | Mineasahi and Chubu 111 | 2007 | 4 | 32.68 | SSR | China | Liu et al. [215] |
| 40 | Pi-39 | Q-15 and Chubu 111 | 2007 | 12 | 10.61 | SSR | China | Liu et al. [215] |
| 41 | Pi-34 | Chubu-32 | 2007 | 11 | 19.96 | SSR | Japan | Zenbayashi et al. [216] |
| 42 | Pi-40(t) | IR65482, Co39, and O. australiensis (W) | 2007 | 6 | 9.86 | STS, SSR | Philippines | Jeung et al. [217] |
| 43 | Piz-5 | C101A51_CO39 | 2006 | 6 | – | – | – | Deng et al. [218] |
| 44 * | Pi9 | Cultivar TP309 | 2006 | 6 | 10.39 | – | – | Qu et al. [219] and Koide et al. [195] |
| 45 * | Pid2 | Digu | 2006 | 6 | 17.16 | CAPS | China | Chen et al. [220] |
| 46 * | Pigm(t) | Gumei 4 | 2006 | 6 | 10.36 | CAPS, InDel | China | Deng et al. [218] |
| 47 | Pi51(t) | Tianjingyeshengdao | 2006 | 12 | – | – | China | Qu et al. [219] |
| 48 | Pi2-1 | Tianjingyeshengdao | 2006 | 6 | 10.08 | SSR, SFP | China | Wang et al. [203] and Qu et al. [219] |
| 49 | Pi24(t) | Azuenca | 2006 | 1 | 5.24 | SSR | France | Nguyen et al. [221] |
| 50 | Pi-38 | Tadukan | 2006 | 11 | 22.48 | SSR, AFLP | India | Gowda et al. [222] |
| 51 * | Pi35(t) | Hokkai 188 | 2006 | 1 | 32.1 | SSR | Japan | Nguyen et al. [221] |
| 52 * | Pi-b | Tohoku, Koshihikari | 2006 | 2 | 35.1 | SNP | Japan | Hayashi et al. [223] |
| 53 * | Piz-t | Toride No. 1 | 2006 | 6 | 10.39 | STS | Japan | Zhou et al. [224] |
| 54 | Pi59(t) | Haoru_US-2 | 2006 | 6 | 10.82 | SSR | Myanmar | Koide et al. [195] and Zhou et al. [224] |
| 55 | Pi-9(t) | IR31917 | 2006 | 6 | 10.38 | STS | Philippines | Qu et al. [219] |
| 56 | Pi-Da(t) | Dacca 6 | 2005 | 2 | 2.21 | SSR | – | Lei et al. [225] |
| 57 * | Pi 37(t) | Cultivar St. No. 1 | 2005 | 1 | 33.1 | SSR | China | Chen et al. [226] |
| 58 | Pi26(t) | Gumei 2 | 2005 | 6 | 11.06 | RFLP, SSR | China | Wu et al. [227] |
| 59 * | Pi-36(t) | Q61 | 2005 | 8 | 2.87 | SSR, CRG | China | Liu et al. [228] |
| 60 * | Pi54 | Tetep | 2005 | 11 | 25.26 | SSR | India | Sharma et al. [121] |
| 61 | PiGD-2(t) | Sanhuangzhan 2 | 2004 | 10 | – | SSR, RFLP, RGA | – | Liu et al. [229] |
| 62 | Pi-d1(t) | Digu | 2004 | 2 | 34.94 | SSR, RFLP | China | Chen et al. [230] |
| 63 | Pi-dt(2) | Digu | 2004 | 6 | 17.16 | SSR, RGA | China | Chen et al. [230] |
| 64 | Pig(t) | Guangchangzhan | 2004 | 2 | 34.34 | SSR | China | Zhou et al. [231] |
| 65 | Pi15 | Q61 and GA25 | 2004 | 9 | 9.61 | SSR, CRG | China | Liu et al. [229] |
| 66 | PiGD-1(t) | Sanhuangzhan 2 | 2004 | 8 | 16.37 | SSR, RFLP, RGA | China | Liu et al. [229] and He et al. [201] |
| 67 | PiGD-3(t) | Sanhuangzhan 2 | 2004 | 12 | 14.45 | SSR, RFLP, RGA | China | Liu et al. [229] |
| 68 | Pi-y2(t) | Yanxian No. 1 | 2004 | 2 | 35.03 | SSR | China | Fukuta [232] and Lei et al. [225] |
| 69 | Pi-y1(t) | Yanxian No. 1 | 2004 | 2 | 35.03 | SSR | China | Fukuta [232] and Lei et al. [225] |
| 70 | Pi27(t) | Q14 and Q61 | 2004 | 1 | 5.55 | SSR | France | Zhu et al. [233] |
| 71 * | Pi-tp(t) | CO39 and Tetep | 2004 | 1 | 25.13 | SSR | India | Barman et al. [234] |
| 72 | Pi-sh | Akihikari and Shin 2 | 2004 | 1 | 33.3 | SSR | Japan | Fukuta [232] |
| 73 * | Pik-s | Shin 2 | 2004 | 11 | 27.31 | SSR | Japan | Fjellstrom et al. [235] |
| 74 | Pi28(t) | Azucena, IR64 | 2003 | 10 | 21.04 | RFLP, RAPD | – | Sallaud et al. [236] |
| 75 * | Pi56(t) | SHZ-2 | 2003 | 9 | 9.77 | SSR, CRG, SNP | – | Jeon et al. [237] |
| 76 | Pizh | Zhai-Ya-Quing8 | 2003 | 8 | 4.38 | – | China | Sallaud et al. [236] |
| 77 | Pi-25(t) | IR64 | 2003 | 2 | 34.36 | QTL | France | Sallaud et al. [236] and Nguyen et al. [221] |
| 78 | Pi27(t) | IR64 | 2003 | 6 | 6.92 | RFLP | France | Sallaud et al. [236] |
| 79 | Pi26(t) | IR64 | 2003 | 5 | 2.78 | RFLP, RAPD | France | Sallaud et al. [236] |
| 80 | Pi-32(t) | IR64 | 2003 | 12 | 21.24 | RFLP, RAPD | France | Sallaud et al. [236] |
| 81 | Pi-31(t) | IR64 | 2003 | 12 | 11.93 | RFLP, RAPD, | France | Sallaud et al. [236] |
| 82 | Pi-29(t) | IR64 | 2003 | 8 | 13.93 | RFLP, RAPD, Isozyme | France | Sallaud et al. [236] and Nguyen et al. [221] |
| 83 | Pi-30(t) | IR64 | 2003 | 11 | 4.41 | RFLP, RAPD, Isozyme | France | Sallaud et al. [236] and Nguyen et al. [221] |
| 84 | Pi-33 | IR64, Bala | 2003 | 8 | 7.56 | SSR, RFLP | France | Berruyer et al. [238] and Sallaud et al. [236] |
| 85 | Pii2 | Ishikari Shiroke | 2003 | 9 | 1.03 | – | Japan | Pan et al. [239], Kinoshita and Kiyosawa [240] |
| 86 * | Pi-5(t) | RIL249, Moroberekan | 2003 | 9 | 9.77 | AFLP, RFLP, CAPS | Philippines | Jeon et al. [237] |
| 87 * | Pi2 | 5173, C101A51 | 2002 | 6 | 10.39 | SSR, STS, RFLP | – | Jiang and Wang [241] and Zhou et al. [224] |
| 88 | Pi-24(t) | Zhong 156 | 2002 | 12 | 10.6 | RFLP, RAPD, RGA | – | Zhuang et al. [242] |
| 89 * | Pi-CO39(t) | Co39 | 2002 | 11 | 6.66 | SSR, RFLP | USA | Chauhan et al. [243] and Huang et al. [205] |
| 90 * | Pi25 | Gumei 2 | 2001 | 6 | 18.09 | – | China | Zhuang et al. [242] |
| 91 | Pi-25(t) | Gumei 2 | 2001 | 6 | 12.33 | RFLP, RGA, SSR | China | Wu et al. [199] and Zhuang et al. [242] |
| 92 | PBR | St. No. 1 | 2001 | 11 | – | RFLP, SSR | Japan | Fukuoka and Okuno [244] |
| 93 | Pi-47 | Xiangzi 3150 | 2000 | 11 | 27.67 | SSR | China | Huang et al. [205] and Ahn et al. [245] |
| 94 | Pi18 | Suweon365 | 2000 | 11 | 28.93 | RFLP | Korea | Ahn et al. [245] |
| 95 | Pi-lm2 | Lemont, Teqing | 2000 | 11 | 28.93 | RFLP | USA | Tabien et al. [246] |
| 96 | Pi-tq5 | Teqing | 2000 | 2 | 34.61 | RFLP | USA | Tabien et al. [246] Tabien et al. [247] and Zhou et al. [231] |
| 97 | Pi-tq1 | Teqing | 2000 | 6 | 29.02 | RFLP | USA | Tabien et al. [246] |
| 98 | Pi-tq6 | Teqing | 2000 | 12 | 7.73 | RFLP | USA | Tabien et al. [246] |
| 99 | Pi49 | Mowanggu | 1999 | 11 | 28.8 | SSR | – | Sun et al. [193] and Chen et al. [248] |
| 100 | Pi-16(t) | AUS373 | 1999 | 2 | 34.94 | RFLP, Isozyme | Japan | Pan et al. [249] and Zhou et al. [231] |
| 101 * | Pb1 | Modan | 1999 | 11 | 21.71 | – | Japan | Fujii et al. [250] and Hayashi et al. [251] |
| 102 | Pi-44(t) | Moroberekan | 1999 | 11 | 28.93 | RFLP, STS, AFLP | USA | Chen et al. [248] and Chauhan et al. [243] |
| 103 | Pi12 | Hong Jiao Zhan K80-R-Hang Jiao-Zhan | 1998 | 12 | 7.73 | RFLP | Japan | Zhuang et al. [252] |
| 104 | Pi-19(t) | IRBL19-A and Aichi Asahi | 1998 | 12 | 10.73 | SSR | Japan | Koide et al. [195] and Hayashi et al. [253] |
| 105 | Pi-14(t) | Maowangu | 1998 | 2 | 34.94 | RFLP, Isozyme | Japan | Pan et al. [254] and Zhou et al. [231] |
| 106 * | Pi3(t) | Pai-kan-tao | 1997 | 9 | 7.8 | – | – | Kinoshita and Kiyosawa [240] |
| 107 * | pi-21 | Owarihatamochi | 1997 | 4 | 19.81 | RFLP, SSR | Japan | Fukuoka and Okuno [244], Ahn et al. [255], and Pan et al. [254] |
| 108 | Pita-2 | Yashiromochi, Pi No. 4 | 1997 | 12 | 10.6 | RFLP, RAPD, SNP | Japan | Hayashi et al. [223] |
| 109 | Pi22 | Suweon 365 | 1997 | 6 | 4.89 | RFLP | Korea | Ahn et al. [255], Terashima et al. [256] |
| 110 | Pi23 | Suweon 365 | 1997 | 5 | 10.75 | RFLP, SSR | Korea | Ahn et al. [255], Rybka et al. [257] |
| 111 | Pi-20(t) | IR64 | 1997 | 12 | 12.95 | SSR | Philippines | Li et al. [258] and Imbe et al. [259] |
| 112 * | Pita | Tadukan, Yashiromochi | 1997 | 12 | 10.6 | RFLP, RAPD, SNP | USA | Rybka et al. [257], Hayashi et al. [223] and Bryan et al. [119] |
| 113 * | Pi-k | Kusabue, Kanto 51 | 1996 | 11 | 28.01 | RFLP, InDel, SNP | China | Hayasaka et al. [260] and Hayashi et al. [223] |
| 114 * | Pik-m | Tohoku IL4, Tsuyuake | 1996 | 11 | 28 | RFLP, SSR | China | Kaji and Ogawa [261] |
| 115 | Pi157 | Moroberekan | 1996 | 12 | 12.37 | RFLP | India | Naqvi et al. [262] |
| 116 * | Pii1 | Fujisaka 5 | 1996 | 6 | 2.29 | – | Japan | Pan et al. [263] |
| 117 | Pikg | GA20 | 1996 | 11 | 27.31 | – | Japan | Pan et al. [263] |
| 118 * | Pit | K-59, Tjahaja, K-59 | 1996 | 1 | 2.27 | RFLP, SNP | Japan | Kaji and Ogawa [261] and Hayashi et al. [223] |
| 119 | Pi8 | Kasalath | 1996 | 6 | 11.36 | leucine aminopeptidase, phosphoglucose isomerase, RFLP | Japan | Pan et al. [263] |
| 120 | Pi62(t) | Yashiromochi | 1996 | 12 | 7.73 | RAPD, RFLP | Japan | Wu et al. [264] |
| 121 | Pi62(t) | Yashiromochi | 1996 | 12 | 7.73 | RAPD, RFLP | Japan | Wu et al. [264] |
| 122 | Pi-17(t) | DJ 123 | 1996 | 7 | 22.25 | leucine aminopeptidase, phosphoglucose isomerase | Philippines | Pan et al. [263] and Zhu et al. [197] |
| 123 | Pib2 | Lemont | 1996 | 11 | 26.79 | – | Philippines | Tabien et al. [265] (1996) |
| 124 | Pitq3 | Teqing | 1996 | 3 | – | USA | Tabien et al. [265] | |
| 125 | Pitq2 | Teqing | 1996 | 2 | – | USA | USA | Tabien et al. [265] |
| 126 | Pitq4 | Teqing | 1996 | 4 | USA | USA | Tabien et al. [265] | |
| 127 | Pik-l | Liziangxintuanheigu, Kusabue | 1995 | 11 | 27.69 | SSR, STS, CAPS | China | Hayasaka et al. [260] and Hayashi et al. [223] |
| 128 | Pi-10(t) | Tongil | 1995 | 5 | 14.52 | RAPD | India | Naqvi et al. [265] and Wu et al. [227] |
| 129 * | Pi-1(t) | Apura, C101LAC | 1995 | 11 | 28 | STS, RFLP, SSR, CAPS | USA | Parco [266], Yu et al. [267] |
| 130 | Pi(t) | P167 | 1994 | 4 | 2.27 | – | – | Causse et al. [268] |
| 131 | Pi-11(t) | Zhai-Ye-Quing | 1994 | 8 | 13.93 | RFLP, RAPD | China | Causse et al. [268] |
| 132 | Pi-6(t) | Apura | 1994 | 12 | 7.73 | RFLP | USA | McCouch et al. [269] |
| 133 | Pi-7(t) | RIL29 (Moroberekan) | 1994 | 11 | 18.64 | 12.37 | USA | Wang et al. [270] |
| 134 | Pi26(t) | Azucena/Gumei 2 | 1993 | 5 | 2.07 | – | France | Wu and Tanksley [271] Ahn et al. [255] and Nguyen et al. [221] |
| 135 | Pi-13 | O. minuta (W), Kasalath | 1992 | 6 | 15.83 | SSR | Philippines | Amante- Bordeos et al. [272] |
| 136 | Pi3(t) | Pai-kan-tao | 1992 | 6 | – | – | Philippines | Mackill and Bonman [273] |
| 137 | Pi1 | LAC23 | 1991 | 11 | 26.49 | RFLP | Philippines | Yu et al. [274] |
| 138 | Pikur2 | Kuroka | 1988 | 11 | 2.84 | – | Japan | Goto [275] |
| 139 * | Pish | Nipponbare | 1985 | 11 | 33.38 | – | Japan | Imbe and Matsumoto [276] |
| 140 | Mpiz | Zenith | 1976 | 11 | 4.07 | – | Japan | Goto [277] |
| 141 | Piz | Zenith, Fukunishiki, Toride 1, Tadukan | 1976 | 6 | 10.39 | STS | Japan | Goto [277] and Zhou et al. [224] |
| 142 | Pif | Chugoku 31-1 | 1971 | 11 | 24.69 | – | Japan | Shinoda et al. [278] |
| 143 | Pii | Ishikari Shiroke | 1971 | 9 | 2.29 | – | Japan | Ise [279] and Shinoda et al. [278] |
| 144 | Piis1 | Imochi Shirazu | 1970 | 11 | 2.84 | – | Japan | Goto [280] |
| 145 | Pikur 1 | Kuroka | 1970 | 4 | 24.61 | Isozyme | Japan | Fukuoka et al. [281] and Goto [280] |
| 146 | Pise | Sensho | 1970 | 11 | 5.74 | – | Japan | Goto [280] |
| S. No. | Name of Blast Resistance Genes | Proteins Encoded by R Genes | Donor Rice Lines/Genotypes | Chromosome No. | Year of Cloning | Cloning Approach Used for Isolation of R Genes | Reference Serial No. of Table 2 | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | Pb2 | NBS-LRR protein with NB-ARC domain and LRR domain | Jiangnanwan | 11 | 2022 | Map-based cloning | 1 | Yu et al. [15] |
| 2 | Pid3-I1 | CC-NBS-LRR | MC276 | 6 | 2019 | Gene Mapping | 31 | Inukai et al. [282] |
| 3 | Pitr | A typical protein with an armadillo repeat (Putative E3 ligase) | Katy | 12 | 2018 | Map-based cloning | 71 | Zhao et al. [283] |
| 4 | Pigm | NBS-LRR | Gumei 4 | 6 | 2017 | Map-based cloning | 46 | Deng et al. [35] |
| 5 | Pi64 | CC–NBS–LRR | Yangmaogu | 1 | 2015 | Map-based cloning | 12 | Ma et al. [192] |
| 6 | Pi50 | NBS-LRR | Er-Ba-zhan (EBZ) | 6 | 2015 | - | 18 | Su et al. [113] |
| 7 | Pik-e | CC-NBS-LRR | Xiangzao 143 | 11 | 2015 | Map-based cloning | 8 | Chen et al. [190] |
| 8 | Pi35 | NBS-LRR | Hokkai-188 | 1 | 2014 | Map-based cloning | 51 | Fukuoka et al. [284] |
| 9 | Pi63/Pikahei-1(t) | NBS-LRR | Kahei | 4 | 2014 | Map-based cloning | 34 | Xu et al. [285] |
| 10 | PiK-h | NBS-LRR | K3 | 11 | 2014 | Positional cloning | 35 | Zhai et al. [127] |
| 11 | Pi54of | NBS–LRR | Oryza officinalis (nrcpb004) | 11 | 2014 | Map-based cloning | 60 | Devanna et al. [28] |
| 12 | Pii | NBS-LRR | Hitomebore | 9 | 2013 | MutMap-Gap | 116 | Takagi et al. [132] |
| 13 | Pi-CO39 | CC-NBS-LRR | CO39 | 11 | 2013 | -- | 89 | Cesari et al. [82] |
| 14 | Pi56 | NBS–LRR | Sanhuangzhan No. 2 | 9 | 2012 | Map-based cloning | 75 | Liu et al. [200] |
| 15 | Pi1 | CC–NBS–LRR | C101LAC | 11 | 2012 | Map-based cloning | 129 | Hua et al. [286] |
| 16 | Pi54rh | NBS-LRR | Oryza rhizomatis (nrcpb 002) | 11 | 2012 | Map-based cloning | - | Das et al. [287] |
| 17 | Pi25 | CC-NBS-LRR | Gumei2 | 6 | 2011 | Map-based cloning | 10 | Chen et al. [288] |
| 18 | Pia | CC-NBS-LRR | Aichi Asahi | 11 | 2011 | MB and mutant screening | 26 | Okuyama et al. [128] |
| 19 | Pik-p | CC-NBS-LRR | K60 | 11 | 2011 | Map-based cloning | 32 | Yuan et al. [289] |
| 20 | Pik | CC-NBS-LRR | Kusabue | 11 | 2011 | Map-based cloning | 113 | Zhai et al. [180] |
| 21 | Pish | NBS–LRR | Shin-2 | 1 | 2010 | Mutant Screening | 139 | Takahashi et al. [290] |
| 22 | Pb1 | CC-NBS-LRR | Modan | 11 | 2010 | Map-based cloning | 101 | Hayashi et al. [251] |
| 23 | Pi54/Pi-kh | NBS-LRR | Tetep | 11 | 2010 | Map-based cloning | 73 | Sharma et al. [122] |
| 24 | Pit | CC-NBS-LRR | K59 | 1 | 2009 | Map-based cloning | 118 | Hayashi and Yoshida [291] |
| 25 | pi21 | Proline-rich heavy metal binding protein | Owarihatamochi | 4 | 2009 | Map-based cloning | 107 | Fukuoka et al. [281] |
| 26 | Pi-d3 | CC-NBS-LRR | Digu | 6 | 2009 | In silico analysis | 106 | Shang et al. [210] |
| 27 | Pi5 | CC-NBS-LRR | Moroberekan | 9 | 2009 | Map-based cloning | 86 | Lee et al. [292] |
| 28 | Pik-m | NBS-LRR | Tsuyuake | 11 | 2008 | Map-based cloning | 114 | Ashikawa et al. [293] |
| 29 | Pi37 | NBS-LRR | St. No. 1 | 1 | 2007 | Map-based cloning | 57 | Lin et al. [294] |
| 30 | Pi36 | CC-NBS-LRR | Q61 | 8 | 2007 | Map-based cloning | 59 | Liu et al. [215] |
| 31 | Pi-d2 | B-lectin receptor kinase | Digu | 6 | 2006 | Map-based cloning | 45 | Chen et al. [220] |
| 32 | Pi9 | NBS-LRR | 75-1-127 | 6 | 2006 | Map-based cloning | 44 | Qu et al. [219] |
| 33 | Pi-2 | NBS-LRR | C101A51 | 6 | 2006 | Map-based cloning | 87 | Zhou et al. [224] |
| 34 | Piz-t | NBS-LRR | Toride 1 | 6 | 2006 | Map-based cloning | 53 | Zhou et al. [224] |
| 35 | Pi-ta | NBS-LRR | Yashiro-mochi | 12 | 2000 | Map-based cloning | 112 | Bryan et al. [119] |
| 36 | Pib | NBS-LRR | Tohoku IL9 | 2 | 1999 | Map-based cloning | 52 | Wang et al. [295] |
| S. No. | Gene/QTL | Trait/Resistance | Type of Molecular Marker | Technique/Approach Used | Application/Lines Developed | References |
|---|---|---|---|---|---|---|
| Single gene for blast resistance | ||||||
| 1. | Pi1 | Resistance to blast disease | ISSR and SSR | MABB | Marker-assisted backcross breeding for improvement of variety Zhenshan97 A | Liu et al. [185] |
| 2. | Piz | Resistance to blast disease | SSR | MAS | Tightly linked markers with Pi-z locus was applied for screening of germplasm for blast resistance in rice | Fjellstrom et al. [332] |
| 3. | Pita | Resistance to blast disease | Gene-specific to gene | MAS | Applied for detection of Pita gene in 141 rice germplasms and introduction of gene through advanced breeding approaches | Wang et al. [333] |
| 4. | Pi9 | Resistance to blast disease | Gene-specific | Marker aided selection | Introgressed Pi-9(t) resistance gene in the cultivar Luhui 17 | Wen and Gao [334] |
| 5. | Pi39 | Resistance to blast disease | InDel | MABB | Introgressed into Chinese cultivar Q15 | Hua et al. [335] |
| 6. | Pikh | Resistance to blast disease | SSR | MABB | Improvement of Malaysian Cultivar, MR264 by Introgression of Pikh gene | Hasan et al. [336] |
| 7. | Pi40 | Resistance to blast disease | SSR | MABB | Introgressed into elite cultivars Turkish, Halilbey and Osmancik-97 | Beser et al. [337] |
| 8. | Pi-ar | Resistance to blast disease | RAPD | MAS | Introgression of Pi-ar gene using double haploid technique | Araujo et al. [338] |
| Two genes for Blast resistance | ||||||
| 9. | Piz-5, Pi54 | Resistance to blast disease | SSR | MABB | Blast disease resistance genes transferred to develop Pusa 1602 and Pusa 1603 | Singh et al. [356] |
