OsVTC1-1 Gene Silencing Promotes a Defense Response in Rice and Enhances Resistance to Magnaporthe oryzae
Abstract
:1. Introduction
2. Results
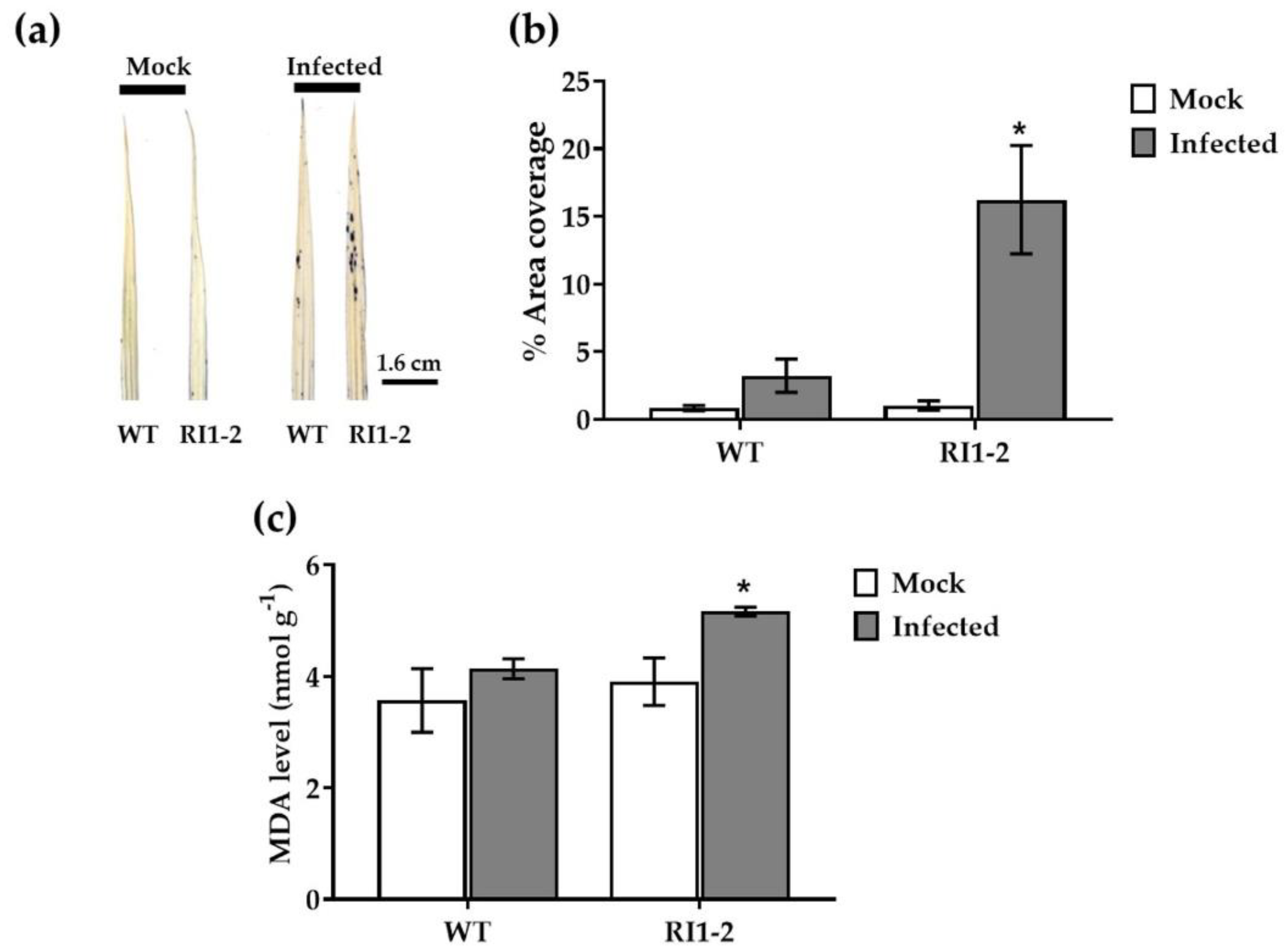
2.1. Pathogenicity Assay
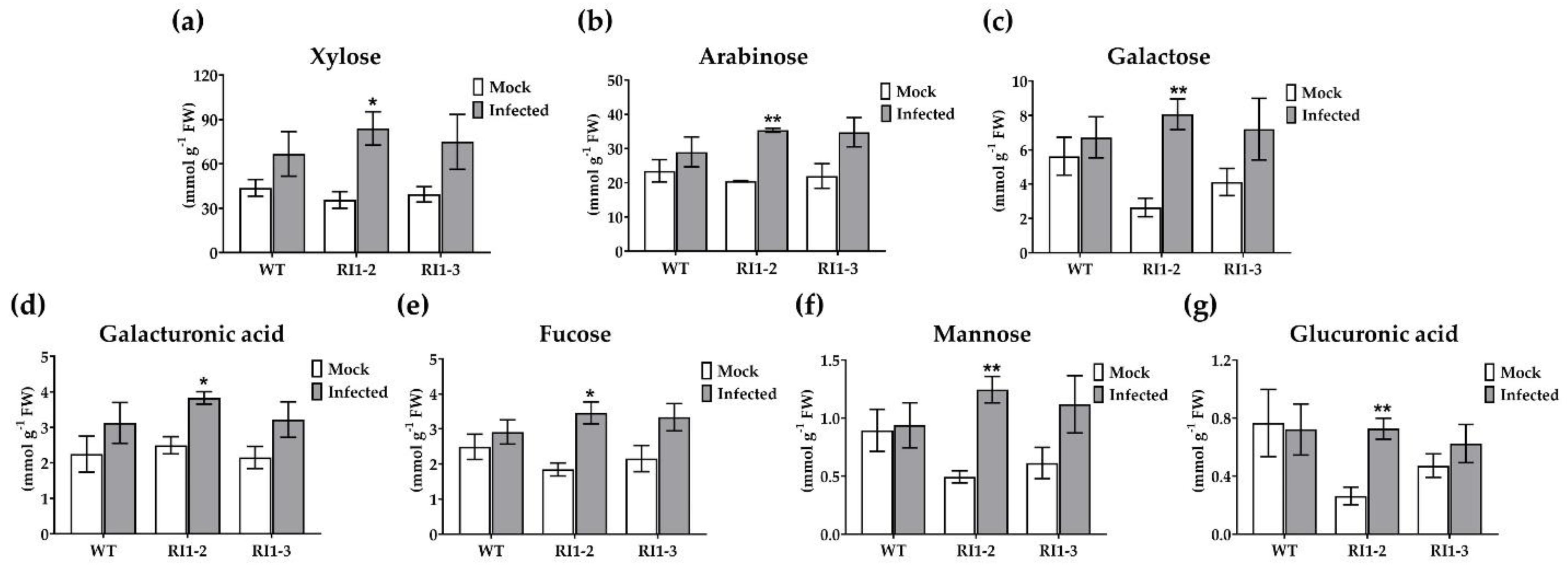
2.2. Cell Wall Sugar Composition Analysis
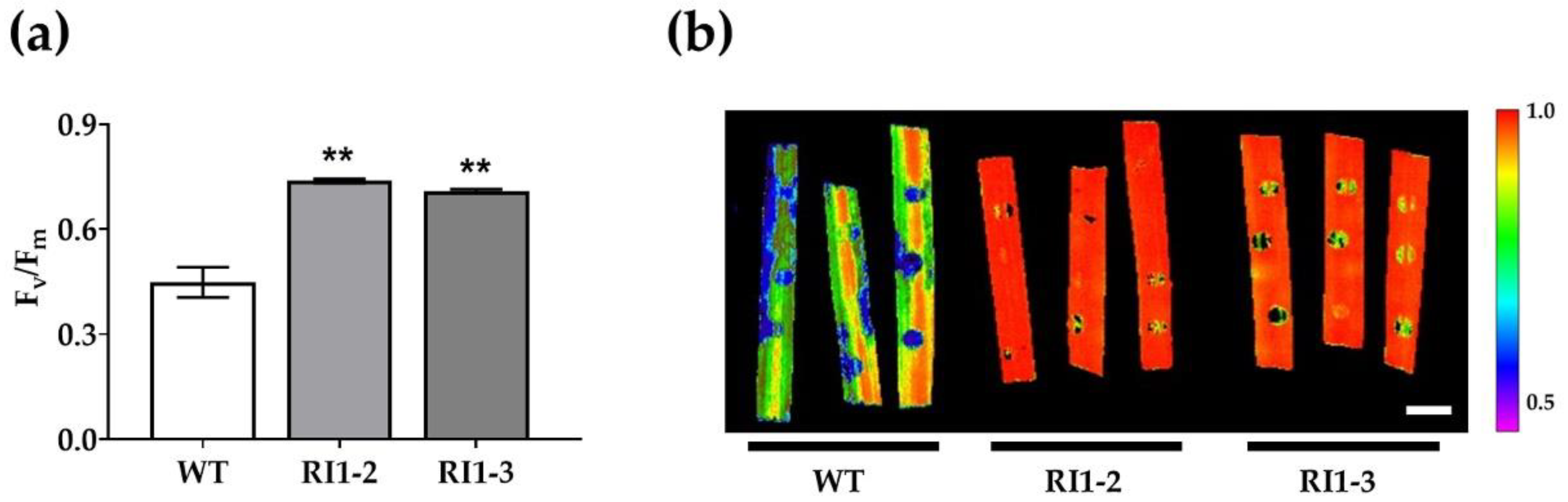
2.3. Photosynthetic Efficiency Analysis
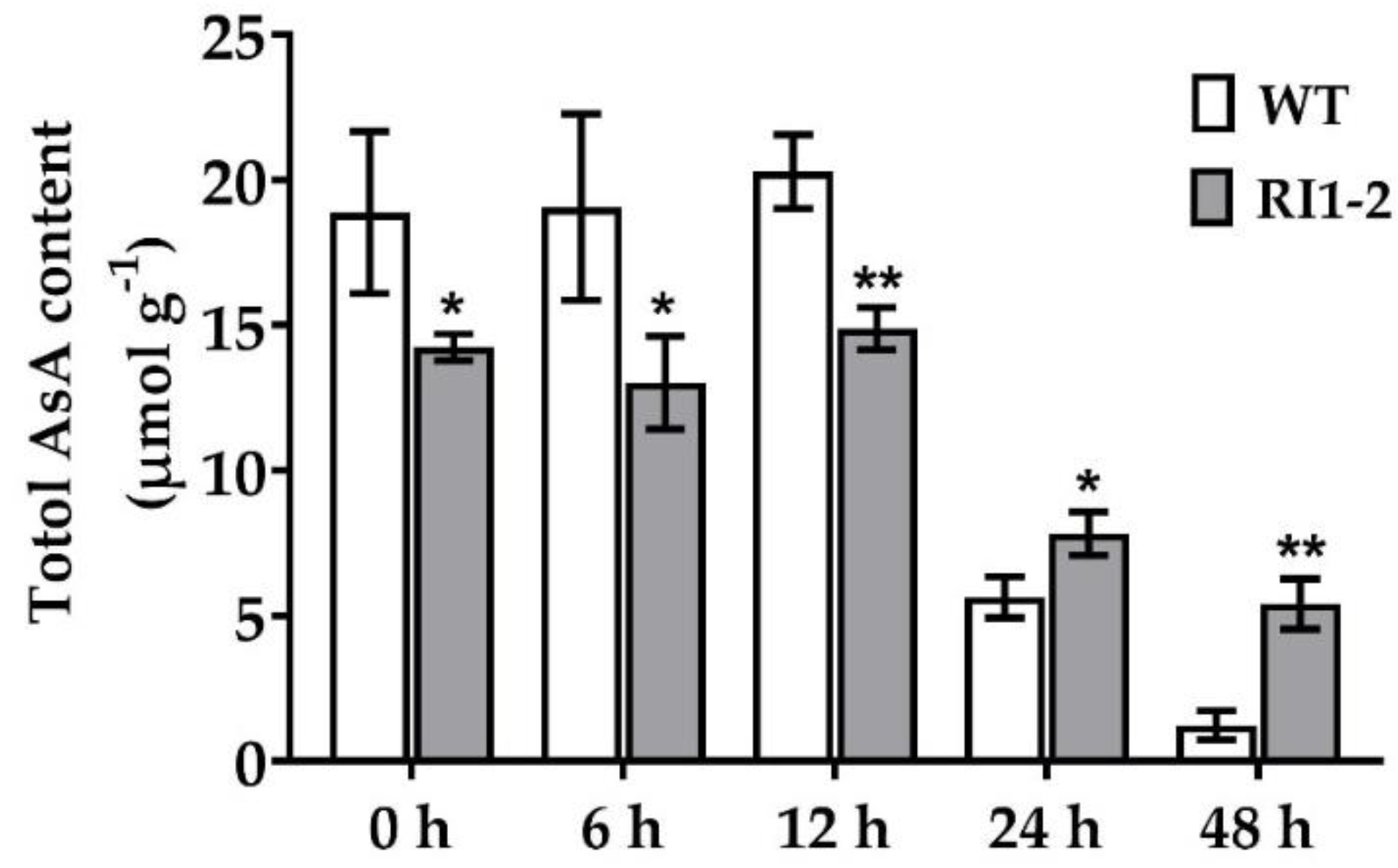
2.4. Measurement of Ascorbic Acid Content
2.5. Detection of Superoxide Anion by Nitroblue Tetrazolium (NBT) Staining and Measurement of Malondialdehyde (MDA) Content
2.6. Transcriptome Analysis
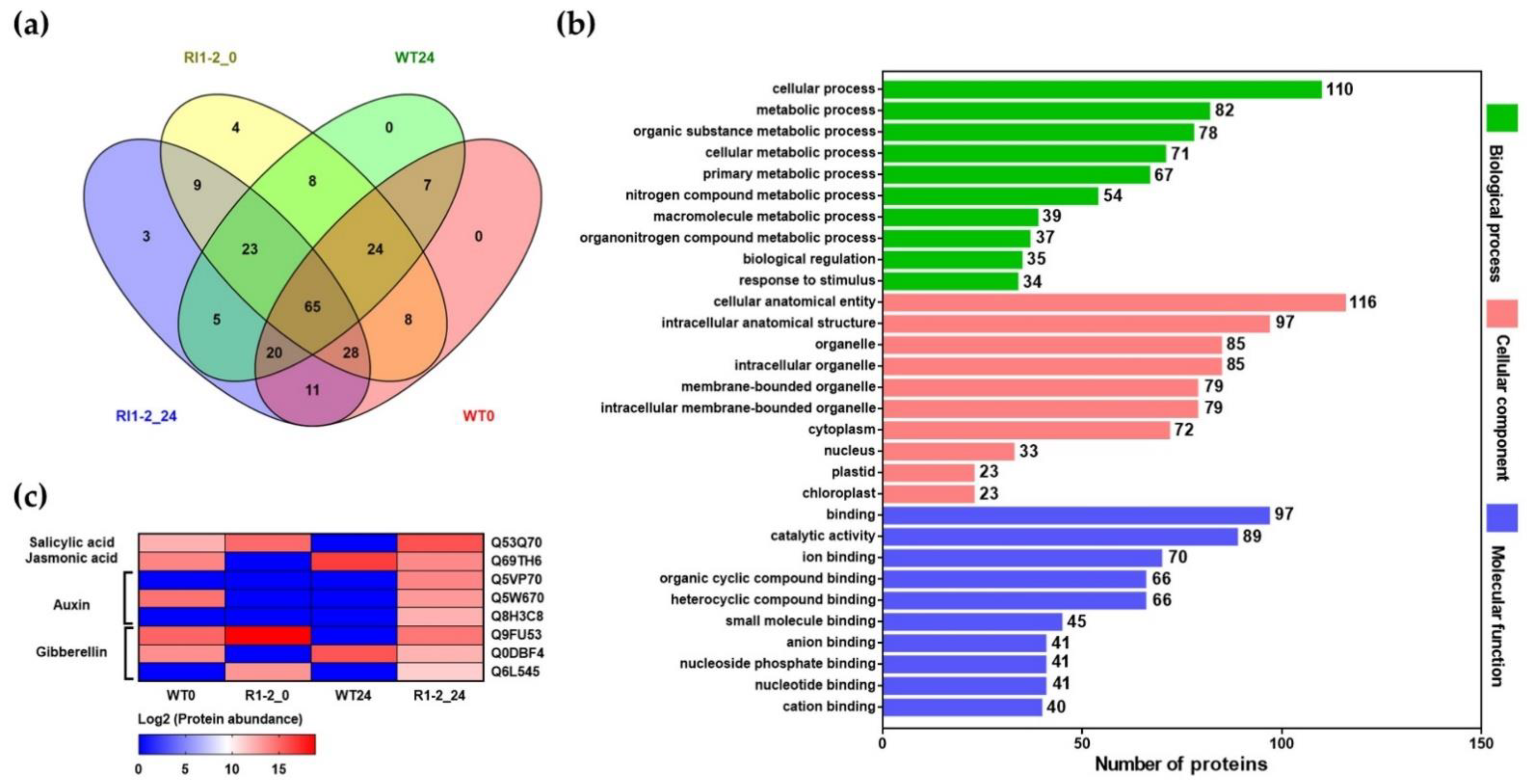
2.7. Proteome Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Fungal Materials and Culture Conditions
4.3. Rice Inoculation and Pathogenicity assay
4.4. Cell Wall Sugar Composition Analysis
4.5. Photosynthetic Efficiency Analysis
4.6. Measurement of Ascorbic Acid Content
4.7. Detection of Superoxide Anion by Nitroblue Tetrazolium (NBT) Staining
4.8. Measurement of Malondialdehyde (MDA) Content
4.9. Transcriptome Analysis
4.10. Proteome Analysis
4.11. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Talbot, N.J. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 2003, 57, 177–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- TeBeest, D.O.; Guerber, C.; Ditmore, M. Rice Blast. Available online: https://www.apsnet.org/edcenter/disandpath/fungalasco/pdlessons/Pages/RiceBlast.aspx (accessed on 20 May 2022).
- Andersen, E.J.; Ali, S.; Byamukama, E.; Yen, Y.; Nepal, M.P. Disease resistance mechanisms in plants. Genes 2018, 9, 339. [Google Scholar] [CrossRef] [Green Version]
- Jain, D.; Khurana, J.P. Role of pathogenesis-related (PR) proteins in plant defense Mechanism. In Molecular Aspects of Plant-Pathogen Interaction; Singh, A., Singh, I.K., Eds.; Springer: Singapore, 2018; pp. 265–281. [Google Scholar]
- Maleck, K.; Lawton, K. Plant strategies for resistance to pathogens. Curr. Opin. Biotechnol. 1998, 9, 208–213. [Google Scholar] [CrossRef]
- Bolwell, G.P.; Bindschedler, L.V.; Blee, K.A.; Butt, V.S.; Davies, D.R.; Gardner, S.L.; Gerrish, C.; Minibayeva, F. The apoplastic oxidative burst in response to biotic stress in plants: A three-component system. J. Exp. Bot. 2002, 53, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Balint-Kurti, P. The plant hypersensitive response: Concepts, control and consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morel, J.-B.; Dangl, J.L. The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 1997, 4, 671–683. [Google Scholar] [CrossRef] [Green Version]
