Location and Identification on Chromosome 3B of Bread Wheat of Genes Affecting Chiasma Number
Abstract
1. Introduction
2. Results
2.1. Chromosome 3B Is Essential for Chiasma Number but Cannot Be Compensated by Chromosomes 3A or 3D
2.2. The Long Arm of Chromosome 3B Is Essential for Chiasma Number in Wheat
2.3. The Long Arm of Chromosome 3B Bears at Least Two Genes Involved in Chiasma Number
2.4. Comparative Analysis of Meiotic Behaviour in CS, 3bl7 and dt3bs Lines
2.5. In Silico Mapping of Genes Expressed during Wheat Meiosis or Known as Involved in Meiosis in Arabidopsis
3. Discussion
3.1. Variation in Chiasma Number in the Aneuploid Stock from Chromosome 3B
3.2. Identification of Candidate Genes Located in the Two Deletion Bins Concerned
3.3. Conclusions
4. Materials and Methods
4.1. Plant Material
4.2. Chiasma Observation and Statistical Analysis
4.3. Rna-Seq Data Production and In Silico Gene Mapping
4.4. Immunostaining
4.5. Fluorescence DNA In Situ Hybridization
4.6. Microscopy and Imaging
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balfourier, F.; Bouchet, S.; Robert, S.; De Oliveira, R.; Rimbert, H.; Kitt, J.; Choulet, F.; Paux, E.; International Wheat Genome Sequencing Consortium; BreedWheat Consortium. Worldwide phylogeography and history of wheat genetic diversity. Sci. Adv. 2019, 5, eaav0536. [Google Scholar] [CrossRef]
- He, F.; Pasam, R.; Shi, F.; Kant, S.; Keeble-Gagnere, G.; Kay, P.; Forrest, K.; Fritz, A.; Hucl, P.; Wiebe, K.; et al. Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet. 2019, 51, 896–904. [Google Scholar] [CrossRef]
- Pont, C.; Leroy, T.; Seidel, M.; Tondelli, A.; Duchemin, W.; Armisen, D.; Lang, D.; Bustos-Korts, D.; Goué, N.; Balfourier, F.; et al. Tracing the ancestry of modern bread wheats. Nat. Genet. 2019, 51, 905–911. [Google Scholar] [CrossRef]
- Rasmussen, S.W. Meiosis in Bombyx mori Females. Phil. Trans. R. Soc. Lond. 1977, 277, 343–350. [Google Scholar]
- Tsai, J.H.; McKee, B.D. Homologous pairing and the role of pairing centers in meiosis. J. Cell Sci. 2011, 124, 1955–1963. [Google Scholar] [CrossRef] [PubMed]
- Cabral, G.; Marques, A.; Schubert, V.; Pedrosa-Harand, A.; Schlögelhofer, P. Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat. Comm. 2014, 5, 5070. [Google Scholar] [CrossRef]
- Mercier, R.; Mézard, C.; Jenczewski, E.; Macaisne, N.; Grelon, M. The Molecular Biology of Meiosis in Plants. Annu. Rev. Plant Biol. 2015, 66, 297–327. [Google Scholar] [CrossRef] [PubMed]
- Blake, N.K.; Lehfeldt, B.R.; Lavin, M.; Talbert, L.E. Phylogenetic reconstruction based on low copy DNA sequence data in an allopolyploid: The B genome of wheat. Genome 1999, 42, 351–360. [Google Scholar] [CrossRef]
- Feldman, M.; Lupton, F.; Miller, T. Wheats. In Evolution of Crop Plants; Smartt, J., Simmonds, N.W., Eds.; Longman Group: London, UK, 1995; pp. 184–192. [Google Scholar]
- Huang, S.; Sirikhachornkit, A.; Faris, J.D.; Su, X.; Gill, B.S.; Haselkorn, R.; Gornicki, P. Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol. Biol. 2002, 48, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Glémin, S.; Scornavacca, C.; Dainat, J.; Burgarella, C.; Viader, V.; Ardisson, M.; Sarah, G.; Santoni, S.; David, J.; Ranwez, V. Pervasive hybridizations in the history of wheat relatives. Sci. Adv. 2019, 5, eaav9188. [Google Scholar] [CrossRef]
- Li, L.-F.; Zhang, Z.-B.; Wang, Z.-H.; Li, N.; Sha, Y.; Wang, X.-F.; Ding, N.; Li, Y.; Zhao, J.; Wu, Y.; et al. Genome sequences of five Sitopsis species of Aegilops and the origin of polyploid wheat B subgenome. Mol. Plant 2022, 15, 488–503. [Google Scholar] [CrossRef]