| 10. | Pi1, Piz | Resistance to blast disease | SSR | MABB | Pyramiding of Pi1 and Piz-5 genes into PRR78 | Gouda et al. [340] |
| 11. | Pi1, Pi2 | Resistance to blast disease | SSR | MABB | Introgressed into Intan variety and BPT5204 | Hegde et al. [344] |
| 12. | Pi46, Pita | Resistance to blast disease | SSR | MABB | Introgression of resistance genes into Hang hui 179 (HH179) | Xiao et al. [343] |
| 13. | Pi2, Pi9 | Resistance to blast disease | SNP | MABB | Introgression of blast resistance genes into R179 | Luo et al. [345] |
| 14. | Pi-b and Pik-h | Resistance to blast disease | SSR, RM 208, RM 206 | MABB | Pyramided two blast resistance genes into MR219Malaysian rice variety | Tanweer et al. [327] |
| 15. | Piz-5 and Pi54 | Resistance to blast disease | SSR | MABB | Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line | Singh et al. [297] |
| 16. | Pi-2 and Pi-54 | Resistance to blast disease | SSR | MABB | Introgression of blast resistance genes into the genetic background of elite, bacterial blight resistant indica rice variety, Improved Samba Mahsuri | Madhavi et al. [342] |
| 17. | Pi54 and Pi1 | Resistance to blast disease | SSR | MAS | Introgression of blast resistance genes into cold tolerant variety Tellahamsa | Oddin et al. [341] |
| 18. | Pi46 and Pita | Resistance to blast disease | SSR | MABB | Blast resistance genes were introgressed into an elite restorer line Hang-Hui-179 (HH179) | Xiao et al. [343] |
| More than Two genes for Blast resistance | ||||||
| 19. | Pi1, Piz-5, Pita | Resistance to blast disease | RFLP | MAS | Pyramiding of three NILs namely (C101LAC, C101A51 and C101PKT) for blast resistance into a single cultivar CO39, each linecarrying resistance genes Pi1, Piz-5 and Pita, respectively. | Korinsak et al. [169] |
| 20. | Pi1, Pi2, Pi33 | Resistance to blast disease | SSR | MABB | Introgression of multiple blast disease resistance genes into Jin23B | Chen et al. [302] |
| 21. | Pi1, Pi2, Pi33 | Resistance to blast disease | SSR | MAS | Improvement of Russian rice varieties by pyramiding of blast disease resistance genes | Usatov et al. [357] |
| 22. | Pi9, Pizt, Pi54 | Resistance to blast disease | SNP | MABB | Introgression of Pi9, Pizt, Pi54 blast resistance genes into japonica rice 07GY31 | Xiao et al. [343] |
| 23. | Pi1,Pi2, Pi33 | Resistance to blast disease | SSR | MABB | Improving blast resistance in Indian rice variety ADT43 by pyramiding three blast resistance genes | Divya et al. [346] |
| 24. | Pid1, Pib, Pita, Pi2 | Resistance to blast disease | SSR | MAS | Pid1, Pib and Pita genes were introduced into G46B, while Pi2 was introduced into Zhenshan 97B | Chen et al. [230] |
| 25. | Pizt, Pi2, Pigm, Pi40, Pi9, Piz | Resistance to blast disease | SSR | MAS | Introgression of multiple blast resistance genes into Yangdao 6 | Wu et al. [350] |
| Multiple stress tolerance | ||||||
| 26. | Xa21, Piz | Resistance to blast and bacterial leaf blight disease | SSR | MAS | Introgression of Blast and Bacterial leaf blight disease resistance gene | Narayanan et al. [351] |
| 27. | Pi2 and Xa23 | Resistance to blast and bacterial leaf blight disease | SSR | MAS | Introgression of broad-spectrum disease resistance genes into, elite thermo-sensitive genic male-sterile rice line-GZ63-4S | Jiang et al. [182] |
| 28. | Xa21 and Pi54 | Resistance to blast and bacterial leaf blight disease | SSR | MABB | Introgression of BLB and blast resistance into DRR17B, an elite, fine-grain type maintainer line of rice | Balachiranjeevi et al. [348] |
| 29. | Pi1, Pi2, Xa23 | Resistance to blast and bacterial leaf blight disease | SSR | MABB | Introgression of bacterial blight and blast resistance into variety Rongfeng B | Fu et al. [358] |
| 30. | Pi2, Xa21, Xa33 | Resistance to blast and bacterial leaf blight disease | SSR | MABB | Introgressed bacterial blight and blast diseases resistance genes into RPHR-1005 | Kumar et al. [359] |
| 31. | Pi9Drought | Resistance to blast and drought tolerance | Gene linked markers | MAS | Pi9 has been introgressed into different genetic backgrounds of cultivated varieties, such as indica cultivar Swarna + drought | Dixit et al. [331] |
| 32. | Pi2, Pi54, xa13 and Xa21 | Resistance to blast and bacterial leaf blight disease | SSR | MABB | Introgressed of bacterial blight and blast diseases resistance genes for improving disease resistance traits in Basmati rice varieties | Ellur et al. [352] |
| 33. | Xa21,xa13 and Pi54 | Resistance to blast and bacterial leaf blight disease | Gene-specific | MABB | Pyramiding of bacterial blight and blast diseases resistance into Indian rice variety MTU1010 | ArunaKumari et al. [353] |
| 34. | Xa21, xa13 and Pi54 | Resistance to blast and bacterial leaf blight disease | SSR | MAS | Improvement of Vallabh Basmati 22 by Introgression of Xa21, xa13 genes for Bacterial Blight and Pi54 for Blast disease resistant genes | Srikanth et al. [349] |
| 35. | Xa21, Xa33, Pi2, Rf3 and Rf4 | Resistance to blast and bacterial leaf blight disease | SSR | MAS | Marker-assisted improvement of the elite restorer line of rice, RPHR-1005 for resistance against diseases | Kumar et al. [360] |
| 36. | Xa 5 and 4 blast QTLs | Resistance to blast and bacterial leaf blight disease | SSR | MAS | Introgression of bacterial leaf blight and blast resistance genes into rice cultivar RD6 | Pinta et al. [361] |
| 37. | Xa13, Xa21, Pi54, qSBR11 | Resistance to blast and sheath bight disease | SSR | MAS | Transfer of multiple disease resistance genes for bacterial blight, blast and sheath blight disease in rice | Singh et al. [297] |
| 38. | Pi54, qSBR11-1, qSBR11-2 and qSBR7-1 | Resistance to blast and sheath bight disease | SSR and QTLs | MABB | Introgression of multiple disease resistance genes into a maintainer of Basmati rice CMS line | Singh et al. [347] |
| 39. | Pi2, Pi9, Gm1, Gm4, Sub1, and Saltol | Blast disease, Gall Midge Submergence and Salinity tolereance | SSR and gene linked markers | MAS | Pyramiding of genes/QTLs to confer resistance/tolerance to blast, gall Midge, submergence, and salinity in a released rice variety CRMAS2621-7-1 as Improved Lalat | Das and Rao [362] |
| 40. | Pi9, Xa4, xa5, xa13, Xa21, Bph3, Bph17, Gm4, Gm8 and qDTY1.1 and qDTY3.1 | Blast Bacterial leaf blight Brown planthopperGall midge and QTLs for drought tolerance | Gene based/linked markers | Marker-assisted forward breeding | MAS in combining tolerance to multiple biotic and abiotic stresses in Swarna + drought recurrent parent | Dixit et al. [331] |
| 41. | Pi9, Xa21, Gm8, qDTY1.1, qDTY2.2 and qDTY4.1 | Blast, Bacterial blight (BB), Gall midge (GM) and QTLSs drought tolerance | Gene based/linked markers | Marker-assisted forward(MAFB) and back cross (MABC) breeding | Introgressed in to Indian elite rice variety, Naveen | Janaki Ramayya et al. [354] |
| 42. | BPH3, BPH24, Pi2, Pi9, Pita, Pib, Xa21Pimh, and badh2 | Brown Plant hopperblast disease, bacterial blight and Aroma | SSR and gene linked markers | MAS | Brown planthopper (BPH), blast, and bacterial leaf blight (BLB) resistance and aroma genes into elite rice maintainers and restorers | Wang et al. [27] |
| 43. | Xa21, Pi54, Pup1 | BB resistance gene, the blast resistance gene, and low soil phosphorous tolerance QTL/gene, | Gene/QTL linked markers | Marker-assisted pedigree breeding | BB resistance gene, the blast resistance gene, and low soil phosphorous tolerance QTL/gene in to MTU 1010 (CottondoraSannalu) | LaxmiPrasanna et al. [355] |
7.4. Transgenic Breeding for Rice Blast Resistance
7.5. Genome Editing Tools for Developing the Blast Resistance in Rice
7.5.1. CRISPR Cas9 Based Resistance for Rice Blast
7.5.2. Transcription Activator-like Effector Nucleases (TALENs)
7.5.3. Meganucleases (MNs)
7.5.4. Zinc Finger Nucleases (ZFNs)
8. Problems Associated with Breeding for Resistance to Rice Blast
9. Conclusions and Future Outlooks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simkhada, K.; Thapa, R. Rice Blast, A Major Threat to the Rice Production and Its Various Management Techniques. Turkish J. Agric. Food Sci. Technol. 2022, 10, 147–157. [Google Scholar] [CrossRef]
- Asibi, E.A.; Chai, Q.; Coulter, J.A. Rice Blast: A Disease with Implications for Global Food Security. Agronomy 2019, 9, 451. [Google Scholar] [CrossRef]
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An Overview of Global Rice Production, Supply, Trade, and Consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, M.G.C.; Martin, F.; Variar, M.; Bhatt, J.C.; Perez-Quintero, A.; Leung, H.; Leach, J.E.; Vera Cruz, C.M. Accumulating Candidate Genes for Broad-Spectrum Resistance to Rice Blast in a Drought-Tolerant Rice Cultivar. Sci. Rep. 2021, 11, 21502. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Burr, B. International rice genome sequencing project: The effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 2000, 3, 138–142. [Google Scholar] [CrossRef]
- Thapa, R.; Bhusal, N. Designing Rice for the 22nd Century: Towards Rice with an Enhanced Productivity and Efficient Photosynthetic Pathway. Turkish J. Agric. Food Sci. Technol. 2020, 8, 2623–2634. [Google Scholar] [CrossRef]
- Acharya, B.; Shrestha, S.M.; Manandhar, H.K.; Chaudhary, B. Screening of Local, Improved and Hybrid Rice Genotypes against Leaf Blast Disease (Pyricularia oryzae) at Banke District, Nepal. J. Agric. Nat. Resour. 2019, 2, 36–52. [Google Scholar] [CrossRef]
- Wilson, R.A.; Talbot, N.J. Under Pressure: Investigating the Biology of Plant Infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 2009, 7, 185–195. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; DI Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 Fungal Pathogens in Molecular Plant Pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Agrios, G. Plant Pathology, 5th ed.; Elsevier Academic Press: Cambridge, MA, USA, 2005; p. 165. [Google Scholar]
- Nasruddin, A.; Amin, N. Effects of Cultivar, Planting Period, and Fungicide Usage on Rice Blast Infection Levels and Crop Yield. J. Agric. Sci. 2012, 5, P160. [Google Scholar] [CrossRef]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Asfaliza, R.; Latif, M.A. Blast resistance in rice: A review of conventional breeding to molecular approaches. Mol. Biol. Rep. 2013, 40, 2369–2388. [Google Scholar] [CrossRef] [PubMed]
- Musiime, O.; Tenywa, M.M.; Majaliwa, M.J.G.; Lufafa, A.; Nanfumba, D.; Wasige, J.E.; Woomer, P.L.; Kyondha, M. Constraints to rice production in Bugiri District. Afr. Crop Sci. Conf. Proc. 2005, 7, 1495–1499. [Google Scholar]
- Kato, H. Rice blast disease. Pestic. Outlook 2001, 12, 23–25. [Google Scholar] [CrossRef]
- Yu, Y.; Ma, L.; Wang, X.; Zhao, Z.; Wang, W.; Fan, Y.; Liu, K.; Jiang, T.; Xiong, Z.; Song, Q.; et al. Genome-Wide Association Study Identifies a Rice Panicle Blast Resistance Gene, Pb2, Encoding NLR Protein. Int. J. Mol. Sci. 2022, 23, 5668. [Google Scholar] [CrossRef]
- Yadav, M.K.; Aravindan, S.; Ngangkham, U.; Raghu, S.; Prabhukarthikeyan, S.R.; Keerthana, U.; Marndi, B.C.; Adak, T.; Munda, S.; Deshmukh, R.; et al. Blast Resistance in Indian Rice Landraces: Genetic Dissection by Gene Specific Markers. PLoS ONE 2019, 14, e0211061. [Google Scholar] [CrossRef]
- Ashkani, S.; Rafii, M.Y.; Shabanimofrad, M.; Ghasemzadeh, A.; Ravanfar, S.A.; Latif, M.A. Molecular Progress on the Mapping and Cloning of Functional Genes for Blast Disease in Rice (Oryza sativa L.): Current Status and Future Considerations. Crit. Rev. Biotechnol. 2016, 36, 353–367. [Google Scholar] [CrossRef]
- Wang, G.L.; Valent, B. Durable Resistance to Rice Blast. Science 2017, 355, 906–907. [Google Scholar] [CrossRef]
- Srivastava, D.; Shamim, M.; Kumar, M.; Mishra, A.; Pandey, P.; Kumar, D.; Yadav, P.; Siddiqui, M.H.; Singh, K.N. Current Status of Conventional and Molecular Interventions for Blast Resistance in Rice. Rice Sci. 2017, 24, 299–321. [Google Scholar] [CrossRef]
- Singh, B.D. Plant Breeding: Principles and Methods; Kalyani Publishers: Ludhiana, India, 2018; Volume 11, pp. 1–907. [Google Scholar]
- Lo, K.-L.; Chen, Y.-N.; Chiang, M.-Y.; Chen, M.-C.; Panibe, J.P.; Chiu, C.-C.; Liu, L.-W.; Chen, L.-J.; Chen, C.-W.; Li, W.-H.; et al. Two Genomic Regions of a Sodium Azide Induced Rice Mutant Confer Broad-Spectrum and Durable Resistance to Blast Disease. Rice 2022, 15, 2. [Google Scholar] [CrossRef]
- Ning, X.; Yunyu, W.; Aihong, L. Strategy for Use of Rice Blast Resistance Genes in Rice Molecular Breeding. Rice Sci. 2020, 27, 263–277. [Google Scholar] [CrossRef]
- Annegowda, D.C.; Prasannakumar, M.K.; Mahesh, H.B.; Siddabasappa, C.B.; Devanna, P.; Banakar, S.N.; Manijkumar, H.B.; Prassad, S.R. Rice Blast Disease in India: Present Status and Future Challenges. In Integrative Advances in Rice Research; IntechOpen Limited: London, UK, 2022; pp. 1–42. [Google Scholar]
- Sharma, S.K.; Sharma, D.; Meena, R.P.; Yadav, M.K.; Hosahatti, R.; Dubey, A.K.; Sharma, P.; Kumar, S.; Pramesh, D.; Nabi, S.U.; et al. Recent Insights in Rice Blast Disease Resistance. In Blast Disease of Cereal Crops; Nayaka, S.C., Hosahatti, R., Prakash, G., Satyavathi, C.T., Sharma, R., Eds.; Fungal Biology; Springer Nature Switzerland AG: Cham, Switzerland, 2021; pp. 89–124. [Google Scholar]
- Mao, T.; Zhu, M.; Ahmad, S.; Ye, G.; Sheng, Z.; Hu, S.; Jiao, G.; Xie, L.; Tang, S.; Wei, X.; et al. Superior Japonica Rice Variety YJ144 with Improved Rice Blast Resistance, Yield, and Quality Achieved Using Molecular Design and Multiple Breeding Strategies. Mol. Breed. 2021, 41, 65. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Das, A.; Devanna, B.N.; Thakur, S.; Singh, P.K.; Singh, N.K.; Sharma, T.R. Mining of Rice Blast Resistance Gene Pi54 Shows Effect of Single Nucleotide Polymorphisms on Phenotypic Expression of the Alleles. Eur. J. Plant Pathol. 2013, 137, 55–65. [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.; Ma, X.; Luo, L.; Fang, Y.; Zhao, N.; Han, Y.; Wei, Z.; Liu, F.; Qin, B.; et al. Development of New Rice (Oryza. Sativa L.) Breeding Lines through Marker-Assisted Introgression and Pyramiding of Brown Planthopper, Blast, Bacterial Leaf Blight Resistance, and Aroma Genes. Agronomy 2021, 11, 2525. [Google Scholar] [CrossRef]
- Devanna, N.B.; Vijayan, J.; Sharma, T.R. The Blast Resistance Gene Pi54of Cloned from Oryza officinalis Interacts with Avr-Pi54 through Its Novel Non-LRR Domains. PLoS ONE 2014, 9, e104840. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Han, Q.; Zi, Q.; Lv, S.; Qiu, D.; Zeng, H. Enhanced Disease Resistance and Drought Tolerance in Transgenic Rice Plants Overexpressing Protein Elicitors from Magnaporthe oryzae. PLoS ONE 2017, 12, e0175734. [Google Scholar] [CrossRef]
- Li, X.; Pan, L.; Bi, D.; Tian, X.; Li, L.; Xu, Z.; Wang, L.; Zou, X.; Gao, X.; Yang, H.; et al. Generation of Marker-Free Transgenic Rice Resistant to Rice Blast Disease Using Ac/Ds Transposon-Mediated Transgene Reintegration System. Front. Plant Sci. 2021, 12, 644437. [Google Scholar] [CrossRef]
- Peng, M.; Lin, X.; Xiang, X.; Ren, H.; Fan, X.; Chen, K. Characterization and Evaluation of Transgenic Rice Pyramided with the Pi Genes Pib, Pi25 and Pi54. Rice 2021, 14, 78. [Google Scholar] [CrossRef]
- Viana, V.E.; Pegoraro, C.; Busanello, C.; Costa de Oliveira, A. Mutagenesis in Rice: The Basis for Breeding a New Super Plant. Front. Plant Sci. 2019, 10, 1326. [Google Scholar] [CrossRef]
- Ijaz, S.; Ul Haq, I. Genome Editing Technologies for Resistance Against Phytopathogens: Principles, Applications and Future Prospects. In Plant Disease Management Strategies for Sustainable Agriculture through Traditional and Modern Approaches; Springer Nature Switzerland AG: Cham, Switzerland, 2020; pp. 237–245. [Google Scholar]
- Nizolli, V.O.; Pegoraro, C.; Oliveira, A.C. Rice blast: Strategies and challenges for improving genetic resistance. Crop. Breed. Appl. Biotechnol. 2021, 21, e387721S9. [Google Scholar] [CrossRef]
- Deng, Y.; Zhai, K.; Xie, Z.; Yang, D.; Zhu, X.; Liu, J.; Wang, X.; Qin, P.; Yang, Y.; Zhang, G.; et al. Epigenetic Regulation of Antagonistic Receptors Confers Rice Blast Resistance with Yield Balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef]