- Dangl, J.L.; Dietrich, R.A.; Richberg, M.H. Death don’t haveno mercy: Cell death programs in plant-microbe interactions. Plant Cell 1996, 8, 1793–1807. [Google Scholar] [CrossRef]
- Pennell, R.I.; Lamb, C. Programmed cell death in plants. Plant Cell 1997, 9, 1157–1168. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 26. [Google Scholar] [CrossRef] [Green Version]
- Smirnoff, N.; Wheeler, G.L. Ascorbic Acid in plants: Biosynthesis and function. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 291–314. [Google Scholar] [CrossRef]
- Zhang, Y. Biological role of ascorbate in plants. In Ascorbic Acid in Plants; Springer Briefs in Plant Science; Springer: New York, NY, USA, 2013; pp. 7–33. [Google Scholar] [CrossRef]
- Davey, M.; Van Montagu, M.; Inzé, D.; Sanmartin, M.; Kanellis, A.; Smirnoff, N.; Benzie, I.; Strain, J.; Favell, D.; Fletcher, J. Plant L-ascorbic acid: Chemistry, function, metabolism, bioavailability and effects of processing. J. Sci. Food Agric. 2000, 80, 825–860. [Google Scholar] [CrossRef]
- Wheeler, G.L.; Jones, M.A.; Smirnoff, N. The biosynthetic pathway of vitamin C in higher plants. Nature 1998, 393, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Conklin, P.L.; Loewus, F.A. Biosynthesis of ascorbic acid in plants: A renaissance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 437–467. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N. Ascorbic acid: Metabolism and functions of a multi-facetted molecule. Curr. Opin. Plant Biol. 2000, 3, 229–235. [Google Scholar] [CrossRef]
- Qin, H.; Deng, Z.; Zhang, C.; Wang, Y.; Wang, J.; Liu, H.; Zhang, Z.; Huang, R.; Zhang, Z. Rice GDP-mannose pyrophosphorylase OsVTC1-1 and OsVTC1-3 play different roles in ascorbic acid synthesis. Plant Mol. Biol. 2016, 90, 317–327. [Google Scholar] [CrossRef]
- Qin, H.; Wang, Y.; Wang, J.; Liu, H.; Zhao, H.; Deng, Z.; Zhang, Z.; Huang, R.; Zhang, Z. Knocking down the expression of gmpase gene OsVTC1-1 decreases salt tolerance of rice at seedling and reproductive stages. PLoS ONE 2016, 11, e0168650. [Google Scholar] [CrossRef]
- Wang, H.-S.; Yu, C.; Zhu, Z.-J.; Yu, X.-C. Overexpression in tobacco of a tomato GMPase gene improves tolerance to both low and high temperature stress by enhancing antioxidation capacity. Plant Cell Rep. 2011, 30, 1029–1040. [Google Scholar] [CrossRef]