- Avni, R.; Lux, T.; Minz-Dub, A.; Millet, E.; Sela, H.; Distelfeld, A.; Deek, J.; Yu, G.; Steuernagel, B.; Pozniak, C.; et al. Genome sequences of three Aegilops species of the section Sitopsis reveal phylogenetic relationships and provide resources for wheat improvement. Plant J. 2022, 110, 179–192. [Google Scholar] [CrossRef]
- Sears, E.R. Cytogenetic Studies with Polyploid Species of Wheat. I. Chromosomal Aberrations in the Progeny of a Haploid of Triticum vulgare. Genetics 1939, 24, 509–523. [Google Scholar] [CrossRef]
- Sears, E.R. Cytogenetic studies with polyploid species of wheat. II. Additional chromosomal aberrations in Triticum vulgare. Genetics 1944, 29, 232–246. [Google Scholar] [CrossRef]
- Sears, E.R.; Sears, L. The telocentric chromosomes of common wheat. In Proceedings of the 5th Wheat Genetics Symposium, New Delhi, India, 23–28 February 1978. [Google Scholar]
- Endo, T.R.; Gill, B.S. The Deletion Stocks of Common Wheat. J. Hered. 1996, 87, 295–307. [Google Scholar] [CrossRef]
- Sears, E.R. The Aneuploids of Common Wheat; University of Missouri College of Agriculture, Agricultural Experiment Station: Columbia, MO, USA, 1954; pp. 1–58. [Google Scholar]
- Sharp, P.J.; Chao, S.; Desai, S.; Gale, M.D. The isolation, characterization and application in the Triticeae of a set of wheat RFLP probes identifying each homoeologous chromosome arm. Theor. Appl. Genet. 1989, 78, 342–348. [Google Scholar] [CrossRef]
- Anderson, J.A.; Churchill, G.A.; Autrique, J.E.; Tanksley, S.D.; Sorrells, M.E. Optimizing parental selection for genetic linkage maps. Genome 1993, 36, 181–186. [Google Scholar] [CrossRef]
- Najanjo, T. Forcing the shift of the crossover site to proximal regions in wheat chromosomes. Theor. Appl. Genet. 2015, 128, 1855–1863. [Google Scholar] [CrossRef]
- Šafár, J.; Bartoš, J.; Janda, J.; Bellec, A.; Kubaláková, M.; Valárik, M.; Pateyron, S.; Weiserová, J.; Tušková, R.; Cíhalíková, J.; et al. Dissecting large and complex genomes: Flow-sorting and BAC cloning of individual chromosomes from bread wheat. Plant J. 2004, 39, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Paux, E.; Sourdille, P.; Salse, J.; Saintenac, C.; Choulet, F.; Leroy, P.; Korol, A.; Michalak, M.; Kianian, S.; Spielmeyer, W.; et al. A physical map of the 1-gigabase bread wheat chromosome 3B. Science 2008, 322, 101–104. [Google Scholar] [CrossRef]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.M.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E.; et al. Structural and Functional Partitioning of Bread Wheat Chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef] [PubMed]
- Daron, J.; Glover, N.M.; Pingault, L.; Theil, S.; Jamilloux, V.; Paux, E.; Barbe, V.; Mangenot, S.; Alberti, A.; Wincker, P.; et al. Organization and evolution of transposable elements along the bread wheat chromosome 3B. Genome Biol. 2014, 15, 546. [Google Scholar] [CrossRef] [PubMed]
- Glover, N.M.; Daron, J.; Pingault, L.; Vandepoele, K.; Paux, E.; Feuillet, C.; Choulet, F. Small-scale gene duplications played a major role in the recent evolution of wheat chromosome 3B. Genome Biol. 2015, 16, 188. [Google Scholar] [CrossRef]
- Pingault, L.; Choulet, F.; Alberti, A.; Glover, N.M.; Wincker, P.; Feuillet, C.; Paux, E. Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biol. 2015, 16, 29. [Google Scholar] [CrossRef]
- Mello-Sampayo, T. Genetic Regulation of Meiotic Chromosome Pairing by Chromosome 3D of Triticum aestivum. Nat. New Biol. 1971, 230, 22–23. [Google Scholar] [CrossRef]
- Mello-Sampayo, T.; Canas, P. Suppressors of meiotic chromosome pairing in common wheat. In Proceedings of the 4th International Wheat Genetics Symposium, Columbia, MO, USA, 6–11 August 1973; pp. 709–713. [Google Scholar]