- Couch, B.C.; Fudal, I.; Lebrun, M.-H.; Tharreau, D.; Valent, B.; van Kim, P.; Nottéghem, J.-L.; Kohn, L.M. Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 2005, 170, 613–630. [Google Scholar] [CrossRef]
- Padmanabhan, S.Y. Recent advances in the study of blast disease of rice. Madras Agri. J. 1965, 564–583. [Google Scholar]
- Waller, J.M. Rice Diseases. By S. H. Ou Slough, UK: Commonwealth Agricultural Bureaux (1985) 2nd Ed., pp. 380, UK £38.00, USA $70, Elsewhere £41.00. Exp. Agric. 1987, 23, 357. [Google Scholar] [CrossRef]
- Khush, G.S. Breeding methods and procedures employed at IRRI for developing rice germplasm with multiple resistance to diseases and insects. In Symposium on Methods of Crop Breeding; Tropical Agricultural Research Series; Japan International Research Center for Agricultural Sciences: Tsukuba, Japan, 1978; Volume 11, pp. 69–76. [Google Scholar]
- Padmanabhan, S.Y.; Chakravarti, N.K.; Mathur, S.C.; Veeraraghavan, J. Identification of pathogenic races of Pyricularia Oryzae in India. Phytopathology 1970, 60, 1574–1577. [Google Scholar] [CrossRef]
- Piotti, E.; Rigano, M.M.; Rodino, D.; Rodolfi, M.; Castiglione, S.; Picco, A.M.; Sala, F. Genetic Structure of Pyricularia Grisea (Cooke) Sacc. Isolates from Italian Paddy Fields. J. Phytopathol. 2005, 153, 80–86. [Google Scholar] [CrossRef]
- Devanna, B.N.; Jain, P.; Solanke, A.U.; Das, A.; Thakur, S.; Singh, P.K.; Kumari, M.; Dubey, H.; Jaswal, R.; Pawar, D.; et al. Understanding the Dynamics of Blast Resistance in Rice-Magnaporthe oryzae Interactions. J. Fungi 2022, 8, 584. [Google Scholar] [CrossRef] [PubMed]
- Nalley, L.; Tsiboe, F.; Durand-Morat, A.; Shew, A.; Thoma, G. Economic and Environmental Impact of Rice Blast Pathogen (Magnaporthe oryzae) Alleviation in the United States. PLoS ONE 2016, 11, e0167295. [Google Scholar] [CrossRef] [PubMed]
- Parker, D.; Beckmann, M.; Enot, D.P.; Overy, D.P.; Rios, Z.C.; Gilbert, M.; Talbot, N.; Draper, J. Rice Blast Infection of BrachypodiumDistachyon as a Model System to Study Dynamic Host/Pathogen Interactions. Nat. Protoc. 2008, 3, 435–445. [Google Scholar] [CrossRef]
- Howard, R.J.; Ferrari, M.A.; Roach, D.H.; Money, N.P. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA 1991, 88, 11281–11284. [Google Scholar] [CrossRef] [PubMed]
- Staskawicz, B.J.; Ausubel, F.M.; Baker, B.J.; Ellis, J.G.; Jones, J.D.G. Molecular Genetics of Plant Disease Resistance. Science 1995, 268, 661–667. [Google Scholar] [CrossRef]
- Hamer, J.E.; Howard, R.J.; Chumley, F.G.; Valent, B. A mechanism for surface attachment in spores of a plant pathogenic fungus. Science 1988, 239, 288–290. [Google Scholar] [CrossRef]
- Jelitto, T.C.; Page, H.A.; Read, N.D. Role of external signals in regulating the pre-penetration phase of infection by the rice blast fungus, Magnaporthe grisea. Planta 1994, 194, 471–477. [Google Scholar] [CrossRef]
- Xiao, J.-Z.; Watanabe, T.; Kamakura, T.; Ohshima, A.; Yamaguchi, I. Studies on Cellular Differentiation of Magnaporthe grisea. Physicochemical Aspects of Substratum Surfaces in Relation to Appressorium Formation. Physiol. Mol. Plant Pathol. 1994, 44, 227–236. [Google Scholar] [CrossRef]
- Jiang, H.; Lin, L.; Tang, W.; Chen, X.; Zheng, Q.; Huang, J.; Wang, Z. Putative interaction proteins of the ubiquitin ligase Hrd1 in Magnaporthe oryzae. Evol. Bioinform. 2018, 14, 1176934318810990. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhou, X.; Xu, J.R. Genetic control of infection-related development in Magnaporthe oryzae. Curr. Opin. Microbiol. 2012, 15, 678–684. [Google Scholar] [CrossRef]
- Gao, J.; Xu, X.; Huang, K.; Liang, Z. Fungal G-Protein-Coupled Receptors: A Promising Mediator of the Impact of Extracellular Signals on Biosynthesis of Ochratoxin A. Front. Microbiol. 2021, 12, 631392. [Google Scholar] [CrossRef]
- DeZwaan, T.M.; Carroll, A.M.; Valent, B.; Sweigard, J.A. Magnaporthe grisea Pth11p Is a Novel Plasma Membrane Protein That Mediates Appressorium Differentiation in Response to Inductive Substrate Cues. Plant Cell 1999, 11, 2013–2030. [Google Scholar] [CrossRef]
- Kou, Y.; Tan, Y.H.; Ramanujam, R.; Naqvi, N.I. Structure–function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 2017, 214, 330–342. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, W.; Liu, K.; Huang, Q.; Zhang, X.; Yan, X.; Chen, Y.; Wang, J.; Qi, Z.; Wang, Z.; et al. Eight RGS and RGS-like Proteins Orchestrate Growth, Differentiation, and Pathogenicity of Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1002450. [Google Scholar] [CrossRef]
- Dean, R.A. signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 1997, 35, 211–234. [Google Scholar] [CrossRef]
- Tucker, S.L.; Talbot, N.J. Surface Attachment and Pre-Penetration Stage Development by Plant Pathogenic Fungi. Annu. Rev. Phytopathol. 2001, 39, 385–417. [Google Scholar] [CrossRef]
- De Jong, J.C.; McCormack, B.J.; Smirnoff, N.; Talbot, N.J. Glycerol Generates Turgor in Rice Blast. Nature 1997, 389, 244. [Google Scholar] [CrossRef]
- Shen, Q.; Liang, M.; Yang, F.; Deng, Y.Z.; Naqvi, N.I. Ferroptosis Contributes to Developmental Cell Death in Rice Blast. New Phytol. 2020, 227, 1831–1846. [Google Scholar] [CrossRef]
- Talbot, N.J.; Kershaw, M.J. The Emerging Role of Autophagy in Plant Pathogen Attack and Host Defence. Curr. Opin. Plant Biol. 2009, 12, 444–450. [Google Scholar] [CrossRef]
- Yin, Z.; Feng, W.; Chen, C.; Xu, J.; Li, Y.; Yang, L.; Wang, J.; Liu, X.; Wang, W.; Gao, C.; et al. Shedding Light on Autophagy Coordinating with Cell Wall Integrity Signaling to Govern Pathogenicity of Magnaporthe oryzae. Autophagy 2020, 16, 900–916. [Google Scholar] [CrossRef] [PubMed]
- Ryder, L.S.; Talbot, N.J. Regulation of Appressorium Development in Pathogenic Fungi. Curr. Opin. Plant Biol. 2015, 26, 8–13. [Google Scholar] [CrossRef]
- Fang, Y.L.; Xia, L.M.; Wang, P.; Zhu, L.H.; Ye, J.R.; Huang, L. The MAPKKK CgMck1 is required for cell wall integrity, appressorium development, and pathogenicity in Colletotrichum gloeosporioides. Genes 2018, 9, 543. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.J.; Ryder, L.S.; Kershaw, M.J.; Talbot, N.J. The Role of Glycerol in the Pathogenic Lifestyle of the Rice Blast Fungus Magnaporthe oryzae. Environ. Microbiol. 2017, 19, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Sweigard, J.A.; Chumley, F.G.; Valent, B. Disruption of a Magnaporthe grisea Cutinase Gene. Mol. Gen. Genet. 1992, 232, 183–190. [Google Scholar] [CrossRef]
- Skamnioti, P.; Gurr, S.J. Magnaporthe grisea cutinase2 mediates appressorium differentiation and host penetration and is required for full virulence. Plant Cell 2007, 19, 2674–2689. [Google Scholar] [CrossRef] [PubMed]
- Quoc, N.B.; Bao Chau, N.N. The role of cell wall degrading enzymes in pathogenesis of Magnaporthe oryzae. Curr. Protein Pept. Sci. 2017, 18, 1019–1034. [Google Scholar] [CrossRef] [PubMed]
- Dagdas, Y.F.; Yoshino, K.; Dagdas, G.; Ryder, L.S.; Bielska, E.; Steinberg, G.; Talbot, N.J. Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 2012, 336, 1590–1595. [Google Scholar] [CrossRef]
- Kleemann, J.; Rincon-Rivera, L.J.; Takahara, H.; Neumann, U.; van Themaat, E.V.L.; van der Does, H.C.; O’Connell, R.J. Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 2012, 8, e1002643. [Google Scholar] [CrossRef]
- Ryder, L.S.; Dagdas, Y.F.; Kershaw, M.J.; Venkataraman, C.; Madzvamuse, A.; Yan, X.; Cruz-Mireles, N.; Soanes, D.M.; Oses-Ruiz, M.; Styles, V.; et al. A Sensor Kinase Controls Turgor-Driven Plant Infection by the Rice Blast Fungus. Nature 2019, 574, 423–427. [Google Scholar] [CrossRef]
- Ryder, L.S.; Dagdas, Y.F.; Mentlak, T.A.; Kershaw, M.J.; Thornton, C.R.; Schuster, M.; Chen, J.; Wang, Z.; Talbot, N.J. NADPH Oxidases Regulate Septin-Mediated Cytoskeletal Remodeling during Plant Infection by the Rice Blast Fungus. Proc. Natl. Acad. Sci. USA 2013, 110, 3179–3184. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.J.; Wang, Z.Y.; Jones, M.A.; Smirnoff, N.; Talbot, N.J. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blastdisease. Proc. Natl. Acad. Sci. USA 2007, 104, 11772–11777. [Google Scholar] [CrossRef]
- Khang, C.H.; Berruyer, R.; Giraldo, M.C.; Kankanala, P.; Park, S.Y.; Czymmek, K.; Valent, B. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 2010, 22, 1388–1403. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.C.; Dagdas, Y.F.; Gupta, Y.K.; Mentlak, T.A.; Yi, M.; Martinez-Rocha, A.L.; Saitoh, H.; Terauchi, R.; Talbot, N.J.; Valent, B. Two Distinct Secretion Systems Facilitate Tissue Invasion by the Rice Blast Fungus Magnaporthe oryzae. Nat. Commun. 2013, 4, 1996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, J.R. Effectors and effector delivery in Magnaporthe oryzae. PLoS Pathog. 2014, 10, e1003826. [Google Scholar] [CrossRef]
- Petit-Houdenot, Y.; Fudal, I. Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management. Front. Plant Sci. 2017, 8, 1072. [Google Scholar] [CrossRef]
- Park, C.H.; Chen, S.; Shirsekar, G.; Zhou, B.; Khang, C.H.; Songkumarn, P.; Wang, G.L. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern–triggered immunity in rice. Plant Cell 2012, 24, 4748–4762. [Google Scholar] [CrossRef]
- Kang, S.; Sweigard, J.A.; Valent, B. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 1995, 8, 939–948. [Google Scholar] [CrossRef]
- Orbach, M.J.; Farrall, L.; Sweigard, J.A.; Chumley, F.G.; Valent, B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 2000, 12, 2019–2032. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, B.; Wu, J.; Lu, G.; Hu, Y.; Zhang, X.; Zhou, B. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant-Microbe Interact. 2009, 22, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Saitoh, H.; Fujisawa, S.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Tosa, Y.; Chuma, I.; Takano, Y.; Win, J.; et al. Association Genetics Reveals Three Novel Avirulence Genes from the Rice Blast Fungal Pathogen Magnaporthe oryzae. Plant Cell 2009, 21, 1573–1591. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y.; et al. The rice resistance protein pair rga4/rga5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wang, L.; Zhang, S.; Li, Z.; Zhang, Y.; Lin, F.; Pan, Q. Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol. Plant-Microbe Interact. 2014, 27, 759–769. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Wu, W.; He, L.; Yang, X.; Pan, Q. Function and Evolution of Magnaporthe oryzae Avirulence Gene AvrPib Responding to the Rice Blast Resistance Gene Pib. Sci. Rep. 2015, 5, 11642. [Google Scholar] [CrossRef]
- Zhang, X.; He, D.; Zhao, Y.; Cheng, X.; Zhao, W.; Taylor, I.A.; Yang, J.; Liu, J.; Peng, Y.-L. A Positive-Charged Patch and Stabilized Hydrophobic Core Are Essential for Avirulence Function of AvrPib in the Rice Blast Fungus. Plant J. 2018, 96, 133–146. [Google Scholar] [CrossRef]
- Mosquera, G.; Giraldo, M.C.; Khang, C.H.; Coughlan, S.; Valent, B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell 2009, 21, 1273–1290. [Google Scholar] [CrossRef]
- Talbot, N.J.; Ebbole, D.J.; Hamer, J.E. Identification and Characterization of MPG1, a Gene Involved in Pathogenicity from the Rice Blast Fungus Magnaporthe grisea. Plant Cell 1993, 5, 1575–1590. [Google Scholar] [CrossRef]
- Ahn, N.; Kim, S.; Choi, W.; Im, K.H.; Lee, Y.H. Extracellular matrix protein gene, EMP1, is required for appressorium formation and pathogenicity of the rice blast fungus, Magnaporthe grisea. Mol. Cells 2004, 17, 166–173. [Google Scholar]
- Kim, S.; Ahn, I.P.; Rho, H.S.; Lee, Y.H. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol. Microbiol. 2005, 57, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Talbot, N.J. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef]
- Saitoh, H.; Fujisawa, S.; Mitsuoka, C.; Ito, A.; Hirabuchi, A.; Ikeda, K.; Irieda, H.; Yoshino, K.; Yoshida, K.; Matsumura, H.; et al. Large-Scale Gene Disruption in Magnaporthe oryzae Identifies MC69, a Secreted Protein Required for Infection by Monocot and Dicot Fungal Pathogens. PLoS Pathog. 2012, 8, e1002711. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, Y.; Zhao, M.; Jing, M.; Liu, X.; Liu, M.; Zhang, Z. Global genome and transcriptome analyses of Magnaporthe oryzae epidemic isolate 98-06 uncover novel effectors and pathogenicity-related genes, revealing gene gain and lose dynamics in genome evolution. PLoS Pathog. 2015, 11, e1004801. [Google Scholar] [CrossRef] [PubMed]
- Mogga, V.; Delventhal, R.; Weidenbach, D.; Langer, S.; Bertram, P.M.; Andresen, K.; Schaffrath, U. Magnaporthe oryzae effectors MoHEG13 and MoHEG16 interfere with host infection and MoHEG13 counteracts cell death caused by Magnaporthe-NLPs in tobacco. Plant Cell Rep. 2016, 35, 1169–1185. [Google Scholar] [CrossRef]
- Sharpee, W.; Oh, Y.; Yi, M.; Franck, W.; Eyre, A.; Okagaki, L.H.; Valent, B.; Dean, R.A. Identification and Characterization of Suppressors of Plant Cell Death (SPD) Effectors from Magnaporthe oryzae. Mol. Plant Pathol. 2017, 18, 850–863. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zeng, H.; Qiu, D.; Guo, L.; Yang, X.; Shi, H.; Zhao, J. Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS ONE 2012, 7, e37654. [Google Scholar] [CrossRef]
- Chen, C.; Lian, B.; Hu, J.; Zhai, H.; Wang, X.; Venu, R.; Liu, E.; Wang, Z.; Chen, M.; Wang, B.; et al. Genome comparison of two Magnaporthe oryzae field isolates reveals genome variations and potential virulence effectors. BMC Genom. 2013, 14, 887. [Google Scholar] [CrossRef]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.-G.; Zhao, K. Enhanced rice blast resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.L.; Peng, Y.-L.; Fan, J. The Nep1-like Protein Family of Magnaporthe oryzae Is Dispensable for the Infection of Rice Plants. Sci. Rep. 2017, 7, 4372. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Yang, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of MoSM1, encoding for an immunity-inducing protein from Magnaporthe oryzae, in rice confers broad-spectrum resistance against fungal and bacterial diseases. Sci. rep. 2017, 7, 41037. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhong, D.; Xie, W.; He, Y.; Zheng, Y.; Lin, Y.; Chen, S. Functional identification of novel cell death-inducing effector proteins from Magnaporthe oryzae. Rice 2019, 12, 59. [Google Scholar] [CrossRef]
- Guillen, K.; Ortiz-Vallejo, D.; Gracy, J.; Fournier, E.; Kroj, T.; Padilla, A. Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi. PLOS Pathog. 2015, 11, e1005228. [Google Scholar] [CrossRef] [PubMed]
- Krause, R.A.; Webster, R.K. The morphology, taxonomy, and sexuality of the rice stem rot fungus, Magnaporthesalvinii (Leptosphaeria salvinii). Mycologia 1972, 64, 103–114. [Google Scholar] [CrossRef]
- Gowrisri, N.; Kamalakannan, A.; Malathi, V.G.; Rajendran, L.; Rajesh, S. Morphological and molecular characterization of Magnaporthe oryzae B. Couch, inciting agent of rice blast disease. Madras Agric. J. 2019, 106, 255–260. [Google Scholar]
- Xue, M.; Yang, J.; Li, Z.; Hu, S.; Yao, N.; Dean, R.A.; Zhao, W.; Shen, M.; Zhang, H.; Li, C.; et al. Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus Magnaporthe oryzae. PLoS Genet. 2012, 8, e1002869. [Google Scholar] [CrossRef]
- Kumar, A.; Sheoran, N.; Prakash, G.; Ghosh, A.; Chikara, S.K.; Rajashekara, H.; Singh, U.D.; Aggarwal, R.; Jain, R.K. Genome sequence of a unique Magnaporthe oryzae RMg-Dl isolate from India that causes blast disease in diverse cereal crops, obtained using PacBio single-molecule and Illumina HiSeq2500 sequencing. Genome Announc. 2017, 5, 1570. [Google Scholar] [CrossRef]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.R.; Pan, H.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Gowda, M.; Shirke, M.D.; Mahesh, H.B.; Chandarana, P.; Rajamani, A.; Chattoo, B.B. Genomeanalysisofrice-blastfungus Magnaporthe oryzae field isolates from southern India. Genom. Data 2015, 5, 284–291. [Google Scholar] [CrossRef][Green Version]