- Barth, C.; Moeder, W.; Klessig, D.F.; Conklin, P.L. The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin c-1. Plant Physiol. 2004, 134, 1784–1792. [Google Scholar] [CrossRef] [Green Version]
- Pavet, V.; Olmos, E.; Kiddle, G.; Mowla, S.; Kumar, S.; Antoniw, J.; Alvarez, M.; Foyer, C. Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol. 2005, 139, 1291–1303. [Google Scholar] [CrossRef] [Green Version]
- Dzhavakhiya, V.G.; Ozeretskovskaya, O.L.; Zinovyeva, S.V. Chapter 10—Immune response. In Comprehensive and Molecular Phytopathology; Dyakov, Y.T., Dzhavakhiya, V.G., Korpela, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 265–314. [Google Scholar]
- Walters, D.R.; McRoberts, N. Plants and biotrophs: A pivotal role for cytokinins? Trends Plant Sci. 2006, 11, 581–586. [Google Scholar] [CrossRef]
- Hückelhoven, R. Cell wall–associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 2007, 45, 101–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.; Fan, B.; Zhu, C.; Chen, Z. Regulation and function of defense-related callose deposition in plants. Int. J. Mol. Sci. 2021, 22, 2393. [Google Scholar] [CrossRef] [PubMed]
- Morkunas, I.; Ratajczak, L. The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol. Plant 2014, 36, 1607–1619. [Google Scholar] [CrossRef] [Green Version]
- Voiniciuc, C.; Pauly, M.; Usadel, B. Monitoring polysaccharide dynamics in the plant cell wall. Plant Physiol. 2018, 176, 2590–2600. [Google Scholar] [CrossRef] [Green Version]
- Jabs, T. Reactive oxygen intermediates as mediators of programmed cell death in plants and animals. Biochem. Pharmacol. 1999, 57, 231–245. [Google Scholar] [CrossRef]
- Mandal, S.; Rajarammohan, S.; Kaur, J. ROS accumulation and associated cell death mediates susceptibility to Alternaria brassicae in Arabidopsis accessions. Physiol. Mol. Plant Pathol. 2019, 107, 51–59. [Google Scholar] [CrossRef]
- Su, L.J.; Zhang, J.H.; Gomez, H.; Murugan, R.; Hong, X.; Xu, D.; Jiang, F.; Peng, Z.Y. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid. Med. Cell Longev. 2019, 2019, 5080843. [Google Scholar] [CrossRef] [Green Version]
- Dangol, S.; Chen, Y.; Hwang, B.K.; Jwa, N.-S. Iron- and reactive oxygen species-dependent ferroptotic cell death in rice- Magnaporthe oryzae interactions. Plant Cell. 2019, 31, 189–209. [Google Scholar] [CrossRef] [Green Version]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Leon-Reyes, A.; Van der Does, D.; De Lange, E.S.; Delker, C.; Wasternack, C.; Van Wees, S.C.M.; Ritsema, T.; Pieterse, C.M.J. Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 2010, 232, 1423–1432. [Google Scholar] [CrossRef] [Green Version]
- Lamanchai, K.; Salmon, D.L.; Smirnoff, N.; Sutthinon, P.; Roytrakul, S.; Leetanasaksakul, K.; Kittisenachai, S.; Jantasuriyarat, C. OsVTC1-1 RNAi mutant with reduction of ascorbic acid synthesis alters cell wall sugar composition and cell wall-associated proteins. Agronomy 2022, 12, 1272. [Google Scholar] [CrossRef]
- Gallie, D.R. Ascorbic Acid: A multifunctional molecule supporting plant growth and development. Scientifica 2013, 2013, 795964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barón, M.; Pineda, M.; Pérez-Bueno, M.L. Picturing pathogen infection in plants. Z. Naturforsch. C J. Biosci. 2016, 71, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, R.W.; Merchant, A.; Tausz, M. Differences in ascorbate and glutathione levels as indicators of resistance and susceptibility in Eucalyptus trees infected with Phytophthora cinnamomi. Tree Physiol. 2012, 32, 1148–1160. [Google Scholar] [CrossRef] [Green Version]
- Schieber, M.; Chandel, N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, 453–462. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Chen, X.; Zhu, F.; Li, H.; Li, L.; Yang, Q.; Chi, X.; Yu, S.; Liang, X. Characterization of peanut germin-like proteins, AhGLPs in plant development and defense. PLoS ONE 2013, 8, e61722. [Google Scholar] [CrossRef] [Green Version]
- Kawano, T. Roles of the reactive oxygen species-generating peroxidase reactions in plant defense and growth induction. Plant Cell Rep. 2003, 21, 829–837. [Google Scholar] [CrossRef]
- Yang, D.; Li, S.; Xiao, Y.; Lu, L.; Zheng, Z.; Tang, D.; Cui, H. Transcriptome analysis of rice response to blast fungus identified core genes involved in immunity. Plant Cell Environ. 2021, 44, 3103–3121. [Google Scholar] [CrossRef]
- Filipenko, E.A.; Kochetov, A.V.; Kanayama, Y.; Malinovsky, V.I.; Shumny, V.K. PR-proteins with ribonuclease activity and plant resistance against pathogenic fungi. Russ. J. Genet. Appl. 2013, 3, 474–480. [Google Scholar] [CrossRef]
- Chandrashekar, N.; Ali, S.; Grover, A. Exploring expression patterns of PR-1, PR-2, PR-3, and PR-12 like genes in Arabidopsis thaliana upon Alternaria brassicae inoculation. 3 Biotech 2018, 8, 230. [Google Scholar] [CrossRef]