- Suresh, R.; Mosser, D.M. Pattern Recognition Receptors in Innate Immunity, Host Defense, and Immunopathology. Adv. Physiol. Educ. 2013, 37, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Boller, T.; He, S.Y. Innate Immunity in Plants: An Arms Race between Pattern Recognition Receptors in Plants and Effectors in Microbial Pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef]
- Gassmann, W.; Bhattacharjee, S. Effector-Triggered Immunity Signaling: From Gene-for-Gene Pathways to Protein-Protein Interaction Networks. Mol. Plant-Microbe Interact. 2012, 25, 862–868. [Google Scholar] [CrossRef]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wang, W.; Han, J.; Chen, S.; Wang, C.; Zeng, L.; Feng, A.; Yang, J.; Zhou, B.; Zhu, X. Functional Divergence of Duplicated Genes Results in a Novel Blast Resistance Gene Pi50 at the Pi2/9 Locus. Theor. Appl. Genet. 2015, 128, 2213–2225. [Google Scholar] [CrossRef]
- Zheng, W.; Wang, Y.; Wang, L.; Ma, Z.; Zhao, J.; Wang, P.; Zhang, L.; Liu, Z.; Lu, X. Genetic Mapping and Molecular Marker Development for Pi65(t), a Novel Broad-Spectrum Resistance Gene to Rice Blast Using next-Generation Sequencing. Theor. Appl. Genet. 2016, 129, 1035–1044. [Google Scholar] [CrossRef]
- Zhu, D.; Kang, H.; Li, Z.; Liu, M.; Zhu, X.; Wang, Y.; Wang, D.; Wang, Z.; Liu, W.; Wang, G.L. A Genome-Wide Association Study of Field Resistance to Magnaporthe oryzae in Rice. Rice 2016, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.K.; Nayak, S. Functional characterization and signal transduction ability of nucleotide-binding site-leucine-rich repeat resistance genes in plants. Genet. Mol. Res. 2011, 10, 2637–2652. [Google Scholar] [CrossRef]
- Wang, B.H.; Ebbole, D.J.; Wang, Z.H. The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. J. Integr. Agric. 2017, 16, 2746–2760. [Google Scholar] [CrossRef]
- Khang, C.H.; Park, S.Y.; Lee, Y.H.; Valent, B.; Kang, S. Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol. Plant Microbe Interact. 2008, 21, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Bryan, G.T.; Wu, K.S.; Farrall, L.; Jia, Y.; Hershey, H.P.; McAdams, S.A.; Faulk, K.N.; Donaldson, G.K.; Tarchini, R.; Valent, B. A Single Amino Acid Difference Distinguishes Resistant and Susceptible Alleles of the Rice Blast Resistance Gene Pi-Ta. Plant Cell 2000, 12, 2033–2045. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Wang, X.; Wang, F.; Zhao, Z.; Li, G.; Zhu, X.; Chen, L. The fungal effector Avr-pita suppresses innate immunity by increasing COX activity in rice mitochondria. Rice 2021, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.R.; Madhav, M.S.; Singh, B.K.; Shanker, P.; Jana, T.K.; Dalal, V.; Pandit, A.; Singh, A.; Gaikwad, K.; Upreti, H.C.; et al. High-Resolution Mapping, Cloning and Molecular Characterization of the Pi-k h Gene of Rice, Which Confers Resistance to Magnaporthe grisea. Mol. Genet. Genomics 2005, 274, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.R.; Rai, A.K.; Gupta, S.K.; Singh, N.K. Broad-Spectrum Blast Resistance Gene Pi-k h Cloned from Rice Line Tetep Designated as Pi54. J. Plant Biochem. Biotechnol. 2010, 19, 87–89. [Google Scholar] [CrossRef]
- Ray, S.; Singh, P.K.; Gupta, D.K.; Mahato, A.K.; Sarkar, C.; Rathour, R.; Singh, N.K.; Sharma, T.R. Analysis of Magnaporthe oryzae Genome Reveals a Fungal Effector, Which Is Able to Induce Resistance Response in Transgenic Rice Line Containing Resistance Gene, Pi54. Front. Plant Sci. 2016, 7, 1140. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Rai, A.K.; Kanwar, S.S.; Chand, D.; Singh, N.K.; Sharma, T.R. The single functional blast resistance gene Pi54 activates a complex defence mechanism in rice. J. Exp. Bot. 2012, 63, 757–772. [Google Scholar] [CrossRef]
- Maidment, J.H.R.; Franceschetti, M.; Maqbool, A.; Saitoh, H.; Jantasuriyarat, C.; Kamoun, S.; Terauchi, R.; Banfield, M.J. Multiple Variants of the Fungal Effector AVR-Pik Bind the HMA Domain of the Rice Protein OsHIPP19, Providing a Foundation to Engineer Plant Defense. J. Biol. Chem. 2021, 296, 100371. [Google Scholar] [CrossRef]
- Kanzaki, H.; Yoshida, K.; Saitoh, H.; Fujisaki, K.; Hirabuchi, A.; Alaux, L.; Terauchi, R. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 2012, 72, 894–907. [Google Scholar] [CrossRef]
- Zhai, C.; Zhang, Y.; Yao, N.; Lin, F.; Liu, Z.; Dong, Z.; Wang, L.; Pan, Q. Function and Interaction of the Coupled Genes Responsible for Pik-h Encoded Rice Blast Resistance. PLoS ONE 2014, 9, e98067. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, Y.; Kanzaki, H.; Abe, A.; Yoshida, K.; Tamiru, M.; Saitoh, H.; Terauchi, R. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011, 66, 467–479. [Google Scholar] [CrossRef]
- Cesari, S.; Bernoux, M.; Moncuquet, P.; Kroj, T.; Dodds, P.N. A novel conserved mechanism for plant NLR protein pairs: The ‘integrated decoy’ hypothesis. Front. Plant Sci. 2014, 5, 606. [Google Scholar] [CrossRef]
- Miki, S.; Matsui, K.; Kito, H.; Otsuka, K.; Ashizawa, T.; Yasuda, N.; Sone, T. Molecular cloning and characterization of the AVR-Pia locus from a Japanese field isolate of Magnaporthe oryzae. Mol. Plant Pathol. 2009, 10, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Dangol, S.; Chen, Y.; Choi, J.; Cho, Y.S.; Lee, J.E.; Jwa, N.S. Magnaporthe oryzae effector AVR-Pii helps to establish compatibility by inhibition of the rice NADP-malic enzyme resulting in disruption of oxidative burst and host innate immunity. Mol.Cells 2016, 39, 426. [Google Scholar]
- Takagi, H.; Uemura, A.; Yaegashi, H.; Tamiru, M.; Abe, A.; Mitsuoka, C.; Terauchi, R. MutMap-Gap: Whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 2013, 200, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Fujisaki, K.; Abe, Y.; Ito, A.; Saitoh, H.; Yoshida, K.; Kanzaki, H.; Kanzaki, E.; Utsushi, H.; Yamashita, T.; Kamoun, S.; et al. Rice Exo70 Interacts with a Fungal Effector, AVR-Pii, and Is Required for AVR-Pii-triggered Immunity. Plant J. 2015, 83, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Kim, S.G.; Tsuda, K.; Gupta, R.; Park, S.Y.; Kim, S.T.; Kang, K.Y. Magnaporthe oryzae -Secreted Protein MSP1 Induces Cell Death and Elicits Defense Responses in Rice. Mol. Plant-Microbe Interact. 2016, 29, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Ning, Y.; Shu, X.; Dong, B.; Zhang, H.; Wu, D.; Wang, H.; Wang, G.-L.; Zhou, B. The Nup98 Homolog APIP12 Targeted by the Effector AvrPiz-t Is Involved in Rice Basal Resistance Against Magnaporthe oryzae. Rice 2017, 10, 5. [Google Scholar] [CrossRef]
- Park, C.H.; Shirsekar, G.; Bellizzi, M.; Chen, S.; Songkumarn, P.; Xie, X.; Wang, G.L. The E3 ligase APIP10 connects the effector AvrPiz-t to the NLR receptor Piz-t in rice. PLoS Pathog. 2016, 12, e1005529. [Google Scholar] [CrossRef]
- Gupta, D.R.; Surovy, M.Z.; Mahmud, N.U.; Chakraborty, M.; Paul, S.K.; Hossain, M.S.; Bhattacharjee, P.; Mehbub, M.S.; Rani, K.; Yeasmin, R.; et al. Suitable methods for isolation, culture, storage and identification of wheat blast fungus Magnaporthe oryzae Triticum pathotype. Phytopathol. Res. 2020, 2, 30. [Google Scholar] [CrossRef]
- Jamaloddin, M.; Mahender, A.; Gokulan, G.C.; Balachiranjeevi, C.; Maliha, A.; Patel, H.K.; Ali, J. Molecular approaches for disease resistance in rice. In Rice Improvement: Physiological, Molecular Breeding and Genetic Perspectives; Ali, J., Wani, S.H., Eds.; Springer: Berlin/Heidelberg, Germany, 2021; pp. 315–378. [Google Scholar]
- Qin, P.; Hu, X.; Jiang, N.; Bai, Z.; Liu, T.; Fu, C.; Song, Y.; Wang, K.; Yang, Y. A Procedure for Inducing the Occurrence of Rice Seedling Blast in Paddy Field. Plant Pathol. J. 2021, 37, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Jamaloddin, M.; Durga Rani, C.V.; Swathi, G.; Anuradha, C.; Vanisri, S.; Rajan, C.P.D.; Raju, S.K.; Bhuvaneshwari, V.; Jagadeeswar, R.; Laha, G.S.; et al. Marker assisted gene pyramiding (MAGP) for bacterial blight and blast resistance into mega rice variety “Tellahamsa”. PLoS ONE 2020, 15, e0234088. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, K.; Vera Cruz, C.M.; Gruissem, W.; Bhullar, N.K. Large Scale Germplasm Screening for Identification of Novel Rice Blast Resistance Sources. Front. Plant Sci. 2014, 5, 505. [Google Scholar] [CrossRef] [PubMed]
- Srinivas Prasad, M.; Sheshu Madhav, M.; Laha, G.S.; Ladha Lalshmi, D.; Krishnaveni, D.; Mangrauthia, S.K.; Balachandran, S.M.; Sundaram, R.M.; Arunakranthi, B.; Madhan Mohan, K.; et al. Rice Blast Disease and Its Management; Technical Bulletin No. 57; Directorate of Rice Research (ICAR): Hyderabad, India, 2011; Volume 52. [Google Scholar]
- Challagulla, V.; Bhattarai, S.; Midmore, D.J. In-vitro vs in-vivo inoculation: Screening for resistance of australian rice genotypes against blast fungus. Rice Sci. 2015, 22, 132–137. [Google Scholar] [CrossRef]
- IRRI. Standard Evaluation System (SES) 2002 for Rice; International Rice Research Institute (IRRI): Los Baños, Philippine, 2002; pp. 1–45. [Google Scholar]
- Prabhu, A.S.; Filippi, M.C.; Castro, N. Pathogenic variation among isolates of Pyricularia Oryzae affecting rice, wheat, and grasses in Brazil. Int. J. Pest Manag. 1992, 38, 367–371. [Google Scholar]
- Suprun, I.I.; Kovalyev, V.S.; Kharchenko, E.S.; Savenko, E.G. Assessment of breeding lines of rice (Oryza sativa L.) carrying the Pi-40 gene for resistance to rice blast strains from the Krasnodar Region. Vavilov J. Genet. Breed. 2016, 20, 333–336. [Google Scholar] [CrossRef]
- Kim, J.S.; Ahn, S.N.; Kim, C.K.; Shim, C.K. Screening of rice blast resistance genes from aromatic rice germplasms with SNP markers. Plant Pathol. J. 2010, 26, 70–79. [Google Scholar] [CrossRef]
- Kim, J.S.; Ahn, S.N.; Hong, S.J.; Kwon, J.H.; Kim, Y.K.; Jee, H.J.; Shim, C.K. Screening of the Dominant Rice Blast Resistance Genes with PCR-based SNP and CAPS Marker in Aromatic Rice Germplasm. Korean J. Crop Sci. 2011, 56, 329–341. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, P.K.; Arya, M.; Singh, N.K.; Singh, U.S. Molecular Screening of Blast Resistance Genes in Rice using SSR Markers. Plant Pathol. J. 2015, 31, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Imam, J.; Alam, S.; Mandal, N.P.; Variar, M.; Shukla, P. Molecular screening for identification of blast resistance genes in North East and Eastern Indian rice germplasm (Oryza sativa L.) with PCR based makers. Euphytica 2014, 196, 199–211. [Google Scholar] [CrossRef]
- Shikari, A.B.; Khanna, A.; Krishnan, S.G.; Singh, U.D.; Rathour, R.; Tonapi, V.; Sharma, T.R.; Nagarajan, M.; Prabhu, K.V.; Singh, A.K. Molecular Analysis and Phenotypic Validation of Blast Resistance Genes Pita and Pita 2 in Landraces of Rice (Oryza sativa L.). Indian J. Genet. Plant Breed. 2013, 73, 131. [Google Scholar] [CrossRef]
- Yan, L.; Bai-Yuan, Y.; Yun-Liang, P.; Zhi-Juan, J.; Yu-Xiang, Z.; Han-Lin, W.; Chang-Deng, Y. Molecular Scree Ning of Blast Resistance Genes in Rice Germplasms Resistant to Magnaporthe oryzae. Rice Sci. 2017, 24, 41–47. [Google Scholar] [CrossRef]
- Teerasan, W.; Moonsap, P.; Longya, A.; Damchuay, K.; Ito, S.; Tasanasuwan, P.; Kate-Ngam, S.; Jantasuriyarat, C. Rice Blast Resistance Gene Profiling of Thai, Japanese and International Rice Varieties Using Gene-Specific Markers. Plant Genet. Resour. Charact. Util. 2022, 20, 22–28. [Google Scholar] [CrossRef]
- Imbe, T.; Tsunematsu, H.; Kato, H.; Khush, G.S. Genetic Analysis of Blast Resistance in IR Varieties and Resistant Breeding Strategy. In Advances in Rice Blast Research; Springer: Dordrecht, The Netherlands, 2000; pp. 1–8. [Google Scholar]
- Ramalingam, J.; Vera Cruz, C.M.; Kukreja, K.; Chittoor, J.M.; Wu, J.-L.; Lee, S.W.; Baraoidan, M.; George, M.L.; Cohen, M.B.; Hulbert, S.H.; et al. Candidate Defense Genes from Rice, Barley, and Maize and Their Association with Qualitative and Quantitative Resistance in Rice. Mol. Plant-Microbe Interact. 2003, 16, 14–24. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Variar, M.; Cruz, C.M.V.; Carrillo, M.G.; Bhatt, J.C.; Sangar, R.B.S. Rice Blast in India and Strategies to Develop Durably Resistant Cultivars. In Advances in Genetics, Genomics and Control of Rice Blast Disease; Springer: Dordrecht, The Netherlands, 2009; pp. 359–373. [Google Scholar]
- Suwarno; Lubis, E.; Hairmansis, A. Santoso Development of a Package of 20 Varieties for Blast Management on Upland Rice. In Advances in Genetics, Genomics and Control of Rice Blast Disease; Springer: Dordrecht, The Netherlands, 2009; pp. 347–357. [Google Scholar]
- Shindo, K.; Horino, O. Control of rice blast disease by mixed plantings of isogenic lines as multiline cultivars. Bull. Tohoku Natl. Agric. Exp. Stn. 1989, 39, 55–96. [Google Scholar]
- Ise, K. Effect of mixing planting of near isogenic lines of ‘Nipponbare’ rice to reduce blast disease. Breed Sci. 1990, 40, 288. [Google Scholar]
- Koizumi, S.; Fuji, S. Effect of mixtures of isogenic lines developed from rice cv. ‘Sasanishiki and Nipponbare’ on blast development. Res. Bull. Aichi Agric. Res. Ctr. 1994, 26, 87–97. [Google Scholar]
- Koizumi, S. Effect of field resistance on leaf blast development in mixtures of susceptible and resistant rice cultivars. Ann. Phytopathol. Soc. Jpn. 1996, 60, 585–594. [Google Scholar] [CrossRef]
- Nakajima, T.; Sonoda, R.; Yaegashi, H.; Saito, H. Factors related to suppression of leaf blast disease with a multiline ofrice cultivar Sasanishiki and its isogenic lines. Ann. Phytopathol. Soc. Jpn. 1996, 62, 360–364. [Google Scholar] [CrossRef][Green Version]
- Abe, S. Breeding of a Blast Resistant Multiline Variety of Rice, Sasanishiki BL. Jpn. Agric. Res. Q. JARQ 2004, 38, 149–154. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, H.; Fan, J.; Wang, Y.; Li, Y.; Chen, J.; Fan, J.; Yang, S.; Hu, L.; Leung, H.; et al. Genetic Diversity and Disease Control in Rice. Nature 2000, 406, 718–722. [Google Scholar] [CrossRef] [PubMed]
- Persaud, R.; McGowan, D.; Persaud, M. Managing the Imminent Danger of Rice Blast (Pyricularia oryzae Cav.) and Sheath Blight (Rhizoctonia solani Kuhn) Disease: A Critical Review Article. Am. J. Agril. Forestry 2021, 9, 409–423. [Google Scholar] [CrossRef]
- Ahn, S.W. International Collaboration on Breeding for Resistance to Rice Blast. In Rice Blast Disease; Zeigler, R.S., Leong, S.A., Teng, P.S., Eds.; CAB International: Wallingford, UK, 1994; pp. 137–153. [Google Scholar]
- Martínez, C.P.; Victoria, F.C.; Amézquita, M.C.; Tulande, E.; Lema, G.; Zeigler, R.S. Comparison of rice lines derived through anther culture and the pedigree method in relation to blast (Pyricularia grisea Sacc.) resistance. Theor. Appl. Genet. 1996, 92, 583–590. [Google Scholar] [CrossRef]
- Allard, R.W. Principles of Plant Breeding; John Wiley: New York, NY, USA, 1999; ISBN 9780471023098. [Google Scholar]
- Korinsak, S.; Sirithunya, P.; Meakwatanakarn, P.; Sarkarung, S.; Vanavichit, A.; Toojinda, T. Changing Allele Frequencies Associated with Specific Resistance Genes to Leaf Blast in Backcross Introgression Lines of Khao Dawk Mali 105 Developed from a Conventional Selection Program. Field Crops Res. 2011, 122, 32–39. [Google Scholar] [CrossRef]
- Fujimski, H. Recurrent selection by using male sterility for rice improvement. Jpn. Agric. Res. Quart. 1979, 13, 153–156. [Google Scholar]
- Guimarães, E.P.; Correa-Victoria, F.J. Utilización de la selección recurrente para desarrollar resistencia a Pyricularia grisea Sacc. en arroz. In Selección Recurrente en Arroz; Guimarães, E.P., Ed.; CIAT: Cali, Colombia, 1997; pp. 165–175. [Google Scholar]
- Hernandez-Soto, A.; Echeverría-Beirute, F.; Abdelnour-Esquivel, A.; Valdez-Melara, M.; Boch, J.; Gatica-Arias, A. Rice Breeding in the New Era: Comparison of Useful Agronomic Traits. Curr. Plant Biol. 2021, 27, 100211. [Google Scholar] [CrossRef]
- FAO/IAEA MVD. 2022. Available online: https://nucleus.iaea.org/sites/mvd/SitePages/Home.aspx (accessed on 25 June 2022).
- Anonymous. The Gazette of India (S.O. 500(E) Dated 02.02.2021); Ministry of Agriculture and Farmers Welfare, Department of Agriculture, Cooperation and Farmers Welfare, Government of India: New Delhi, India, 2021.