- McQueen-Mason, S.J. Expansins and cell wall expansion. J. Exp. Bot. 1995, 46, 1639–1650. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Chong, K. The novel functions of kinesin motor proteins in plants. Protoplasma 2012, 249 (Suppl. S2), S95–S100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, J.H.; Narváez-Vásquez, J.; Aromdee, D.N.; Pautot, V.; Holzer, F.M.; Walling, L.L. Leucine aminopeptidase regulates defense and wound signaling in tomato downstream of jasmonic acid. Plant Cell. 2009, 21, 1239–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kwon, S.J.; Wu, J.; Choi, J.; Lee, Y.-H.; Agrawal, G.K.; Tamogami, S.; Rakwal, R.; Park, S.-R.; Kim, B.-G.; et al. Transcriptome analysis of early responsive genes in rice during Magnaporthe oryzae infection. Plant Pathol. J. 2014, 30, 343–354. [Google Scholar] [CrossRef] [Green Version]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef] [Green Version]
- Talbot, N.J.; Ebbole, D.J.; Hamer, J.E. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 1993, 5, 1575–1590. [Google Scholar] [CrossRef] [Green Version]
- Conklin, P.L.; Gatzek, S.; Wheeler, G.L.; Dowdle, J.; Raymond, M.J.; Rolinski, S.; Isupov, M.; Littlechild, J.A.; Smirnoff, N. Arabidopsis thaliana VTC4 Encodes L-Galactose-1-P Phosphatase, a plant ascorbic acid biosynthetic enzyme. J. Biol. Chem. 2006, 281, 15662–15670. [Google Scholar] [CrossRef] [Green Version]
- Baker, N.R. Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 2008, 59, 89–113. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, K.; Ainsworth, E. Measurement of reduced, oxidized and total ascorbate content in plants. Nat. Protoc. 2007, 2, 871–874. [Google Scholar] [CrossRef]
- Kumar, D.; Yusuf, M.; Singh, P.; Sardar, M.; Sarin, N. Histochemical detection of superoxide and H2O2 accumulation in Brassica juncea seedlings. Bio-Protocol 2014, 4, e1108. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts. I. kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Merchant, N.; Lyons, E.; Goff, S.; Vaughn, M.; Ware, D.; Micklos, D.; Antin, P. The iPlant collaborative: Cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol. 2016, 14, e1002342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakai, H.; Lee, S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.-C.; Iwamoto, M.; Abe, T.; et al. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Bastide, M.; Hamilton, J.; Kanamori, H.; McCombie, W.; Ouyang, S.; Schwartz, D.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Consortium, T.G.O. The Gene Ontology resource: Enriching a Gold mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Naithani, S.; Gupta, P.; Preece, J.; D’Eustachio, P.; Elser, J.L.; Garg, P.; Dikeman, D.A.; Kiff, J.; Cook, J.; Olson, A.; et al. Plant Reactome: A knowledgebase and resource for comparative pathway analysis. Nucleic Acids Res. 2019, 48, D1093–D1103. [Google Scholar] [CrossRef]
- Extract and Visualize the Results of Multivariate Data Analyses [R Package Factoextra Version 1.0.7]. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 14 June 2022).
- Lowry, O.; Rosebrough, N.; Farr, A.L.; Randall, R. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]

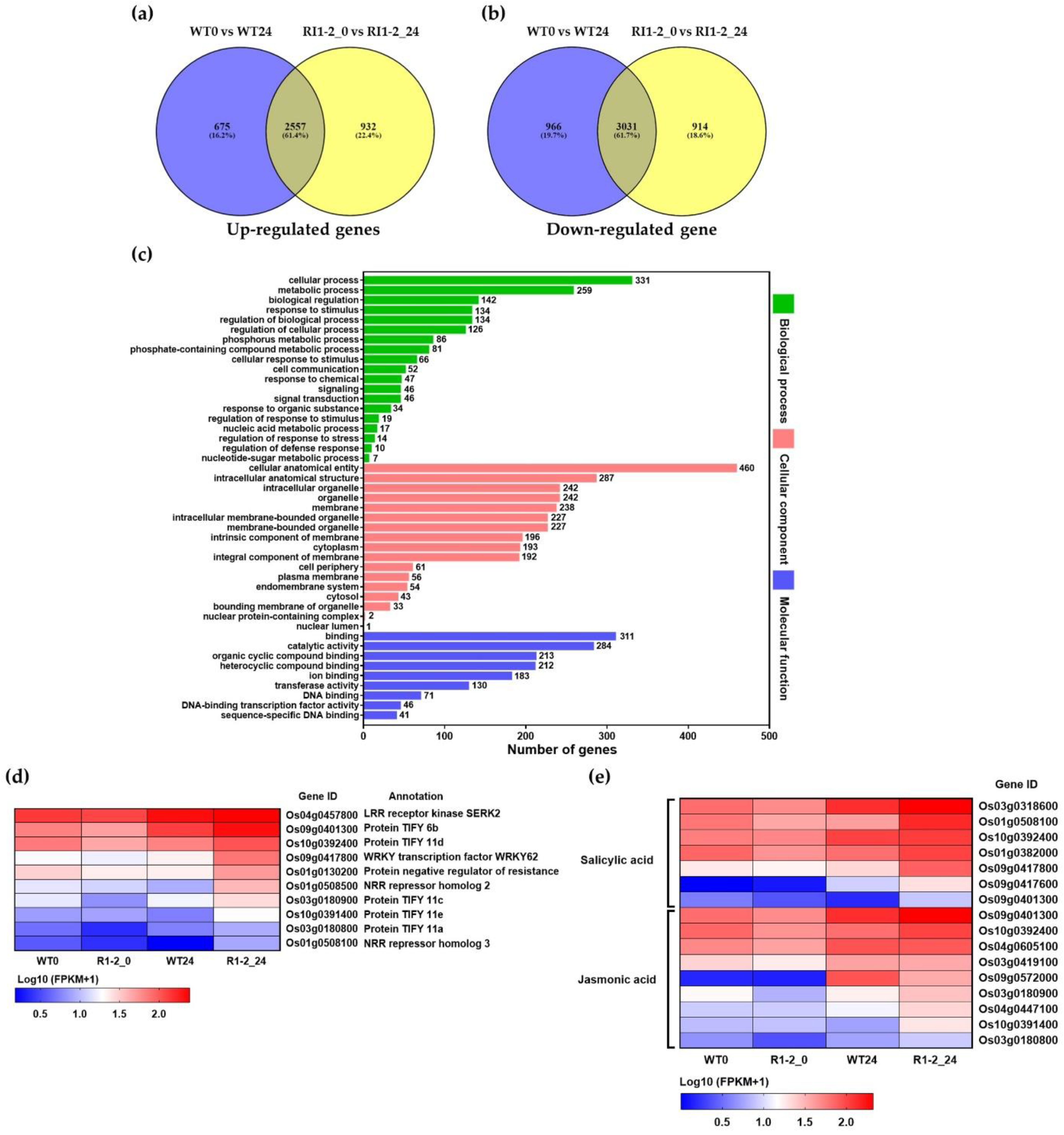
| Up-Regulated DEGs of RI1-2 | |||
| No. | Gene ID | Gene Description | Log2FC |
| 1 | Os08g0189900 | Germin-like protein 8-10, disease resistance | 10.156 |
| 2 | Os12g0503000 | Allantoin transporter | 8.199 |
| 3 | Os03g0363100 | Armadillo-like helical domain containing protein | 8.186 |
| 4 | Os06g0522300 | Haem peroxidase family protein | 8.156 |
| 5 | Os05g0473101 | Hypothetical gene | 7.671 |
| 6 | Os07g0541900 | Similar to KI domain interacting kinase 1 | 7.659 |
| 7 | Os07g0677200 | Peroxidase, negative regulation of rice resistance to blast disease | 7.640 |
| 8 | Os08g0507100 | Cytochrome P450 family protein | 7.599 |
| 9 | Os04g0493400 | Chitinase-4, Pathogenesis-related (PR)-3 | 7.596 |
| 10 | Os07g0539100 | Glycoside hydrolase family 17 protein | 7.476 |
| 11 | Os06g0244000 | Similar to anthranilic acid methyltransferase | 7.395 |
| 12 | Os01g0731150 | Hypothetical protein | 7.374 |
| 13 | Os09g0447300 | Cytochrome P450 family protein | 7.297 |
| 14 | Os01g0615100 | Chymotrypsin protease inhibitor | 7.297 |
| 15 | Os01g0615050 | Chymotrypsin inhibitor | 7.274 |
| 16 | Os09g0400400 | Cinnamyl alcohol dehydrogenase | 7.260 |
| 17 | Os08g0189400 | Germin-like protein 8-5, disease resistance | 7.104 |
| 18 | Os09g0319800 | Terpene synthase-like domain containing protein | 7.067 |
| 19 | Os11g0618700 | Protein of unknown function DUF594 domain containing protein. | 7.001 |
| 20 | Os08g0189600 | Germin-like protein 8-7, disease resistance | 6.956 |
| Down-Regulated DEGs of RI1-2 | |||
| No. | Gene ID | Gene Description | Log2FC |