- Sahu, P.K.; Sao, R.; Mondal, S.; Vishwakarma, G.; Gupta, S.K.; Kumar, V.; Singh, S.; Sharma, D.; Das, B.K. Next Generation Sequencing Based Forward Genetic Approaches for Identification and Mapping of Causal Mutations in Crop Plants: A Comprehensive Review. Plants 2020, 9, 1355. [Google Scholar] [CrossRef]
- Li, C.; Wang, D.; Peng, S.; Chen, Y.; Su, P.; Chen, J.; Zheng, L.; Tan, X.; Liu, J.; Xiao, Y.; et al. Genome-wide association mapping of resistance against rice blast strains in South China and identification of a new Pik allele. Rice 2019, 12, 47. [Google Scholar] [CrossRef]
- Sasaki, R. Existence of strains in rice blast fungus. Int. J. Plant Prot. Tokyo 1922, 9, 631–644. [Google Scholar]
- Kiyosawa, S. Genetic studies on host-pathogen relationship in the rice blast disease. Tokyo Agric. Forest Fish Res. Counc. Trop. Agric. Res. Ser. 1967, 1, 137–153. [Google Scholar]
- Ballini, E.; Morel, J.-B.; Droc, G.; Price, A.; Courtois, B.; Notteghem, J.-L.; Tharreau, D. A Genome-Wide Meta-Analysis of Rice Blast Resistance Genes and Quantitative Trait Loci Provides New Insights into Partial and Complete Resistance. Mol. Plant-Microbe Interact. 2008, 21, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Zhai, C.; Lin, F.; Dong, Z.; He, X.; Yuan, B.; Zeng, X.; Wang, L.; Pan, Q. The Isolation and Characterization of Pik, a Rice Blast Resistance Gene Which Emerged after Rice Domestication. New Phytol. 2011, 189, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.D.; Liu, J.L.; Leach, J.E.; Wang, G.L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014, 52, 213–241. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yang, D.; Ali, J.; Mou, T. Molecular marker-assisted pyramiding of broad-spectrum disease resistance genes, Pi2 and Xa23, into GZ63-4S, an elite thermo-sensitive genic male-sterile line in rice. Mol. Plant Breed. 2015, 35, 83–96. [Google Scholar] [CrossRef]
- Khanna, A.; Sharma, V.; Ellur, R.K.; Shikari, A.B.; Gopala Krishnan, S.; Singh, U.D.; Prakash, G.; Sharma, T.R.; Rathour, R.; Variar, M.; et al. Development and Evaluation of Near-Isogenic Lines for Major Blast Resistance Gene(s) in Basmati Rice. Theor. Appl. Genet. 2015, 128, 1243–1259. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Cheng, H.; Hu, J.; Lei, C.; Zhu, X.; Qian, Q. Effect of introgressed Pigm gene on rice blast resistance and yield traits of Japonica rice in cold area. J Shenyang Agric. Uni. 2016, 47, 520–526. [Google Scholar]
- Liu, S.P.; Li, X.; Wang, C.Y.; Li, X.H.; He, Y.Q. Improvement of resistance to rice blast in Zhenshan 97 by molecular marker aided selection. J. Integr. Plant Biol. 2002, 45, 1346–1350. [Google Scholar]
- Joshi, S.; Dhatwalia, S.; Kaachra, A.; Sharma, K.D.; Rathour, R. Genetic and Physical Mapping of a New Rice Blast Resistance Specificity Pi-67 from a Broad Spectrum Resistant Genotype Tetep. Euphytica 2019, 215. [Google Scholar] [CrossRef]
- Dong, L.; Liu, S.; Xu, P.; Deng, W.; Li, X.; Tharreau, D.; Li, J.; Zhou, J.; Wang, Q.; Tao, D.; et al. Fine Mapping of Pi57(t) Conferring Broad Spectrum Resistance against Magnaporthe oryzae in Introgression Line IL-E1454 Derived from Oryza Longistaminata. PLoS ONE 2017, 12, e0186201. [Google Scholar] [CrossRef]
- Wang, R.; Fang, N.; Guan, C.; He, W.; Bao, Y.; Zhang, H. Characterization and Fine Mapping of a Blast Resistant Gene Pi-Jnw1 from the Japonica Rice Landrace Jiangnanwan. PLoS ONE 2016, 11, e0169417. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liang, Z.; Wang, L.; Pan, Q. A New Recessive Gene Conferring Resistance against Rice Blast. Rice 2016, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Peng, P.; Tian, J.; He, Y.; Zhang, L.; Liu, Z.; Yin, D.; Zhang, Z. Pike, a rice blast resistance allele consisting of two adjacent NBS–LRR genes, was identified as a novel allele at the Pik Locus. Mol. Breed. 2015, 35, 117. [Google Scholar] [CrossRef]
- Xiao, W.; Yang, Q.; Sun, D.; Wang, H.; Guo, T.; Liu, Y.; Zhu, X.; Chen, Z. Identification of Three Major R Genes Responsible for Broad-Spectrum Blast Resistance in an Indica Rice Accession. Mol. Breed. 2015, 35, 49. [Google Scholar] [CrossRef]
- Ma, J.; Lei, C.; Xu, X.; Hao, K.; Wang, J.; Cheng, Z.; Ma, X.; Ma, J.; Zhou, K.; Zhang, X.; et al. Pi64, Encoding a Novel CC-NBS-LRR Protein, Confers Resistance to Leaf and Neck Blast in Rice. Mol. Plant-Microbe Interact. 2015, 28, 558–568. [Google Scholar] [CrossRef]
- Sun, P.; Liu, J.; Wang, Y.; Jiang, N.; Wang, S.; Dai, Y.; Gao, J.; Li, Z.; Pan, S.; Wang, D.; et al. Molecular Mapping of the Blast Resistance Gene Pi49 in the Durably Resistant Rice Cultivar Mowanggu. Euphytica 2012, 192, 45–54. [Google Scholar] [CrossRef]
- Lei, C.; Hao, K.; Yang, Y.; Ma, J.; Wang, S.; Wang, J.; Cheng, Z.; Zhao, S.; Zhang, X.; Guo, X.; et al. Identification and Fine Mapping of Two Blast Resistance Genes in Rice Cultivar 93-11. Crop J. 2013, 1, 2–14. [Google Scholar] [CrossRef]
- Koide, Y.; Telebanco-Yanoria, M.J.; Fukuta, Y.; Kobayashi, N. Detection of Novel Blast Resistance Genes, Pi58(t) and Pi59(t), in a Myanmar Rice Landrace Based on a Standard Differential System. Mol. Breed. 2013, 32, 241–252. [Google Scholar] [CrossRef]
- Xiao, W.; Yang, Q.; Wang, H.; Duan, J.; Guo, T.; Liu, Y.; Zhu, X.; Chen, Z. Identification and Fine Mapping of a Major R Gene to Magnaporthe oryzae in a Broad-Spectrum Resistant Germplasm in Rice. Mol. Breed. 2012, 30, 1715–1726. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, S.; Yang, J.; Zhou, S.; Zeng, L.; Han, J.; Su, J.; Wang, L.; Pan, Q. The Identification of Pi50(t), a New Member of the Rice Blast Resistance Pi2/Pi9 Multigene Family. Theor. Appl. Genet. 2012, 124, 1295–1304. [Google Scholar] [CrossRef]
- Jiang, N.; Li, Z.; Wu, J.; Wang, Y.; Wu, L.; Wang, S.; Wang, D.; Wen, T.; Liang, Y.; Sun, P.; et al. Molecular Mapping of the Pi2/9 Allelic Gene Pi2-2 Conferring Broad-Spectrum Resistance to Magnaporthe oryzae in the Rice Cultivar Jefferson. Rice 2012, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Bao, Y.; Xie, L.; Su, Y.; Chu, R.; He, W.; Huang, J.; Wang, J.; Zhang, H. Fine Mapping and Identification of Blast Resistance Gene Pi-Hk1 in a Broad-Spectrum Resistant Japonica Rice Landrace. Phytopathology 2013, 103, 1162–1168. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, B.; Zhu, X.; Yang, J.; Bordeos, A.; Wang, G.; Leach, J.E.; Leung, H. Fine-Mapping and Molecular Marker Development for Pi56(t), a NBS-LRR Gene Conferring Broad-Spectrum Resistance to Magnaporthe oryzae in Rice. Theor. Appl. Genet. 2012, 126, 985–998. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Liu, X.; Wang, L.; Wang, L.; Lin, F.; Cheng, Y.; Chen, Z.; Liao, Y.; Pan, Q. Identification of the Novel Recessive Gene Pi55(t) Conferring Resistance to Magnaporthe oryzae. Sci. China Life Sci. 2012, 55, 141–149. [Google Scholar] [CrossRef][Green Version]
- Endo, T.; Yamaguchi, M.; Kaji, R.; Nakagomi, K.; Kataoka, T.; Yokogami, N.; Nakamura, T.; Ishikawa, G.; Yonemaru, J.; Nishio, T. Close Linkage of a Blast Resistance Gene, Pias(t), with a Bacterial Leaf Blight Resistance Gene, Xa1-as(t), in a Rice Cultivar ‘Asominori’. Breed. Sci. 2012, 62, 334–339. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Deng, X.; Liu, J.; Sun, P.; Liu, Y.; Huang, H.; Jiang, N.; Kang, H.; Ning, Y.; et al. Molecular Mapping of the Blast Resistance Genes Pi2-1 and Pi51(t) in the Durably Resistant Rice ‘Tianjingyeshengdao’. Phytopathology 2012, 102, 779–786. [Google Scholar] [CrossRef]
- Xiao, W.; Yang, Q.; Wang, H.; Guo, T.; Liu, Y.; Zhu, X.; Chen, Z. Identification and Fine Mapping of a Resistance Gene to Magnaporthe oryzae in a Space-Induced Rice Mutant. Mol. Breed. 2010, 28, 303–312. [Google Scholar] [CrossRef]
- Huang, H.; Huang, L.; Feng, G.; Wang, S.; Wang, Y.; Liu, J.; Jiang, N.; Yan, W.; Xu, L.; Sun, P.; et al. Molecular Mapping of the New Blast Resistance Genes Pi47 and Pi48 in the Durably Resistant Local Rice Cultivar Xiangzi 3150. Phytopathology 2011, 101, 620–626. [Google Scholar] [CrossRef]
- Zeng, X.; Yang, X.; Zhao, Z.; Lin, F.; Wang, L.; Pan, Q. Characterization and Fine Mapping of the Rice Blast Resistance Gene Pia. Sci. China Life Sci. 2011, 54, 372–378. [Google Scholar] [CrossRef][Green Version]
- Kumar, P.; Pathania, S.; Katoch, P.; Sharma, T.R.; Plaha, P.; Rathour, R. Genetic and Physical Mapping of Blast Resistance Gene Pi-42(t) on the Short Arm of Rice Chromosome 12. Mol. Breed. 2009, 25, 217–228. [Google Scholar] [CrossRef]
- Lee, S.; Wamishe, Y.; Jia, Y.; Liu, G.; Jia, M.H. Identification of Two Major Resistance Genes against Race IE-1k of Magnaporthe oryzae in the Indica Rice Cultivar Zhe733. Mol. Breed. 2009, 24, 127–134. [Google Scholar] [CrossRef]
- Yang, Q.; Lin, F.; Wang, L.; Pan, Q. Identification and Mapping of Pi41, a Major Gene Conferring Resistance to Rice Blast in the Oryza sativa Subsp. Indica Reference Cultivar, 93-11. Theor. Appl. Genet. 2009, 118, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Tao, Y.; Chen, X.; Zou, Y.; Lei, C.; Wang, J.; Li, X.; Zhao, X.; Zhang, M.; Lu, Z.; et al. Identification of a New Rice Blast Resistance Gene, Pid3, by Genome wide Comparison of Paired Nucleotide-Binding Site–Leucine-Rich Repeat Genes and Their Pseudogene Alleles Between the Two Sequenced Rice Genomes. Genetics 2009, 182, 1303–1311. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, X.; Lin, F.; Pan, Q. Characterization of Rice Blast Resistance Genes in the Pik Cluster and Fine Mapping of the Pik-p Locus. Phytopathology 2009, 99, 900–905. [Google Scholar] [CrossRef]
- Xu, X.; Chen, H.; Fujimura, T.; Kawasaki, S. Fine Mapping of a Strong QTL of Field Resistance against Rice Blast, Pikahei-1(t), from Upland Rice Kahei, Utilizing a Novel Resistance Evaluation System in the Greenhouse. Theor. Appl. Genet. 2008, 117, 997–1008. [Google Scholar] [CrossRef]
- Xu, X.; Hayashi, N.; Wang, C.T.; Kato, H.; Fujimura, T.; Kawasaki, S. Efficient Authentic Fine Mapping of the Rice Blast Resistance Gene Pik-h in the Pik Cluster, Using New Pik-h-Differentiating Isolates. Mol. Breed. 2008, 22, 289–299. [Google Scholar] [CrossRef]
- Dwinita, W.U.; Sugiono, M.; Hajrial, A.; Asep, S.; Ida, H. Blast resistance genes in wild rice Oryza rufipogon and rice cultivar IR64. Ind J Agric. 2008, 1, 71–76. [Google Scholar]
- Liu, X.; Yang, Q.; Lin, F.; Hua, L.; Wang, C.; Wang, L.; Pan, Q. Identification and Fine Mapping of Pi39(t), a Major Gene Conferring the Broad-Spectrum Resistance to Magnaporthe oryzae. Mol. Genet. Genom. 2007, 278, 403–410. [Google Scholar] [CrossRef]
- Zenbayashi-Sawata, K.; Fukuoka, S.; Katagiri, S.; Fujisawa, M.; Matsumoto, T.; Ashizawa, T.; Koizumi, S. Genetic and Physical Mapping of the Partial Resistance Gene, Pi34, to Blast in Rice. Phytopathology 2007, 97, 598–602. [Google Scholar] [CrossRef]
- Jeung, J.U.; Kim, B.R.; Cho, Y.C.; Han, S.S.; Moon, H.P.; Lee, Y.T.; Jena, K.K. A Novel Gene, Pi40(t), Linked to the DNA Markers Derived from NBS-LRR Motifs Confers Broad Spectrum of Blast Resistance in Rice. Theor. Appl. Genet. 2007, 115, 1163–1177. [Google Scholar] [CrossRef]
- Deng, Y.; Zhu, X.; Shen, Y.; He, Z. Genetic Characterization and Fine Mapping of the Blast Resistance Locus Pigm(t) Tightly Linked to Pi2 and Pi9 in a Broad-Spectrum Resistant Chinese Variety. Theor. Appl. Genet. 2006, 113, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Liu, G.; Zhou, B.; Bellizzi, M.; Zeng, L.; Dai, L.; Han, B.; Wang, G.-L. The Broad-Spectrum Blast Resistance Gene Pi9 Encodes a Nucleotide-Binding Site–Leucine-Rich Repeat Protein and Is a Member of a Multigene Family in Rice. Genetics 2006, 172, 1901–1914. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G.; et al. A B-Lectin Receptor Kinase Gene Conferring Rice Blast Resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Koizumi, S.; La, T.N.; Zenbayashi, K.S.; Ashizawa, T.; Yasuda, N.; Imazaki, I.; Miyasaka, A. Pi35(t), a New Gene Conferring Partial Resistance to Leaf Blast in the Rice Cultivar Hokkai 188. Theor. Appl. Genet. 2006, 113, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Gowda, M.; Roy-Barman, S.; Chattoo, B.B. Molecular Mapping of a Novel Blast Resistance Gene Pi38 in Rice Using SSLP and AFLP Markers. Plant Breed. 2006, 125, 596–599. [Google Scholar] [CrossRef]
- Hayashi, K.; Yoshida, H.; Ashikawa, I. Development of PCR-Based Allele-Specific and InDel Marker Sets for Nine Rice Blast Resistance Genes. Theor. Appl. Genet. 2006, 113, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Qu, S.; Liu, G.; Dolan, M.; Sakai, H.; Lu, G.; Bellizzi, M.; Wang, G.L. The Eight Amino-Acid Differences Within Three Leucine-Rich Repeats Between Pi2 and Piz-t Resistance Proteins Determine the Resistance Specificity to Magnaporthe grisea. Mol. Plant-Microbe Interact. 2006, 19, 1216–1228. [Google Scholar] [CrossRef]
- Lei, C.L.; Huang, D.Y.; Li, W.; Wang, L.J.; Liu, Z.L.; Wang, X.T.; Shi, K.; Cheng, Z.J.; Zhang, X.; Ling, Z.Z.; et al. Molecular mapping of a blast resistance gene in an indica rice cultivar Yanxian No. 1. Rice Genet. Newsl. 2005, 22, 76–77. [Google Scholar]
- Chen, S.; Wang, L.; Que, Z.; Pan, R.; Pan, Q. Genetic and physical mapping of Pi37(t), a new gene conferring resistance to rice blast in the famous cultivar St. No. 1. Theor. Appl. Genet. 2005, 111, 1563–1570. [Google Scholar] [CrossRef]
- Wu, J.L.; Fan, Y.-Y.; Li, D.-B.; Zheng, K.-L.; Leung, H.; Zhuang, J.-Y. Genetic Control of Rice Blast Resistance in the Durably Resistant Cultivar Gumei 2 against Multiple Isolates. Theor. Appl. Genet. 2005, 111, 50–56. [Google Scholar] [CrossRef]
- Liu, X.Q.; Wang, L.; Chen, S.; Lin, F.; Pan, Q.H. Genetic and Physical Mapping of Pi36(t), a Novel Rice Blast Resistance Gene Located on Rice Chromosome 8. Mol. Genet. Genom. 2005, 274, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhang, S.; Zhu, X.; Yang, Q.; Wu, S.; Mei, M.; Mauleon, R.; Leach, J.; Mew, T.; Leung, H. Candidate Defense Genes as Predictors of Quantitative Blast Resistance in Rice. Mol. Plant-Microbe Interact. 2004, 17, 1146–1152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, X.W.; Li, S.G.; Xu, J.C.; Zhai, W.X.; Ling, Z.Z.; Ma, B.T.; Wang, Y.P.; Wang, W.M.; Cao, G.; Ma, Y.Q.; et al. Identification of two blast resistance genes in a rice variety, Digu. J. Phytopathol. 2004, 152, 77–85. [Google Scholar] [CrossRef]
- Zhou, J.H.; Wang, J.L.; Xu, J.C.; Lei, C.L.; Ling, Z.Z. Identification and Mapping of a Rice Blast Resistance Gene Pi-g(t) in the Cultivar Guangchangzhan. Plant Pathol. 2004, 53, 191–196. [Google Scholar] [CrossRef]
- Fukuta, Y. The Pish blast resistance gene of rice (Oryza sativa L.) is located on the long arm of chromosome 1. Japan International Research Center for Agricultural Sciences (JIRCAS). Ann. Rep. 2004, 16, 46–47. [Google Scholar]
- Zhu, M.; Wang, L.; Pan, Q. Identification and Characterization of a New Blast Resistance Gene Located on Rice Chromosome 1 through Linkage and Differential Analyses. Phytopathology 2004, 94, 515–519. [Google Scholar] [CrossRef]
- Barman, S.R.; Gowda, M.; Venu, R.C.; Chattoo, B.B. Identification of a Major Blast Resistance Gene in the Rice Cultivar “Tetep”. Plant Breed. 2004, 123, 300–302. [Google Scholar] [CrossRef]
- Fjellstrom, R.; Conaway-Bormans, C.A.; McClung, A.M.; Marchetti, M.A.; Shank, A.R.; Park, W.D. Development of DNA Markers Suitable for Marker Assisted Selection of Three Pi Genes Conferring Resistance to Multiple Pyricularia grisea Pathotypes. Crop Sci. 2004, 44, 1790–1798. [Google Scholar] [CrossRef]
- Sallaud, C.; Lorieux, M.; Roumen, E.; Tharreau, D.; Berruyer, R.; Svestasrani, P.; Garsmeur, O.; Ghesquiere, A.; Notteghem, J.L. Identification of Five New Blast Resistance Genes in the Highly Blast-Resistant Rice Variety IR64 Using a QTL Mapping Strategy. Theor. Appl. Genet. 2003, 106, 794–803. [Google Scholar] [CrossRef]
- Jeon, J.S.; Chen, D.; Yi, G.-H.; Wang, G.L.; Ronald, P.C. Genetic and Physical Mapping of Pi5(t), a Locus Associated with Broad-Spectrum Resistance to Rice Blast. Mol. Genet. Genom. 2003, 269, 280–289. [Google Scholar] [CrossRef]
- Berruyer, R.; Adreit, H.; Milazzo, J.; Gaillard, S.; Berger, A.; Dioh, W.; Lebrun, M.H.; Tharreau, D. Identification and Fine Mapping of Pi33, the Rice Resistance Gene Corresponding to the Magnaporthe grisea Avirulence Gene ACE1. Theor. Appl. Genet. 2003, 107, 1139–1147. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.H.; Hu, Z.D.; Tanisaka, T.; Wang, L. Fine mapping of the blast resistance gene Pi15, linked to Pii, on rice chromosome 9. Acta Bot. Sin. 2003, 45, 871–877. [Google Scholar]
- Kinoshita, T.; Kiyosawa, S. Some considerations on linkage relationships between Pii and Piz in the blast resistance of rice. Rice Genet. Newsl. 1997, 14, 57–59. [Google Scholar]
- Jiang, J.; Wang, S. Identification of a 118-Kb DNA Fragment Containing the Locus of Blast Resistance Gene Pi-2(t) in Rice. Mol. Genet. Genom. 2002, 268, 249–252. [Google Scholar] [CrossRef]
- Zhuang, J.Y.; Wu, J.L.; Fan, Y.Y.; Rao, Z.M.; Zheng, K.L. Genetic drag between a blast resistance gene and QTL conditioning yield trait detected in a recombinant inbred line population in rice. Rice Genet Newsl. 2001, 18, 69–70. [Google Scholar]
- Chauhan, R.; Farman, M.; Zhang, H.B.; Leong, S. Genetic and Physical Mapping of a Rice Blast Resistance Locus, Pi-CO39(t), That Corresponds to the Avirulence Gene AVR1-CO39 of Magnaporthe grisea. Mol. Genet. Genom. 2002, 267, 603–612. [Google Scholar] [CrossRef]
- Fukuoka, S.; Okuno, K. QTL analysis and mapping of pi21, a recessive gene for field resistance to rice blast in Japanese upland rice. Theor. Appl. Genet. 2001, 103, 185–190. [Google Scholar] [CrossRef]