| 1 | Os09g0537700 | S-like ribonuclease, regulation of photomorphogenesis | −9.326 |
| 2 | Os05g0579600 | Homeodomain-like containing protein. | −8.560 |
| 3 | Os01g0975900 | Similar to tonoplast membrane integral protein ZmTIP1-2 | −7.913 |
| 4 | Os01g0922700 | Conserved hypothetical protein. | −7.627 |
| 5 | Os05g0409500 | Similar to MtN21 protein. | −7.565 |
| 6 | Os01g0973100 | Hypothetical conserved gene | −7.496 |
| 7 | Os06g0107100 | Protein of unknown function DUF819 family protein | −7.267 |
| 8 | Os07g0628900 | Similar to KI domain interacting kinase 1 | −7.125 |
| 9 | Os01g0734600 | UDP-glucuronosyl/UDP-glucosyltransferase family protein | −6.893 |
| 10 | Os03g0371300 | Similar to Cytochrome P450 | −6.689 |
| 11 | Os12g0493900 | Armadillo-like helical domain containing protein. | −6.654 |
| 12 | Os07g0142900 | Aldo/keto reductase family protein. | −6.645 |
| 13 | Os04g0282400 | Flowering-promoting factor 1-like protein 4 | −6.383 |
| 14 | Os05g0500101 | Hypothetical gene | −6.379 |
| 15 | Os07g0111900 | Flavin-containing monooxygenase FMO family protein | −6.326 |
| 16 | Os07g0558300 | Inositol monophosphatase family protein | −6.267 |
| 17 | Os03g0223100 | Cytochrome P450 family protein | −6.242 |
| 18 | Os08g0249900 | Similar to Gibberellin 20 oxidase 2 | −6.239 |
| 19 | Os01g0733500 | Similar to dehydration-induced protein RD22-like protein 1 | −6.185 |
| 20 | Os04g0583900 | MYB-related transcription factor | −6.183 |
| No. | Protein ID | Protein Name | Log2 Protein Abundance | |||
|---|---|---|---|---|---|---|
| WT0 | RI1-2_0 | WT24 | RI1-2_24 | |||
| 1 | Q5VP70 | Auxin efflux carrier component 3a | 0 | 0.00 | 0 | 13.89 |
| 2 | Q942P9 | Probable protein phosphatase 2C 1 | 0 | 0.00 | 0 | 13.40 |
| 3 | Q8H3C8 | IAA-amino acid hydrolase ILR1-like 8 | 0 | 0.00 | 0 | 12.32 |
| 4 | Q75KV9 | Transcription factor BHLH148 | 14.86 | 0.00 | 0.00 | 12.16 |
| 5 | Q53Q70 | Transcription factor TGAL4 | 12.27 | 14.91 | 0.00 | 15.84 |
| 6 | Q9FWX2 | NAC domain-containing protein 7 | 13.61 | 13.03 | 0.00 | 15.28 |
| 7 | Q9FY82 | NAC domain-containing protein 82 | 14.16 | 17.64 | 11.13 | 13.48 |
| 8 | Q6K537 | Dof zinc finger protein 3 | 0.00 | 15.55 | 12.77 | 11.53 |
| 9 | A4LBC0 | B3 domain-containing protein LFL1 | 12.15 | 17.90 | 16.00 | 0.00 |
| 10 | Q7Y0V7 | Homeobox-leucine zipper protein ROC6 | 12.86 | 16.98 | 15.16 | 0.00 |
| 11 | Q6ZBS8 | Transcription factor TGAL10 | 0.00 | 12.66 | 12.05 | 0.00 |