- Ahn, S.N.; Kim, Y.K.; Hong, H.C.; Han, S.S.; Kwon, S.J.; Choi, H.C.; Moon, H.P.; McCouch, S.R. Molecular mapping of a new gene for resistance to rice blast (Pyricularia grisea Sacc.). Euphytica 2000, 116, 17–22. [Google Scholar] [CrossRef]
- Tabien, R.E.; Li, Z.; Paterson, A.H.; Marchetti, M.A.; Stansel, J.W.; Pinson, S.R.M.; Park, W.D. Mapping of Four Major Rice Blast Resistance Genes from ’Lemont’ and ’Teqing’ and Evaluation of Their Combinatorial Effect for Field Resistance. Theor. Appl. Genet. 2000, 101, 1215–1225. [Google Scholar] [CrossRef]
- Tabien, R.; Li, Z.; Paterson, A.; Marchetti, M.; Stansel, J.; Pinson, S. Mapping QTLs for Field Resistance to the Rice Blast Pathogen and Evaluating Their Individual and Combined Utility in Improved Varieties. Theor. Appl. Genet. 2002, 105, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.H.; dela Viña, M.; Inukai, T.; Mackill, D.J.; Ronald, P.C.; Nelson, R.J. Molecular Mapping of the Blast Resistance Gene, Pi44(t), in a Line Derived from a Durably Resistant Rice Cultivar. Theor. Appl. Genet. 1999, 98, 1046–1053. [Google Scholar] [CrossRef]
- Wang, P.; Tanisaka, A. New Blast Resistance Gene Identified in the Indian Native Rice Cultivar Aus373 through Allelism and Linkage Tests. Plant Pathol. 1999, 48, 288–293. [Google Scholar] [CrossRef]
- Fujii, K.; Hayano-Saito, Y.; Sugiura, N.; Hayashi, N.; Saka, N.; Tooyama, T.; Izawa, T.; Shumiya, A. Gene Analysis of Panicle Blast Resistance in Rice Cultivars with Rice Stripe Resistance. Breed. Res. 1999, 1, 203–210. [Google Scholar] [CrossRef]
- Hayashi, N.; Inoue, H.; Kato, T.; Funao, T.; Shirota, M.; Shimizu, T.; Kanamori, H.; Yamane, H.; Hayano-Saito, Y.; Matsumoto, T.; et al. Durable Panicle Blast-Resistance Gene Pb1 Encodes an Atypical CC-NBS-LRR Protein and Was Generated by Acquiring a Promoter through Local Genome Duplication. Plant J. 2010, 64, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.Y.; Lu, J.; Qian, H.R.; Lin, H.X.; Zheng, K.L. Tagging of blast resistance gene(s) to DNA markers and marker-assisted selection (MAS) in rice improvement. In IAEA-TECDOC:1010; Int Atomic Energy Agency (IAEA): Tokyo, Japan, 1998; pp. 55–61. [Google Scholar]
- Hayashi, N.; Ando, I.; Imbe, T. Identification of a New Resistance Gene to a Chinese Blast Fungus Isolate in the Japanese Rice Cultivar Aichi Asahi. Phytopathology 1998, 88, 822–827. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pan, Q.H.; Wang, L.; Ikehashi, H.; Yamagata, H.; Tanisaka, T. Identification of Two New Genes Conferring Resistance to Rice Blast in the Chinese Native Cultivar ‘Maowangu’. Plant Breed. 1998, 117, 27–31. [Google Scholar] [CrossRef]
- Ahn, S.N.; Kim, Y.K.; Hong, H.C.; Han, S.S.; Kwon, S.J.; Choi, H.C.; Moon, H.P.; McCouch, S.R. Molecular mapping of a gene for resistance to a Korean isolate of rice blast. Rice Genet. Newsl. 1996, 13, 74. [Google Scholar]
- Terashima, T.; Fukuoka, S.; Saka, N.; Kudo, S. Mapping of a Blast Field Resistance GenePi39(t) of Elite Rice Strain Chubu 111. Plant Breed. 2008, 127, 485–489. [Google Scholar] [CrossRef]
- Rybka, K.; Miyamoto, M.; Ando, I.; Saito, A.; Kawasaki, S. High Resolution Mapping of the Indica-Derived Rice Blast Resistance Genes II. Pi-Ta2 and Pi-Ta and a Consideration of Their Origin. Mol. Plant-Microbe Interact. 1997, 10, 517–524. [Google Scholar] [CrossRef]
- Li, W.; Lei, C.; Cheng, Z.; Jia, Y.; Huang, D.; Wang, J.; Wang, J.; Zhang, X.; Su, N.; Guo, X.; et al. Identification of SSR Markers for a Broad-Spectrum Blast Resistance Gene Pi20(t) for Marker-Assisted Breeding. Mol. Breed. 2008, 22, 141–149. [Google Scholar] [CrossRef]
- Imbe, T.; Oba, S.; Yanoria, M.J.T. Tsunematsu, H. A new gene for blast resistance in rice cultivar, IR24. Rice Genet. Newsl. 1997, 14, 60–62. [Google Scholar]
- Hayasaka, H.; Miyao, A.; Yano, M.; Matsunaga, K.; Sasaki, T. RFLP mapping of a rice blast resistance gene Pik. Breed Sci. 1996, 46, 68. [Google Scholar]
- Kaji, R.; Ogawa, T. RFLP mapping of a blast resistance gene Pikm in rice. Breed Sci. 1996, 46, 70. [Google Scholar]
- Naqvi, N.I.; Bonman, J.M.; Mackill, D.J.; Nelson, R.J.; Chattoo, B.B. Identification of RAPD Markers Linked to a Major Blast Resistance Gene in Rice. Mol. Breed. 1995, 1, 341–348. [Google Scholar] [CrossRef]
- Pan, Q. Identification of a New Blast Resistance Gene in the Indica Rice Cultivar Kasalath Using Japanese Differential Cultivars and Isozyme Markers. Phytopathology 1996, 86, 1071. [Google Scholar] [CrossRef]
- Wu, K.S.; Martinez, C.; Lentini, Z.; Tohme, J.; Chumley, F.G.; Scolnik, P.A.; Valent, B. Cloning a blast resistance gene by chromosome walking. In Proceedings of the Third International Rice Genetics Symposium, Manila, Philippines, 16–20 October 1995; Khush, G.S., Ed.; IRRI: Los Baños, Philippines, 1996; pp. 669–674. [Google Scholar]
- Tabien, R.E.; Pinson, S.R.M.; Marchetti, M.A.; Li, Z.; Park, W.D.; Paterson, A.H.; Stansel, J.W. Blast resistance genes from Teqing and Lemont. In Rice Genetics III. Proceedings of Third International Rice Genetics Symposium, Manila, Philippines, 16–20 October 1996; Khush, G.S., Ed.; IRRI: Los Baños, Philippines, 1996; pp. 451–455. [Google Scholar]
- Parco, A. STS markers for the blast resistance gene Pi-1(t). Rice Genome 1995, 4, 9. [Google Scholar]
- Yu, Z.H.; Mackill, D.J.; Bonman, J.M.; McCouch, S.R.; Guiderdoni, E.; Notteghem, J.L.; Tanksley, S.D. Molecular Mapping of Genes for Resistance to Rice Blast (Pyricularia grisea Sacc.). Theor. Appl. Genet. 1996, 93, 859–863. [Google Scholar] [CrossRef]
- Causse, M.A.; Fulton, T.M.; Cho, Y.G.; Ahn, S.N.; Chunwongse, J.; Wu, K.; Xiao, J.; Yu, Z.; Ronald, P.C.; Harrington, S.E. Saturated Molecular Map of the Rice Genome Based on an Interspecific Backcross Population. Genetics 1994, 138, 1251–1274. [Google Scholar] [CrossRef]
- McCouch, S.R.; Nelson, R.J.; Tohme, J.; Zeigler, R.S. Mapping of blast resistance genes in rice. In Rice Blast Disease; Zeigler, R.S., Leong, S.A., Teng, P.S., Eds.; C.A.B. International: Wallingford, UK, 1994; pp. 167–186. [Google Scholar]
- Wang, G.L.; Mackill, D.J.; Bonman, J.M.; McCouch, S.R.; Champoux, M.C.; Nelson, R.J. RFLP Mapping of Genes Conferring Complete and Partial Resistance to Blast in a Durably Resistant Rice Cultivar. Genetics 1994, 136, 1421–1434. [Google Scholar] [CrossRef]
- Wu, K.S.; Tanksley, S.D. Abundance, Polymorphism and Genetic Mapping of Microsatellites in Rice. Mol. Gen. Genet. 1993, 241, 225–235. [Google Scholar] [CrossRef]
- Amante-Bordeos, A.; Sitch, L.A.; Nelson, R.; Dalmacio, R.D.; Oliva, N.P.; Aswidinnoor, H.; Leung, H. Transfer of Bacterial Blight and Blast Resistance from the Tetraploid Wild Rice Oryza minuta to Cultivated Rice, Oryza sativa. Theor. Appl. Genet. 1992, 84, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Mackill, D.J. Inheritance of Blast Resistance in Near-Isogenic Lines of Rice. Phytopathology 1992, 82, 746. [Google Scholar] [CrossRef]
- Yu, Z.H.; Mackill, D.J.; Bonman, J.M.; Tanksley, S.D. Tagging Genes for Blast Resistance in Rice via Linkage to RFLP Markers. Theor. Appl. Genet. 1991, 81, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Goto, I. Genetic Studies on Resistance of Rice Plant to Blast Fungus. (VII). Blast Resistance Genes of Kuroka. Jpn. J. Phytopathol. 1988, 54, 460–465. [Google Scholar] [CrossRef]
- Imbe, T.; Matsumoto, S. Inheritance of Resistance of Rice Varieties to the Blast Fungus Strains Virulent to the Variety “Reiho”. Jpn. J. Breed. 1985, 35, 332–339. [Google Scholar] [CrossRef]
- Goto, I. Genetic Studies on Resistance of Rice Plant to Blast Fungus. II. Difference in Resistance to the Blast Disease between Fukunishiki and Its Parental Cultivar, Zenith. Jpn. J. Phytopathol. 1976, 42, 253–260. [Google Scholar] [CrossRef]
- Shinoda, H.; Toriyama, K.; Yunoki, T.; Ezuka, A.; Sakurai, Y. Studies on the varietal resistance of rice to blast. VI. Linkage relationship of blast resistance genes. Bull. Chugoku Nat. Agric. Exp. Stat. A 1971, 20, 1–25. [Google Scholar]
- Ise, K. Linkage analysis of some blast resistance gene in rice, Oryza sativa L. Jpn. J Breed. 1991, 42, 388–389. [Google Scholar]
- Goto, I. Genetic studies on the resistance of rice plant to the blast fungus I. Inheritance of resistance in crosses Sensho x H-79 and Imochishirazu x H-79. Ann. Phytopathol. Soc. Jpn. 1970, 36, 304–312. [Google Scholar] [CrossRef]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K.; et al. Loss of Function of a Proline-Containing Protein Confers Durable Disease Resistance in Rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef]
- Inukai, T.; Nagashima, S.; Kato, M. Pid3-I1 Is a Race-Specific Partial-Resistance Allele at the Pid3 Blast Resistance Locus in Rice. Theor. Appl. Genet. 2019, 132, 395–404. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, X.; Jia, Y.; Minkenberg, B.; Wheatley, M.; Fan, J.; Jia, M.H.; Famoso, A.; Edwards, J.D.; Wamishe, Y.; et al. The Rice Blast Resistance Gene Ptr Encodes an Atypical Protein Required for Broad-Spectrum Disease Resistance. Nat. Commun. 2018, 9, 2039. [Google Scholar] [CrossRef]
- Fukuoka, S.; Yamamoto, S.-I.; Mizobuchi, R.; Yamanouchi, U.; Ono, K.; Kitazawa, N.; Yasuda, N.; Fujita, Y.; Thi Thanh Nguyen, T.; Koizumi, S.; et al. Multiple Functional Polymorphisms in a Single Disease Resistance Gene in Rice Enhance Durable Resistance to Blast. Sci. Rep. 2014, 4, 4550. [Google Scholar] [CrossRef]
- Xu, X.; Hayashi, N.; Wang, C.-T.; Fukuoka, S.; Kawasaki, S.; Takatsuji, H.; Jiang, C.-J. Rice Blast Resistance Gene Pikahei-1(t), a Member of a Resistance Gene Cluster on Chromosome 4, Encodes a Nucleotide-Binding Site and Leucine-Rich Repeat Protein. Mol. Breed. 2014, 34, 691–700. [Google Scholar] [CrossRef]
- Hua, L.; Wu, J.; Chen, C.; Wu, W.; He, X.; Lin, F.; Wang, L.; Ashikawa, I.; Matsumoto, T.; Wang, L.; et al. The Isolation of Pi1, an Allele at the Pik Locus Which Confers Broad Spectrum Resistance to Rice Blast. Theor. Appl. Genet. 2012, 125, 1047–1055. [Google Scholar] [CrossRef]
- Das, A.; Soubam, D.; Singh, P.K.; Thakur, S.; Singh, N.K.; Sharma, T.R. A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct. Integr. Genom. 2012, 12, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shi, Y.; Liu, W.; Chai, R.; Fu, Y.; Zhuang, J.; Wu, J. A Pid3 Allele from Rice Cultivar Gumei2 Confers Resistance to Magnaporthe oryzae. J. Genet. Genom. 2011, 38, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Zhai, C.; Wang, W.; Zeng, X.; Xu, X.; Hu, H.; Lin, F.; Wang, L.; Pan, Q. The Pik-p Resistance to Magnaporthe oryzae in Rice Is Mediated by a Pair of Closely Linked CC-NBS-LRR Genes. Theor. Appl. Genet. 2011, 122, 1017–1028. [Google Scholar] [CrossRef]
- Takahashi, A.; Hayashi, N.; Miyao, A.; Hirochika, H. Unique Features of the Rice Blast Resistance Pish Locus Revealed by Large Scale Retrotransposon-Tagging. BMC Plant Biol. 2010, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Yoshida, H. Refunctionalization of the Ancient Rice Blast Disease Resistance Gene Pit by the Recruitment of a Retrotransposon as a Promoter. Plant J. 2009, 57, 413–425. [Google Scholar] [CrossRef]
- Lee, S.K.; Song, M.Y.; Seo, Y.-S.; Kim, H.K.; Ko, S.; Cao, P.-J.; Suh, J.P.; Yi, G.; Roh, J.-H.; Lee, S.; et al. Rice Pi5-Mediated Resistance to Magnaporthe oryzae Requires the Presence of Two Coiled-Coil–Nucleotide-Binding–Leucine-Rich Repeat Genes. Genetics 2009, 181, 1627–1638. [Google Scholar] [CrossRef]
- Ashikawa, I.; Hayashi, N.; Yamane, H.; Kanamori, H.; Wu, J.; Matsumoto, T.; Ono, K.; Yano, M. Two Adjacent Nucleotide-Binding Site–Leucine-Rich Repeat Class Genes Are Required to Confer Pikm-Specific Rice Blast Resistance. Genetics 2008, 180, 2267–2276. [Google Scholar] [CrossRef]
- Lin, F.; Chen, S.; Que, Z.; Wang, L.; Liu, X.; Pan, Q. The Blast Resistance GenePi37Encodes a Nucleotide Binding Site–Leucine-Rich Repeat Protein and Is a Member of a Resistance Gene Cluster on Rice Chromosome 1. Genetics 2007, 177, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.X.; Yano, M.; Yamanouchi, U.; Iwamoto, M.; Monna, L.; Hayasaka, H.; Katayose, Y.; Sasaki, T. The Pib Gene for Rice Blast Resistance Belongs to the Nucleotide Binding and Leucine-Rich Repeat Class of Plant Disease Resistance Genes. Plant J. 1999, 19, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Hittalmani, S.; Parco, A.; Mew, T.V.; Zeigler, R.S.; Huang, N. Fine Mapping and DNA Marker-Assisted Pyramiding of the Three Major Genes for Blast Resistance in Rice. Theor. Appl. Genet. 2000, 100, 1121–1128. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.; Singh, S.P.; Ellur, R.K.; Choudhary, V.; Sarkel, S.; Singh, D.; Krishnan, S.G.; Nagarajan, M.; Vinod, K.K.; et al. Incorporation of Blast Resistance into “PRR78”, an Elite Basmati Rice Restorer Line, through Marker Assisted Backcross Breeding. Field Crop. Res. 2012, 128, 8–16. [Google Scholar] [CrossRef]
- Skamnioti, P.; Gurr, S.J. Against the Grain: Safeguarding Rice from Rice Blast Disease. Trends Biotechnol. 2009, 27, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Zenbayashi, K.; Ashizawa, T.; Tani, T.; Koizumi, S. Mapping of the QTL (Quantitative Trait Locus) Conferring Partial Resistance to Leaf Blast in Rice Cultivar Chubu 32. Theor. Appl. Genet. 2002, 104, 547–552. [Google Scholar] [CrossRef]
- Sirithunya, P.; Tragoonrung, S.; Vanavichit, A.; Pa-In, N.; Vongsaprom, C.; Toojinda, T. Quantitative trait loci associated with leaf and neck blast resistance in recombinant inbred line population of rice (Oryza sativa). DNA Res. 2002, 9, 79–88. [Google Scholar] [CrossRef][Green Version]
- Talukder, Z.I.; Tharreau, D.; Price, A.H. Quantitative Trait Loci Analysis Suggests That Partial Resistance to Rice Blast Is Mostly Determined by Race–Specific Interactions. New Phytol. 2004, 162, 197–209. [Google Scholar] [CrossRef]
- Cho, Y.C.; Kwon, S.W.; Suh, J.P.; Kim, J.J.; Lee, J.H.; Roh, J.H.; Oh, M.K.; Kim, M.K.; Ahn, S.N.; Koh, H.J.; et al. QTLs identification and confirmation of field resistance to leaf blast in temperate japonica rice (Oryza sativa L.). J. Crop Sci. Biotechnol. 2008, 11, 269–276. [Google Scholar]
- Koide, Y.; Kobayashi, N.; Xu, D.; Fukuta, Y. Resistance genes and selection DNA markers for blast disease in rice (Oryza sativa L.). Jpn. Agril. Res. Q. 2009, 43, 255–280. [Google Scholar] [CrossRef]
- Jia, Y.; Liu, G. Mapping quantitative trait Loci for resistance to rice blast. Phytopathology 2011, 101, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Ashkani, S.; Rafii, M.Y.; Rahim, H.A.; Latif, M.A. Genetic dissection of rice blast resistance by QTL mapping approach using an F3 population. Mol. Biol. Rep. 2013, 40, 2503–2515. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, T.; Hayano-Saito, Y.; Oide, S.; Ebana, K.; La, N.T.; Hayashi, K.; Ashizawa, T.; Suzuki, F.; Koizumi, S. Quantitative trait locus analysis of resistance to panicle blast in the rice cultivar Miyazakimochi. Rice 2014, 7, 2. [Google Scholar] [CrossRef]
- Fang, N.; Wang, R.; He, W.; Yin, C.; Guan, C.; Chen, H.; Huang, J.; Wang, J.; Bao, Y.; Zhang, H. QTL mapping of panicle blast resistance in japonica landrace heikezijing and its application in rice breeding. Mol. Breed. 2016, 36, 171. [Google Scholar] [CrossRef]
- Nagaoka, I.; Sasahara, H.; Tabuchi, H.; Shigemune, A.; Matsushita, K.; Maeda, H.; Goto, A.; Fukuoka, S.; Ando, T.; Miura, K. Quantitative trait loci analysis of blast resistance in Oryza sativa L. ‘Hokuriku 193’. Breed. Sci. 2017, 67, 159–164. [Google Scholar] [CrossRef]
- Fang, N.; Wei, X.; Shen, L.; Yu, Y.; Li, M.; Yin, C.; He, W.; Guan, C.; Chen, H.; Zhang, H.; et al. Fine mapping of a panicle blast resistance gene Pb-bd1 in japonica landrace Bodao and its application in rice breeding. Rice 2019, 12, 18. [Google Scholar] [CrossRef]
- Devi, S.J.S.R.; Singh, K.; Umakanth, B.; Vishalakshi, B.; Rao, K.V.S.; Suneel, B.; Sharma, S.K.; Kadambari, G.K.M.; Prasad, M.S.; Senguttvel, P.; et al. Identification and Characterization of a Large Effect QTL from Oryza glumaepatula Revealed Pi68(t) as Putative Candidate Gene for Rice Blast Resistance. Rice 2020, 13, 17. [Google Scholar] [CrossRef]
- Korinsak, S.; Wongsaprom, C.; Jamboonsri, W.; Sriprakhon, S.; Sirithunya, K.; Vanavichit, A.; Toojinda, T. Identification of broad-spectrum resistance QTLs against rice blast fungus and their application in different rice genetic backgrounds. J. Genet. 2022, 101, 16. [Google Scholar] [CrossRef]
- Tian, T.; Chen, L.; Ai, Y.; He, H. Selection of Candidate Genes Conferring Blast Resistance and Heat Tolerance in Rice through Integration of Meta-QTLs and RNA-Seq. Genes 2022, 13, 224. [Google Scholar] [CrossRef]
- Zhao, K.; Tung, C.-W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-Wide Association Mapping Reveals a Rich Genetic Architecture of Complex Traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Wang, C.; Yang, Y.; Yuan, X.; Xu, Q.; Feng, Y.; Yu, H.; Wang, Y.; Wei, X. Genome-Wide Association Study of Blast Resistance in Indica Rice. BMC Plant Biol. 2014, 14, 311. [Google Scholar] [CrossRef]
- Shinada, H.; Yamamoto, T.; Sato, H.; Yamamoto, E.; Hori, K.; Yonemaru, J.; Sato, T.; Fujino, K. Quantitative Trait Loci for Rice Blast Resistance Detected in a Local Rice Breeding Population by Genome-Wide Association Mapping. Breed. Sci. 2015, 65, 388–395. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, X.; Jia, M.H.; Ghai, P.; Lee, F.N.; Jia, Y. Genome-Wide Association of Rice Blast Disease Resistance and Yield-Related Components of Rice. Mol. Plant-Microbe Interact. 2015, 28, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.Y.; Guo, W.; Zhao, H.W.; Wang, J.G.; Liu, H.L.; Sun, J.; Zheng, H.L.; Sha, H.J.; Zou, D.T. Association mapping and resistant alleles’ analysis for japonica rice blast resistance. Plant Breed. 2015, 134, 646–652. [Google Scholar] [CrossRef]