| 12 | Q40636 | Expansin-A2 | 14.33 | 0.00 | 0.00 | 13.32 |
| 13 | B9FFA3 | Kinesin-like protein KIN-7E | 15.97 | 14.95 | 15.44 | 17.06 |
| 14 | Q5Z7L0 | Beta-galactosidase 9 | 13.81 | 0.00 | 12.71 | 13.43 |
| 15 | Q7XPY5 | Putative beta-glucosidase 15 | 11.39 | 18.11 | 10.45 | 15.45 |
| 16 | Q6K669 | Leucine aminopeptidase 2 | 0.00 | 12.06 | 15.04 | 17.61 |
| 17 | Q3EBC8 | Endoribonuclease dicer homolog 2 | 13.73 | 18.60 | 14.93 | 16.27 |
| 18 | Q0DV66 | Pheophorbide a oxygenase | 15.81 | 0.00 | 14.65 | 16.10 |
| 19 | Q0D5R3 | Cysteine-rich receptor-like protein kinase 6 | 14.21 | 19.46 | 0.00 | 14.71 |
| 20 | Q6Z8P4 | Plant intracellular Ras-group-related LRR protein 4 | 14.85 | 15.68 | 13.29 | 14.80 |
| 21 | Q7X7X4 | Cytochrome P450 | 13.27 | 17.21 | 0.00 | 13.74 |
| 22 | P48642 | Glutathione reductase | 15.75 | 15.39 | 15.06 | 0.00 |
| 23 | P93407 | Superoxide dismutase | 0.00 | 14.93 | 12.14 | 11.05 |
| 24 | Q65XA0 | Probable glutathione S-transferase DHAR1 | 0.00 | 13.86 | 16.46 | 11.51 |
| 25 | B9FSC8 | Putative 12-oxophytodienoate reductase 11 | 13.27 | 15.73 | 12.51 | 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamanchai, K.; Smirnoff, N.; Salmon, D.L.; Ngernmuen, A.; Roytrakul, S.; Leetanasaksakul, K.; Kittisenachai, S.; Jantasuriyarat, C. OsVTC1-1 Gene Silencing Promotes a Defense Response in Rice and Enhances Resistance to Magnaporthe oryzae. Plants 2022, 11, 2189. https://doi.org/10.3390/plants11172189
Lamanchai K, Smirnoff N, Salmon DL, Ngernmuen A, Roytrakul S, Leetanasaksakul K, Kittisenachai S, Jantasuriyarat C. OsVTC1-1 Gene Silencing Promotes a Defense Response in Rice and Enhances Resistance to Magnaporthe oryzae. Plants. 2022; 11(17):2189. https://doi.org/10.3390/plants11172189
Chicago/Turabian StyleLamanchai, Kanyanat, Nicholas Smirnoff, Deborah L. Salmon, Athipat Ngernmuen, Sittiruk Roytrakul, Kantinan Leetanasaksakul, Suthathip Kittisenachai, and Chatchawan Jantasuriyarat. 2022. "OsVTC1-1 Gene Silencing Promotes a Defense Response in Rice and Enhances Resistance to Magnaporthe oryzae" Plants 11, no. 17: 2189. https://doi.org/10.3390/plants11172189
APA StyleLamanchai, K., Smirnoff, N., Salmon, D. L., Ngernmuen, A., Roytrakul, S., Leetanasaksakul, K., Kittisenachai, S., & Jantasuriyarat, C. (2022). OsVTC1-1 Gene Silencing Promotes a Defense Response in Rice and Enhances Resistance to Magnaporthe oryzae. Plants, 11(17), 2189. https://doi.org/10.3390/plants11172189