- Raboin, L.-M.; Ballini, E.; Tharreau, D.; Ramanantsoanirina, A.; Frouin, J.; Courtois, B.; Ahmadi, N. Association Mapping of Resistance to Rice Blast in Upland Field Conditions. Rice 2016, 9, 59. [Google Scholar] [CrossRef]
- McCouch, S.R.; Wright, M.H.; Tung, C.-W.; Maron, L.G.; McNally, K.L.; Fitzgerald, M.; Singh, N.; DeClerck, G.; Agosto-Perez, F.; Korniliev, P.; et al. Open access resources for genome wide association mapping in rice. Nat. Commun. 2016, 7, 10532. [Google Scholar] [CrossRef]
- Kang, H.; Wang, Y.; Peng, S.; Zhang, Y.; Xiao, Y.; Wang, D.; Qu, S.; Li, Z.; Yan, S.; Wang, Z.; et al. Dissection of the genetic architecture of rice resistance to the blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 2016, 17, 959–972. [Google Scholar] [CrossRef]
- Mgonja, E.M.; Park, C.H.; Kang, H.; Balimponya, E.G.; Opiyo, S.; Bellizzi, M.; Mutiga, S.K.; Rotich, F.; Ganeshan, V.D.; Mabagala, R.; et al. Genotyping-by-Sequencing-Based Genetic Analysis of African Rice Cultivars and Association Mapping of Blast Resistance Genes Against Magnaporthe oryzae Populations in Africa. Phytopathology 2017, 107, 1039–1046. [Google Scholar] [CrossRef]
- Lin, H.A.; Chen, S.Y.; Chang, F.Y.; Tung, C.W.; Chen, Y.C.; Shen, W.C.; Chen, R.S.; Wu, C.W.; Chung, C.L. Genome-wide association study of rice genes and loci conferring resistance to Magnaporthe oryzae isolates from Taiwan. Bot. Stud. 2018, 59, 32. [Google Scholar] [CrossRef] [PubMed]
- Korinsaka, S.; Tangphatsornruangb, S.; Pootakhamb, W.; Wanchanab, S.; Plabplac, A.; Jantasuriyaratd, C.; Patarapuwadole, S.; Vanavichitf, S. Genome-wide association mapping of virulence gene in rice blast fungus Magnaporthe oryzae using a genotyping by sequencing approach. Genomics 2019, 111, 661–668. [Google Scholar] [CrossRef]
- Volante, A.; Tondelli, A.; Desiderio, F.; Abbruscato, P.; Menin, B.; Biselli, C.; Casella, L.; Singh, N.; McCouch, S.R.; Tharreau, D.; et al. Genome Wide Association Studies for Japonica Rice Resistance to Blast in Field and Controlled Conditions. Rice 2020, 13, 71. [Google Scholar] [CrossRef]
- Jiang, H.; Feng, Y.; Qiu, L.; Gao, G.; Zhang, Q.; He, Y. Identification of Blast Resistance QTLs Based on Two Advanced Backcross Populations in Rice. Rice 2020, 13, 1. [Google Scholar] [CrossRef]
- Frontini, M.; Boisnard, A.; Frouin, J.; Ouikene, M.; Morel, J.B.; Ballini, E. Genome-wide association of rice response to blast fungus identifies loci for robust resistance under high nitrogen. BMC Plant Biol. 2021, 21, 99. [Google Scholar] [CrossRef]
- Tanweer, F.A.; Rafii, M.Y.; Sijam, K.; Rahim, H.A.; Ahmed, F.; Ashkani, S.; Latif, M.A. Introgression of Blast Resistance Genes (Putative Pi-b and Pi-Kh) into Elite Rice Cultivar MR219 through Marker-Assisted Selection. Front. Plant Sci. 2015, 6, 1002. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, M. Pyramiding resistance-conferring gene sequences in crops. Curr. Opin. Virol. 2017, 26, 36–42. [Google Scholar] [CrossRef]
- Das, G.; Patra, J.K.; Baek, K.H. Insight into MAS: A molecular tool for development of stress resistant and quality of rice through gene stacking. Front. Plant Sci. 2017, 8, 985. [Google Scholar] [CrossRef]
- Xiao, W.; Yang, Q.; Huang, M.; Guo, T.; Liu, Y.; Wang, J.; Yang, G.; Zhou, J.; Yang, J.; Zhu, X.; et al. Improvement of Rice Blast Resistance by Developing Monogenic Lines, Two-Gene Pyramids and Three-Gene Pyramid through MAS. Rice 2019, 12, 78. [Google Scholar] [CrossRef]
- Dixit, S.; Singh, U.M.; Singh, A.K.; Alam, S.; Venkateshwarlu, C.; Nachimuthu, V.V.; Yadav, S.; Abbai, R.; Selvaraj, R.; Devi, M.N.; et al. Marker Assisted Forward Breeding to Combine Multiple Biotic-Abiotic Stress Resistance/Tolerance in Rice. Rice 2020, 13, 29. [Google Scholar] [CrossRef] [PubMed]
- Fjellstrom, R.; McClung, A.M.; Shank, A.R. SSR Markers Closely Linked to the Pi-z Locus Are Useful for Selection of Blast Resistance in a Broad Array of Rice Germplasm. Mol. Breed. 2006, 17, 149–157. [Google Scholar] [CrossRef]
- Wang, Z.; Jia, Y.; Rutger, J.N.; Xia, Y. Rapid Survey for Presence of a Blast Resistance Gene Pi-Ta in Rice Cultivars Using the Dominant DNA Markers Derived from Portions of the Pi-Ta Gene. Plant Breed. 2007, 126, 36–42. [Google Scholar] [CrossRef]
- Wen, S.; Gao, B. Introgressing Blast Resistant Gene Pi-9(t) into Elite Rice Restorer Luhui17 by Marker-Assisted Selection. Rice Genom. Genet. 2012, 2, 31–36. [Google Scholar] [CrossRef][Green Version]
- Hua, L.X.; Liang, L.-Q.; He, X.-Y.; Wang, L.; Zhang, W.-S.; Liu, W.; Liu, X.-Q.; Lin, F. Development of a Marker Specific for the Rice Blast Resistance Gene Pi39 in the Chinese Cultivar Q15 and Its Use in Genetic improvement. Biotechnol. Biotechnol. Equip. 2015, 29, 448–456. [Google Scholar] [CrossRef]
- Hasan, N.; Rafii, M.; Rahim, H.; Syd Ali, N.; Mazlan, N.; Abdullah, S. Introgression of Pi-Kh Resistance Gene into a Malaysian Cultivar, MR264 Using Marker-Assisted Backcrossing (MABC). Int. J. Agric. Biol. 2015, 17, 1172–1178. [Google Scholar] [CrossRef]
- Beser, N.; Del Valle, M.M.; Kim, S.M.; Vinarao, B.R.; Surek, H.; Jena, K.K. Marker-assisted introgression of a broad-spectrum resistance gene, Pi40 improved blast resistance of two elite rice (Oryza sativa L.). Mol. Plant Breed. 2016, 7, 1–15. [Google Scholar]
- Araujo, L.; Soares, J.M.; de Filippi, M.C.C.; Rodrigues, F.Á. Cytological Aspects of Incompatible and Compatible Interactions between Rice, Wheat and the Blast Pathogen Pyricularia oryzae. Sci. Agric. 2016, 73, 177–183. [Google Scholar] [CrossRef][Green Version]
- Singh, A.; Singh, V.K.; Singh, S.P.; Pandian, R.T.P.; Ellur, R.K.; Singh, D.; Bhowmick, P.K.; Gopala Krishnan, S.; Nagarajan, M.; Vinod, K.K.; et al. Molecular Breeding for the Development of Multiple Disease Resistance in Basmati Rice. AoB Plants 2012, 2012, pls029. [Google Scholar] [CrossRef]
- Gouda, P.K.; Saikumar, S.; Varma, C.M.K.; Nagesh, K.; Thippeswamy, S.; Shenoy, V.; Ramesha, M.S.; Shashidhar, H.E. Marker-Assisted Breeding of Pi-1 and Piz-5 Genes Imparting Resistance to Rice Blast in PRR78, Restorer Line of Pusa RH-10 Basmati Rice Hybrid. Plant Breed. 2013, 132, 61–69. [Google Scholar] [CrossRef]
- Oddin, J.; Durga Rani, H.V.; Swathi, G.; Anuradha, C.H.; Vanisri, S.; SheshuMadhav, M.; Sundaram, R.M.; ArunPrem Kumar, N. Introgression of blast resistance genes Pi-54 and Pi1 into cold tolerant variety Tellahamsa, by marker assisted selection. Int. J. Curr. Res. 2015, 7, 22348–22353. [Google Scholar]
- Madhavi, R.K.; Rambabu, R.; Abhilash Kumar, V.; Vijay Kumar, S.; Aruna, J.; Ramesh, S.; Sundaram, R.M.; Laha, G.S.; Sheshu Madhav, M.; Ravindra babu, V.; et al. Marker Assisted Introgression of Blast (Pi-2 and Pi-54) Genes in to the Genetic Background of Elite, Bacterial Blight Resistant Indica Rice Variety, Improved Samba Mahsuri. Euphytica 2016, 212, 331–342. [Google Scholar] [CrossRef]
- Xiao, W.; Luo, L.; Wang, H.; Guo, T.; Liu, Y.; Zhou, J.; Zhu, X.; Yang, Q.; Chen, Z. Pyramiding of Pi46 and Pita to Improve Blast Resistance and to Evaluate the Resistance Effect of the Two R Genes. J. Integr. Agric. 2016, 15, 2290–2298. [Google Scholar] [CrossRef]
- Hegde, S.S.; Prashanthi, S.K. Identification of polymorphic markers and introgression of Pi1 and Pi2 genes for blast resistance in rice. J Farm Sci. 2016, 29, 327–331. [Google Scholar]
- Luo, Y.; Ma, T.; Zhang, A.; Ong, K.H.; Li, Z.; Yang, J.; Yin, Z. Marker-assisted breeding of the rice restorer line Wanhui 6725 for disease resistance, submergence tolerance and aromatic fragrance. Rice 2016, 9, 66. [Google Scholar] [CrossRef]
- Divya, B.; Robin, S.; Rabindran, R.; Senthil, S.; Raveendran, M.; Joel, A.J. Marker Assisted Backcross Breeding Approach to Improve Blast Resistance in Indian Rice (Oryza sativa) Variety ADT43. Euphytica 2014, 200, 61–77. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, V.K.; Singh, A.; Ellur, R.K.; Pandian, R.T.P.; Gopala Krishnan, S.; Singh, U.D.; Nagarajan, M.; Vinod, K.K.; Prabhu, K.V. Introgression of Multiple Disease Resistance into a Maintainer of Basmati Rice CMS Line by Marker Assisted Backcross Breeding. Euphytica 2015, 203, 97–107. [Google Scholar] [CrossRef]
- Balachiranjeevi, C.; Bhaskar, N.S.; Abhilash, V.; Akanksha, S.; Viraktamath, B.C.; Madhav, M.S.; Hariprasad, A.S.; Laha, G.S.; Prasad, M.S.; Balachandran, S.M.; et al. Marker-Assisted Introgression of Bacterial Blight and Blast Resistance into DRR17B, an Elite, Fine-Grain Type Maintainer Line of Rice. Mol. Breed. 2015, 35, 151. [Google Scholar] [CrossRef]
- Srikanth, S.; Pandey, M.K.; BalaChiranjeevi, C.H.; Hajira, S.K.; Vijay Kumar, S.; Kousik, M.B.V.N.; Bhadana, V.P.; SheshuMadhav, M.; Suneetha, K.; Subba Rao, L.V.; et al. Introgression of major bacterial blight and blast resistant genes into Vallabh Basmati 22, an elite Basmati variety. Int. J. Dev. Res. 2016, 6, 8366–8370. [Google Scholar]
- Wu, Y.; Yu, L.; Pan, C.; Dai, Z.; Li, Y.; Xiao, N.; Zhang, X.; Ji, H.; Huang, N.; Zhao, B.; et al. Development of Near-Isogenic Lines with Different Alleles of Piz Locus and Analysis of Their Breeding Effect under Yangdao 6 Background. Mol. Breed. 2016, 36, 12. [Google Scholar] [CrossRef]
- Narayanan, N.N.; Baisakh, N.; Vera Cruz, C.M.; Gnanamanickam, S.S.; Datta, K.; Datta, S.K. Molecular Breeding for the Development of Blast and Bacterial Blight Resistance in Rice Cv. IR50. Crop Sci. 2002, 42, 2072–2079. [Google Scholar] [CrossRef]
- Ellur, R.K.; Khanna, A.; Yadav, A.; Pathania, S.; Rajashekara, H.; Singh, V.K.; Gopala Krishnan, S.; Bhowmick, P.K.; Nagarajan, M.; Vinod, K.K.; et al. Improvement of Basmati Rice Varieties for Resistance to Blast and Bacterial Blight Diseases Using Marker Assisted Backcross Breeding. Plant Sci. J. 2016, 242, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Arunakumari, K.; Durgarani, C.V.; Satturu, V.; Sarikonda, K.R.; Chittoor, P.D.R.; Vutukuri, B.; Laha, G.S.; Nelli, A.P.K.; Gattu, S.; Jamal, M.; et al. Marker-Assisted Pyramiding of Genes Conferring Resistance Against Bacterial Blight and Blast Diseases into Indian Rice Variety MTU1010. Rice Sci. 2016, 23, 306–316. [Google Scholar] [CrossRef]
- Janaki Ramayya, P.; Vinukonda, V.P.; Singh, U.M.; Alam, S.; Venkateshwarlu, C.; Vipparla, A.K.; Dixit, S.; Yadav, S.; Abbai, R.; Badri, J.; et al. Marker-assisted forward and backcross breeding for improvement of elite Indian rice variety Naveen for multiple biotic and abiotic stress tolerance. PLoS ONE 2021, 16, e0256721. [Google Scholar]
- Laxmi Prasanna, B.; Dangi, K.S.; Damodar Raju, C.H.; Jagadeeshwar, R.; Rekha, G.; Sinha, P.; Aleena, D.; Harika, G.; Mastanbee, S.K.; Swapnil Ravindra, K.; et al. Marker Assisted Pedigree Breeding Based Improvement of the Indian Mega Variety of Rice MTU1010 for Resistance against Bacterial Blight and Blast and Tolerance to Low Soil Phosphorus. PLoS ONE 2022, 17, e0260535. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.; Singh, S.P.; Ellur, R.K.; Singh, D.; Gopala Krishnan, S.; Bhowmick, P.K.; Nagarajan, M.; Vinod, K.K.; Singh, U.D.; et al. Marker-Assisted Simultaneous but Stepwise Backcross Breeding for Pyramiding Blast Resistance Genes Piz5 and Pi54 into an Elite Basmati Rice Restorer Line ‘PRR78’. Plant Breed. 2013, 132, 486–495. [Google Scholar] [CrossRef]
- Usatov, A.V.; Kostylev, P.I.; Azarin, K.V.; Markin, N.V.; Makarenko, M.S.; Khachumov, V.A.; Bibov, M.Y. Introgression of the Rice Blast Resistance Genes Pi1, Pi2 and Pi33 into Russian Rice Varieties by Marker-Assisted Selection. Indian J. Genet. Plant Breed. 2016, 76, 18–23. [Google Scholar] [CrossRef][Green Version]
- Fu, C. Genetic Improvement of Resistance to Blast and Bacterial Blight of the Elite Maintainer Line Rongfeng B in Hybrid Rice (Oryza sativa L.) by Using Marker-Assisted Selection. Afr. J. Biotechnol. 2012, 11, 13104–13114. [Google Scholar] [CrossRef]
- Kumar, V.A.; Balachiranjeevi, C.H.; Naik, S.B.; Rambabu, R.; Rekha, G.; Madhavi, K.R.; Harika, G.; Vijay, S.; Pranathi, K.; Hajira, S.K.; et al. Marker-Assisted Introgression of the Major Bacterial Blight Resistance Gene, Xa21 and Blast Resistance Gene, Pi54 into RPHR-1005, the Restorer Line of the Popular Rice Hybrid, DRRH3. J. Plant Biochem. Biotechnol. 2016, 25, 400–409. [Google Scholar] [CrossRef]
- Kumar, V.A.; Balachiranjeevi, C.H.; Naik, S.B.; Rambabu, R.; Rekha, G.; Harika, G.; Hajira, S.K.; Pranathi, K.; Vijay, S.; Anila, M.; et al. Marker-Assisted Improvement of the Elite Restorer Line of Rice, RPHR-1005 for Resistance against Bacterial Blight and Blast Diseases. J. Genet. 2016, 95, 895–903. [Google Scholar] [CrossRef]
- Pinta, W.; Toojinda, T.; Thummabenjapone, P.; Sanitchon, J. Pyramiding of Blast and Bacterial Leaf Blight Resistance Genes into Rice Cultivar RD6 Using Marker Assisted Selection. Afr. J. Biotechnol. 2013, 12, 4432–4438. [Google Scholar] [CrossRef]
- Das, G.; Rao, G.J.N. Molecular Marker Assisted Gene Stacking for Biotic and Abiotic Stress Resistance Genes in an Elite Rice Cultivar. Front. Plant Sci. 2015, 6, 698. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, Y.; Nishio, Z.; Nakazono, K.; Soma, M.; Nakajima, E.; Ugaki, M.; Hibi, T. Enhanced Resistance to Blast (Magnaporthe grisea) in Transgenic Japonica Rice by Constitutive Expression of Rice Chitinase. Theor. Appl. Genet. 1999, 99, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Gandikota, M.; De Kochko, A.; Chen, L.; Ithal, N.; Fauquet, C.; Reddy, A.R. Development of transgenic rice plants expressing maize anthocyanin genes and increased blast resistance. Mol. Plant Breed. 2001, 7, 73–83. [Google Scholar]
- Nishizawa, Y.; Saruta, M.; Nakazono, K.; Nishio, Z.; Soma, M.; Yoshida, T.; Nakajima, E.; Hibi, T. Characterization of Transgenic Rice Plants Over-Expressing the Stress-Inducible β-Glucanase Gene Gns1. Plant Mol. Biol. 2003, 51, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Coca, M.; Penas, G.; Gómez, J.; Campo, S.; Bortolotti, C.; Messeguer, J.; Segundo, B.S. Enhanced Resistance to the Rice Blast Fungus Magnaporthe grisea Conferred by Expression of a Cecropin A Gene in Transgenic Rice. Planta 2006, 223, 392–406. [Google Scholar] [CrossRef]
- Zhang, S.; Wei, Y.; Pan, H. Transgenic Rice Plants Expressing a Novel Antifreeze Glycopeptide Possess Resistance to Cold and Disease. Zeitschrift für Naturforschung C 2007, 62, 583–591. [Google Scholar] [CrossRef]
- Imamura, T.; Yasuda, M.; Kusano, H.; Nakashita, H.; Ohno, Y.; Kamakura, T.; Taguchi, S.; Shimada, H. Acquired Resistance to the Rice Blast in Transgenic Rice Accumulating the Antimicrobial Peptide Thanatin. Transgenic Res. 2010, 19, 415–424. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, D.; Chen, X.; Lei, C.; Ma, B.; Wang, Y.; Li, S. Rice Blast Resistance of Transgenic Rice Plants with Pi-D2 Gene. Rice Sci. 2010, 17, 179–184. [Google Scholar] [CrossRef]
- Helliwell, E.E.; Wang, Q.; Yang, Y. Transgenic Rice with Inducible Ethylene Production Exhibits Broad-Spectrum Disease Resistance to the Fungal Pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol. J. 2013, 11, 33–42. [Google Scholar] [CrossRef]
- Feng, D.R.; Wei, J.W.; Xu, X.P.; Xu, Y.; Li, B.J. Introduction of multiple antifungal protein genes into rice and preliminary study on resistance to Pyricularia oryzae of transgenic rices. Acta Sci. Nat. Univ. Sunyatsen 1999, 38, 62–66, (In Chinese with English Abstract). [Google Scholar]
- Zhu, C.; Bortesi, L.; Baysal, C.; Twyman, R.M.; Fischer, R.; Capell, T.; Schillberg, S.; Christou, P. Characteristics of Genome Editing Mutations in Cereal Crops. Trends Plant Sci. 2017, 22, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome Editing in Rice: Recent Advances, Challenges, and Future Implications. Front. Plant Sci. 2018, 9, 1361. [Google Scholar] [CrossRef] [PubMed]
- Pingoud, A.; Silva, G.H. Precision Genome Surgery. Nat. Biotechnol. 2007, 25, 743–744. [Google Scholar] [CrossRef]
- Li, T.; Huang, S.; Jiang, W.Z.; Wright, D.; Spalding, M.H.; Weeks, D.P.; Yang, B. TAL Nucleases (TALNs): Hybrid Proteins Composed of TAL Effectors and FokI DNA-Cleavage Domain. Nucleic Acids Res. 2011, 39, 359–372. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, Z.; Wang, C.; Li, Y.; Xu, J.-R. Germination and Infectivity of Microconidia in the Rice Blast Fungus Magnaporthe oryzae. Nat. Commun. 2014, 5, 4518. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, S.; Jiang, N.; Zhao, X.; Bai, Z.; Liu, J.; Yao, W.; Tang, Q.; Xiao, G.; Lv, C.; et al. Engineering of Rice Varieties with Enhanced Resistances to Both Blast and Bacterial Blight Diseases via CRISPR/Cas9. Plant Biotechnol. J. 2022, 20, 876–885. [Google Scholar] [CrossRef]
- Tao, H.; Shi, X.; He, F.; Wang, D.; Xiao, N.; Fang, H.; Wang, R.; Zhang, F.; Wang, M.; Li, A.; et al. Engineering Broad-spectrum Disease-resistant Rice by Editing Multiple Susceptibility Genes. J. Integr. Plant Biol. 2021, 63, 1639–1648. [Google Scholar] [CrossRef]
- Ma, J.; Chen, J.; Wang, M.; Ren, Y.; Wang, S.; Lei, C.; Cheng, Z. Sodmergen Disruption of OsSEC3A Increases the Content of Salicylic Acid and Induces Plant Defense Responses in Rice. J. Exp. Bot. 2018, 69, 1051–1064. [Google Scholar] [CrossRef]
- Xie, K.; Yang, Y. RNA-Guided Genome Editing in Plants Using a CRISPR–Cas System. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Nemudryi, A.A.; Valetdinova, K.R.; Medvedev, S.P.; Zakian, S.M. TALEN and CRISPR/Cas genome editing systems: Tools of discovery. Acta Nat. 2014, 6, 19–40. [Google Scholar] [CrossRef]
- Schornack, S.; Meyer, A.; Römer, P.; Jordan, T.; Lahaye, T. Gene-for-Gene-Mediated Recognition of Nuclear-Targeted AvrBs3-like Bacterial Effector Proteins. J. Plant Physiol. 2006, 163, 256–272. [Google Scholar] [CrossRef] [PubMed]
- Moscou, M.J.; Bogdanove, A.J. A Simple Cipher Governs DNA Recognition by TAL Effectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef]
- Arazoe, T.; Ogawa, T.; Miyoshi, K.; Yamato, T.; Ohsato, S.; Sakuma, T.; Yamamoto, T.; Arie, T.; Kuwata, S. Tailor-Made TALEN System for Highly Efficient Targeted Gene Replacement in the Rice Blast Fungus. Biotechnol. Bioeng. 2015, 112, 1335–1342. [Google Scholar] [CrossRef] [PubMed]
- Gogarten, J.P.; Hilario, E. Inteins, Introns, and Homing Endonucleases: Recent Revelations about the Life Cycle of Parasitic Genetic Elements. BMC Evol. Biol. 2006, 6, 94. [Google Scholar] [CrossRef]
- Orlowski, J.; Boniecki, M.; Bujnicki, J.M. I-Ssp6803I: The First Homing Endonuclease from the PD-(D/E)XK Superfamily Exhibits an Unusual Mode of DNA Recognition. Bioinformatics 2007, 23, 527–530. [Google Scholar] [CrossRef]
- Maeder, M.L.; Gersbach, C.A. Genome-Editing Technologies for Gene and Cell Therapy. Mol. Ther. 2016, 24, 430–446. [Google Scholar] [CrossRef]
- Gupta, A.; Meng, X.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Zinc Finger Protein-Dependent and -Independent Contributions to the in Vivo off-Target Activity of Zinc Finger Nucleases. Nucleic Acids Res. 2011, 39, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Paschon, D.E.; Lussier, S.; Wangzor, T.; Xia, D.F.; Li, P.W.; Hinkley, S.J.; Scarlott, N.A.; Lam, S.C.; Waite, A.J.; Truong, L.N.; et al. Diversifying the Structure of Zinc Finger Nucleases for High-Precision Genome Editing. Nat. Commun. 2019, 10, 1133. [Google Scholar] [CrossRef] [PubMed]
- Gammage, P.A.; Viscomi, C.; Simard, M.-L.; Costa, A.S.H.; Gaude, E.; Powell, C.A.; Van Haute, L.; McCann, B.J.; Rebelo-Guiomar, P.; Cerutti, R.; et al. Genome Editing in Mitochondria Corrects a Pathogenic MtDNA Mutation in Vivo. Nat. Med. 2018, 24, 1691–1695. [Google Scholar] [CrossRef] [PubMed]
- Cantos, C.; Francisco, P.; Trijatmiko, K.R.; Slamet-Loedin, I.; Chadha-Mohanty, P.K. Identification of ‘safe harbor’ Loci in Indica rice genome by harnessing the property of zinc-finger nucleases to induce DNA damage and repair. Front. Plant Sci. 2014, 5, 302. [Google Scholar] [CrossRef] [PubMed]
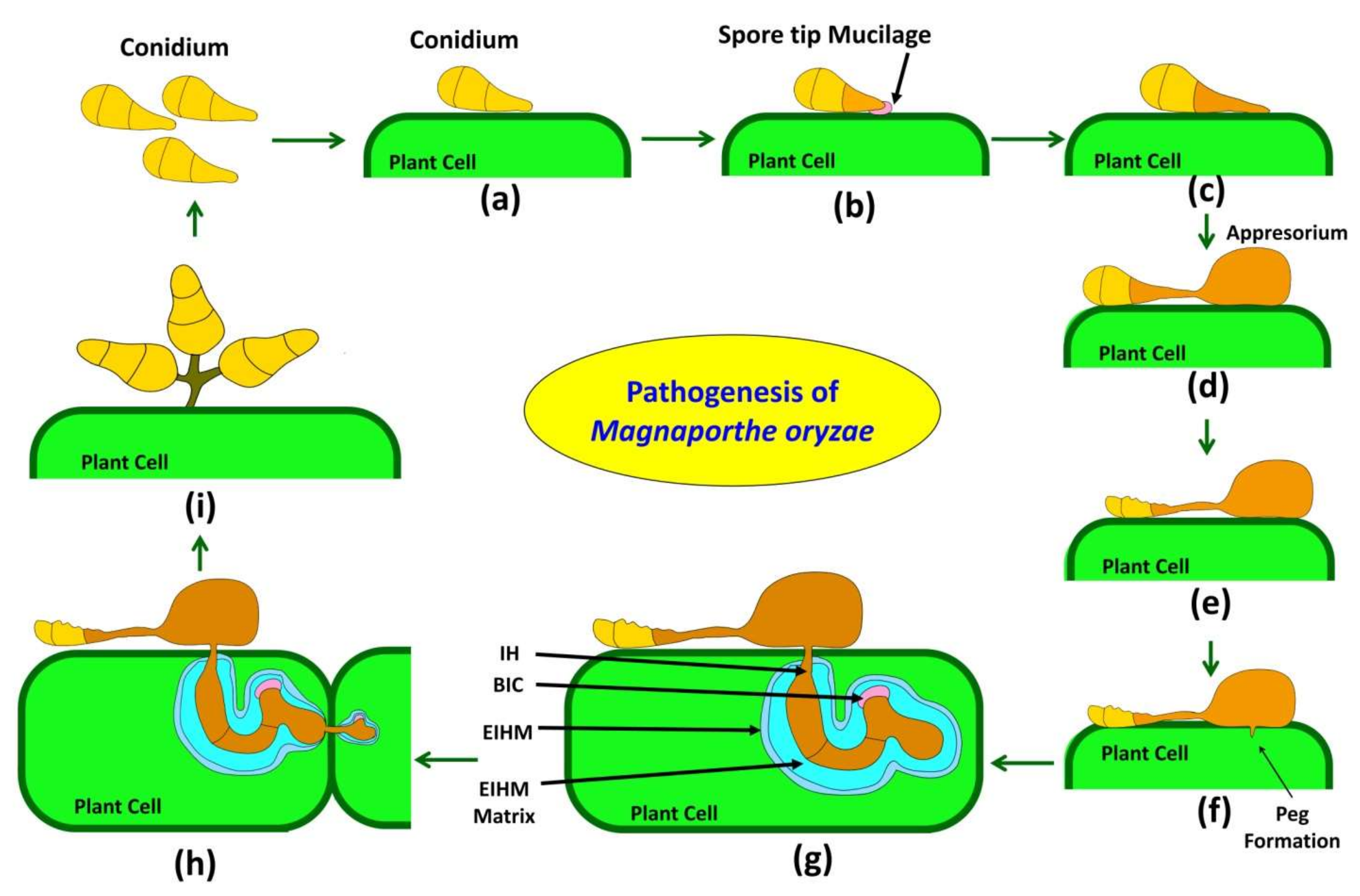



| S. No. | Name of Mutant Variety * | Registration Year in MVD | Country | Mutagen Used and Dose | Character Improvement Details |
|---|---|---|---|---|---|
| 1 | Xiongyue 613 | 1965 | China | Gamma rays (200 Gy) | Moderate resistance to blast, higher yield and good quality |
| 2 | Fulianai | 1966 | China | Gamma rays (200 Gy). | Short culm, resistance to blast, early maturity and high yield |
| 3 | Aifu 9 | 1966 | China | Gamma rays (350 Gy) | Short culm, resistance to blast and higher yield |
| 4 | Liaofeng 5 | 1969 | China | Gamma rays (250 Gy) | Early maturity, short culm and resistance to blast |
| 5 | Fuxuan 3 | 1970 | China | Gamma rays (300 Gy) | Good tillering and resistance to blast |
| 6 | Fushe 94 | 1971 | China | Neutrons | Early maturity, good tillering and resistance to blast |
| 7 | Nucleoryza | 1972 | Hungary | Fast neutrons (25 Krad) | Early maturity, maintained blast resistance and improved yield |
| 8 | Fuxuan 124 | 1972 | China | Gamma rays (350 Gy) | Resistance to blast and intermediate maturity |
| 9 | Yifunuo 1 | 1973 | China | Gamma rays (100 Gy) | Resistance to blast, long panicles and higher grain number |
| 10 | Fulgente | 1973 | Italy | X-rays (250 Gy) | Blast resistance and high productivity |
| 11 | Fushe 410 | 1974 | China | Gamma rays (350 Gy) | Intermediate resistance to blast |
| 12 | Wangeng 257 | 1975 | China | Gamma rays (300 Gy) | Tolerance to fertilizers, resistance to blast and higher yield |
| 13 | Nongshi 4 | 1975 | China | Neutrons | Early maturity, resistance to low temperature, resistance to blast and xantomonas oryzae |
| 14 | Xiangfudao | 1976 | China | Gamma rays (300 Gy) | Resistance to low temperature, resistance to blast and xantomonas |
| 15 | RD 6 | 1977 | Thailand | Gamma rays (200 Gy) | Glutinous endosperm and improved resistance to blast |
| 16 | Guifu 3 | 1977 | China | Gamma rays (300 Gy) | Early maturity, resistance to low temperatures and resistance to blast |
| 17 | 7404 | 1977 | China | Gamma rays (350 Gy) | Short culm, higher yield, resistance to bacterial blight and blast |
| 18 | Wanfu 33 | 1978 | China | Gamma rays (300 Gy) | Early maturity, resistance to low temperatures and resistance to blast |
| 19 | Zhuqin 40 | 1978 | China | Gamma rays (300 Gy) | Resistance to blast and suitable for mountain areas |
| 20 | Juangyebai | 1978 | China | Neutrons | Good tillering and resistance to blast |
| 21 | Fuzhu | 1979 | China | Gamma rays (350 Gy) | Early maturity, resistance to low temperatures, resistance to blast |
| 22 | Jagannath (BSS-873) | 1979 | India | X-r (300 Gy) | Wide adaptability, semi dwarf, resistance to blast and sheath blast |
| 23 | Mutashali | 1980 | Hungary | Fast neutrons (20 Gy) | Resistance to blast and shattering of grains and high yield |
| 24 | Atomita 1 | 1982 | Indonesia | Gamma rays (200 Gy) | Early maturity, resistance to bph (biotype 1), green leaf hopper and blast |
| 25 | CNM 31 | 1982 | India | X-rays (300 Gy) | Early maturity, semidwarf, higher yield, resistance blast |
| 26 | Atomita 2 | 1983 | Indonesia | Gamma rays (200 Gy) | Tolerance to salt, early maturity, resistance to brown plant hopper (biotype 1), higher protein content and resistance to blast |
| 27 | Danau atas | 1988 | Indonesia | Gamma rays (400 Gy) | High yield, resistance to blast, drought and low ph |
| 28 | Xiangjing 832 | 1989 | China | X-rays | Short straw, high resistance to blast and bacterial blight, high yield |
| 29 | CRM 49 | 1989 | India | 0.001 m Sodium azide (Nan3) | Resistance to blast disease |
| 30 | Quannuo 101 | 1990 | China | Gamma rays (200 Gy) | High grain yield, wine making rice, moderately resistant to blast and bacterial leaf blight |
| 31 | Jinfu 1 | 1990 | China | Gamma rays (300 Gy) | Early maturity (7 days earlier) and resistance to blast |
| 32 | Ejingnuo 6 | 1991 | China | Gamma rays (350 Gy) | Resistance to blast and blight, good grain quality and higher grain yield |
| 33 | Xiushui 04 | 1991 | China | Physical mutagen | Resistance to blast and bacterial blight, high yield, good grain quality and altered maturity |
| 34 | Zhenuo 2 | 1993 | China | Gamma rays (300 Gy) | High grain yield, good cooking quality, resistance to rice blast and bacterial blight |
| 35 | Fuyou 63 | 1993 | China | Physical mutagen | High grain yield, altered maturity, blast resistance, 21.98% amylose content |
| 36 | Zhefu 762 | 1993 | China | Physical mutagen | High grain yield, high resistance to blast and bacterial blight |
| 37 | Zhefu 7 | 1994 | China | Gamma rays (300 Gy) | Early maturity, resistance to low temperature, resistance to blast and sheath blight |
| 38 | II You 838 | 1995 | China | Physical mutagen. | High grain yield, plant height (120 cm), resistance to leaf blast and panicle blast, amylose content (22.8%) |
| 39 | Shengxianggeng No. 4 | 1996 | China | Gamma rays (180 Gy) | Short stem, high yield and good quality, high resistance to rice blast |
| 40 | Camago-8 | 1996 | Costa Rica | Gamma rays (250 Gy) | Resistance to blast and resistance to viruses |
| 41 | CRM 53 | 1997 | India | 0.66% EMS | Resistance to blast disease |
| 42 | VND 95-19 | 1999 | Viet Nam | Gamma rays (200 Gy) | Strong tolerance to acid sulphate soil, high yield (5–10 t/ha), resistance to brown plant hopper and blast disease |
| 43 | VND 95-20 | 1999 | Viet Nam | Gamma rays (200 Gy) | Short duration (90–95 days), wide adaptation, intermediate resistant to brown plant hopper, blast disease |
| 44 | CNM 25 | 1999 | India | X-rays (300 Gy) | Early maturity, increased tillering, higher yield, moderately resistant to blast |
| 45 | CNM 6 (Lakshmi) | 1999 | India | X-rays (300 Gy) | Early maturity (15–23 days), resistance to drought, dwarf (85 cm), and moderately resistant to blast |
| 46 | Yueyou 938 | 2000 | China | Gamma rays | High yield, semi dwarf plant height, resistance to bacterial blight and blast |
| 47 | Radhi | 2000 | India | Gamma rays (250 Gy) | Tolerance to blast and bph, good yield and early maturity (120 days) |
| 48 | IACuba 28 | 2001 | Cuba | Fast neutrons (20 Gy) | Large grain size, high yield, resistance to blast |
| 49 | CRM 51 | 2003 | India | 0.001 m Sodium azide (Nan3) | Resistance to blast disease |
| 50 | Woncheongbyeo | 2003 | Korea | Gamma rays (300 Gy) | Short stature, resistance to blast and early maturity |
| 51 | Zhongzao 21 | 2003 | China | Na | Medium maturity, tillering ability, good grain quality, blast resistance |
| 52 | Yangfujing4901 | 2004 | China | Gamma rays | Strong resistance to blast, bacterial leaf blight |
| 53 | Pooya | 2004 | Iran | Gamma rays (150 Gy) | Resistance to lodging and blast and higher yield |
| 54 | Tabesh | 2004 | Iran | Gamma rays (150 Gy) | Resistance to lodging, short culm, tolerance to blast and higher yield |
| 55 | VND99-3 | 2004 | Viet Nam | Gamma rays (200 Gy) | Short duration, high yield, resistant to brown plant hopper, blast disease, |
| 56 | Yangfuxian 9850 | 2004 | China | Gamma rays (300 Gy) | High yield, medium maturity, excellent eating quality, resistance to blast, |
| 57 | Chiyou S162 | 2005 | China | Gamma rays (300 Gy) | Moderate plant height, moderate tillering ability, resistance to blast and bacterial blight |
| 58 | Zhe 101 | 2005 | China | Treatment of seeds in aerospace | Late maturity, high yield, resistance to blast and bacterial blight |
| 59 | Pusa-NR-546 | 2006 | India | Gamma Rays (300 Gy) | Grain quality, semi dwarf (100 cm), super fine grain, tolerance to brown spot and leaf blast, |
| 60 | Hangtian 36 | 2006 | China | Treatment of seeds in aerospace | Early maturity, high grain quality and blast resistance |
| 61 | Huahang-simiao | 2006 | China | Treatment of seeds in aerospace | Resistance to blast and good quality |
| 62 | Minami-yutaka | 2007 | Japan | Gamma rays (200 Gy) | Late maturity, resistance to lodging, leaf blast and panicle blast |
| 63 | Jahesh | 2008 | Iran | 0.001% EMS | Short stature, early maturity, high yield and tolerant to stem borer and blast disease |
| 64 | Partou | 2008 | Iran | Gamma rays (350 Gy) | Short stature, early maturity, high yield, tolerant to stem borer and blast disease |
| 65 | Guangyinruanzhan | 2008 | China | Physical mutagen | High yield, resistance to blast and bacterial leaf blight |
| 66 | Liangyouhang 2 | 2008 | China | Treatment of seeds in aerospace | High yield, resistance to blast and bacterial blight and good grain quality |
| 67 | Neiyouhang 148 | 2008 | China | Treatment of seeds in aerospace | High yield, blast resistance and late maturity |
| 68 | Zhejing 41 | 2009 | China | Physical mutagen | Medium maturity, high yield, the resistance to blast and bacterial leaf blight |
| 69 | SCS118 Marques | 2013 | Brazil | Gamma rays (300 Gy) | Moderate resistance to blast, high yield potential, |
| 70 | NMR 151 | 2015 | Malaysia | Gamma rays (300 Gy) | Minimal water requirement, tolerant to blast disease, and high yield |
| 71 | Roshan | 2019 | Iran | Gamma rays (250 Gy) | Short stature, early maturity, tolerant to stem borer and blast disease |
| 72 | Vikram-TCR | 2021 | India | Gamma rays (300 Gy) | Semidwarf, Mid-early Maturity, High Yielding and Resistant to Blast Disease |
| S. No. | Donor Organism | Transferred Gene | Gene Function | Technique Used | Host Organism | Reference |
|---|---|---|---|---|---|---|
| 1 | Wild rice | MoHrip1 and MoHrip2 | Imparts resistance against blast and improvement in agronomic traits | Agrobacterium tumefaciens mediated transfer | Oryza sativa L. | Wang et al. [29] |
| 2 | Wild rice | Cht-2 and Cht-3 | Formation of chitin | Agrobacterium tumefaciens mediated transfer | Oryza sativa L. japonica (Nipponbare and Koshihikari varieties) | Nishizawa et al. [363] |
| 3 | Wild rice | Gns1 | Hydrolysesglucosidic bonds in cell walls | Introduced through vectors by incooperating CaMV35S as its promoter | Oryza sativa L. | Nishizawa et al. [365] |
| 4 | Wild rice | Pi54of | Confers resistance against blast | Transferred using pET29a vector | Two susceptible rice lines IET16310 (indica) and TP309 (japonica) | Devanna et al. [28] |
| 5 | Wild rice | Pi-d2 | Confers resistance against blast | Vector mediated transformation | Oryza sativa L. | Chen et al. [369] |
| 6 | Oryza rhizomatis | Pi54rh | Confers resistance against blast | - | - | Das et al. [287] |
| 7 | Giant silk moth Hyalophora cecropia | ER-CecA | Produce scecropin A peptides in paddy which are antimicrobial protein | Vector mediated transformation | Oryza sativa L. | Coca et al. [366] |
| 8 | Artificially made | Thanatin | Antimicrobial protein | Vector mediated transformation | Oryza sativa L. | Imamura et al. [368] |
| 9 | Maize | C2 | Flavanoid production | pUOH series plasmids | Oryza sativa L. | Gandikota et al. [364] |
| 10 | Antifreeze glycopeptide gene | Agrobacterium tumefaciens mediated transfer | Oryza sativa L. | Zhang et al. [367] | ||
| 11 | Wild rice | RC24 | An alfalfa glucanase gene, | Biolistics | Oryza sativa L. indica var. Qisiruanzhan, | Feng et al. [371] |
| 12 | Wild rice | Beta-Glu | Beta glucanase | Biolistics | Oryza sativa L. indica var. Qisiruanzhan, | Feng et al. [371] |
| 13 | Barley | B-RIP | Ribosome-inactivating protein | Biolistics | Hardeum vulgare | Feng et al. [371] |
| 14 | Wild rice | Pi21 | Confers resistance against blast | Ac/Ds transposon vectors | Oryza sativa L. | Li et al. [30] |
| 15 | Rice | ACS2 | 1-aminocyclopropane-1-carboxylic acid synthase | Vector mediated transformation | Oryza sativa L. | Helliwell et al. [370] |
| 16 | Rice | Pib, Pi25 and Pi54 | Confers resistance against blast | Vector mediated transformation | indica variety Kasalath and the japonica variety Zhenghan 10 | Peng et al. [31] |
| S. No. | Plant Species | Target Gene | Gene Function | Strategy | Reference |
|---|---|---|---|---|---|
| 1 | Oryza sativa L. japonica | SEC3A | Subunit of the exocyst complex | Protoplast transformation with Cas9/gRNA expression binary vectors | Ma et al. [379] |
| 2 | Oryza sativa L. japonica | ERF922 | Transcription factor implicated in multiple stress responses | Agrobacterium-mediated transformation of embryogenic calli with Cas9/gRNA expression binary vectors | Wang et al. [97] |
| 3 | Oryza sativa L. | ALB1 (MGG_07219). | Polyketide synthase | Poisons the fungus by converting it into albino colour. | Foster et al. [64] |
| 4 | Oryza sativa L. | RSY1(MGG_05059) | Scytalone dehydratase enzyme | Poisons the fungus by converting it into orange-red (rosy) fungal colonies | Foster et al. [64] |
| 5 | Oryza sativa L. | OsERF922 | ABA accumulation | Number of blast lesions formed were less, improving blast resistance | Wang et al. [97] |
| 6 | Oryza sativa L. | OsMPK5 | Responsible for pathogen infection | Improves resistance against blast diseases | Xie and Yang [380] |
| 7 | Oryza sativa L. | OsERF922 | Responsible for pathogen infection | Improves resistance against blast diseases | Wang et al. 2016 [97] |
| 8 | Oryza sativa L. | ALB1, RSY1 | Aids in growth of pathogen | Ribonucleoprotein (RNP) based CRISPR induced, marker free resistant plants against blast | Xie and Yang [380] |
| 9 | Oryza sativa L. | OsSEC3A | Interacts with SNAP25-type t-SNARE protein OsSNAP32 which is responsible for blast resistance | Induces plant defense responses for Magnoporthe oryzae. | Ma et al. [379] |
| 10 | Oryza sativa Longke638S (LK638S) | SA and JA pathway associated genes | Improves plant immunity | Increases resistance against blast. | Zhou et al. [377] |
| 11 | Oryza sativa L. | S genes, Pi21 and Bsr-d1 | Responsible for susceptible reaction in rice for blast. | Increases resistance by knocking out S gene or by causing mutation | Tao et al. [378] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahu, P.K.; Sao, R.; Choudhary, D.K.; Thada, A.; Kumar, V.; Mondal, S.; Das, B.K.; Jankuloski, L.; Sharma, D. Advancement in the Breeding, Biotechnological and Genomic Tools towards Development of Durable Genetic Resistance against the Rice Blast Disease. Plants 2022, 11, 2386. https://doi.org/10.3390/plants11182386
Sahu PK, Sao R, Choudhary DK, Thada A, Kumar V, Mondal S, Das BK, Jankuloski L, Sharma D. Advancement in the Breeding, Biotechnological and Genomic Tools towards Development of Durable Genetic Resistance against the Rice Blast Disease. Plants. 2022; 11(18):2386. https://doi.org/10.3390/plants11182386
Chicago/Turabian StyleSahu, Parmeshwar K., Richa Sao, Devendra K. Choudhary, Antra Thada, Vinay Kumar, Suvendu Mondal, Bikram K. Das, Ljupcho Jankuloski, and Deepak Sharma. 2022. "Advancement in the Breeding, Biotechnological and Genomic Tools towards Development of Durable Genetic Resistance against the Rice Blast Disease" Plants 11, no. 18: 2386. https://doi.org/10.3390/plants11182386
APA StyleSahu, P. K., Sao, R., Choudhary, D. K., Thada, A., Kumar, V., Mondal, S., Das, B. K., Jankuloski, L., & Sharma, D. (2022). Advancement in the Breeding, Biotechnological and Genomic Tools towards Development of Durable Genetic Resistance against the Rice Blast Disease. Plants, 11(18), 2386. https://doi.org/10.3390/plants11182386