- Mello-Sampayo, T.; Lorente, R. The role of chromosome 3D in the regulation of meiotic pairing in hexaploid wheat. EWAC Newsl. 1969, 2, 19–24. [Google Scholar]
- Martinez, M.; Cuñado, N.; Carcelén, N.; Romero, C. The Ph1 and Ph2 loci play different roles in the synaptic behaviour of hexaploid wheat Triticum aestivum. Theor. Appl. Genet. 2001, 103, 398–405. [Google Scholar] [CrossRef]
- Prieto, P.; Moore, G.; Reader, S. Control of conformation changes associated with homologue recognition during meiosis. Theor. Appl. Genet. 2005, 111, 505–510. [Google Scholar] [CrossRef]
- Sears, E.R. Genetic Control of Chromosome Pairing in Wheat. Annu. Rev. Genet. 1976, 10, 31–51. [Google Scholar] [CrossRef]
- Sears, E.R. An induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 1977, 19, 585–593. [Google Scholar] [CrossRef]
- Sears, E.R. A wheat mutation conditioning an intermediate level of homoeologous chromosome pairing. Can. J. Genet. Cytol. 1982, 24, 715–719. [Google Scholar] [CrossRef]
- Dong, C.; Whitford, R.; Langridge, P. A DNA mismatch repair gene links to the Ph2 locus in wheat. Genome 2002, 45, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Sutton, T.; Whitford, R.; Baumann, U.; Dong, C.; Able, J.A.; Langridge, P. The Ph2 pairing homoeologous locus of wheat (Triticum aestivum): Identification of candidate meiotic genes using a comparative genetics approach. Plant J. 2003, 36, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Serra, H.; Svačina, R.; Baumann, U.; Whitford, R.; Sutton, T.; Bartoš, J.; Sourdille, P. Ph2 encodes the mismatch repair protein MSH7-3D that inhibits wheat homoeologous recombination. Nat. Comm. 2021, 12, 803. [Google Scholar] [CrossRef]
- Driscoll, C.J. Genetic suppression of homoeologous chromosome pairing in hexaploid wheat. Can. J. Genet. Cytol. 1972, 14, 39–42. [Google Scholar] [CrossRef]
- Riley, R.; Chapman, V. Genetic Control of the Cytologically Diploid Behaviour of Hexaploid Wheat. Nature 1958, 182, 713–715. [Google Scholar] [CrossRef]
- Griffiths, S.; Sharp, R.; Foote, T.N.; Bertin, I.; Wanous, M.; Reader, S.; Colas, I.; Moore, G. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 2006, 439, 749–752. [Google Scholar] [CrossRef]
- Martín, A.C.; Rey, M.D.; Shaw, P.; Moore, G. Dual effect of the wheat Ph1 locus on chromosome synapsis and crossover. Chromosoma 2017, 126, 669–680. [Google Scholar] [CrossRef]
- Rey, M.D.; Martín, A.C.; Higgins, J.; Swarbreck, D.; Uauy, C.; Shaw, P.; Moore, G. Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol. Breed. 2017, 37, 95. [Google Scholar] [CrossRef]
- Miller, T.E.; Chapman, V. Aneuhaploids in bread wheat. Genet. Res. 1976, 28, 37–45. [Google Scholar] [CrossRef]
- Viegas, W.S. The effect of B-chromosomes of rye on the chromosome association in F1 hybrids Triticum aestivum x Secale cereale in the absence of chromosomes 5B or 5D. Theor. Appl. Genet. 1980, 56, 193–198. [Google Scholar] [CrossRef]
- Denis, J.-B.; Bernard, M.; Arnoux, J.; Cauderon, Y. Analyse statistique des configurations méiotiques lors de la création d’une série monosomique du blé tendre “ Courtot ”. Agronomie 1982, 2, 701–708. [Google Scholar] [CrossRef]
- The International Wheat Genome Sequence Consortium Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [CrossRef]
- Colas, I.; Darrier, B.; Arrieta, M.; Mittmann, S.U.; Ramsay, L.; Sourdille, P.; Waugh, R. Observation of Extensive Chromosome Axis Remodeling during the “Diffuse-Phase” of Meiosis in Large Genome Cereals. Front. Plant Sci. 2017, 8, 1235. [Google Scholar] [CrossRef]
- Boden, S.A.; Langridge, P.; Spangenberg, G.; Able, J.A. TaASY1 promotes homologous chromosome interactions and is affected by deletion of Ph1. Plant J. 2009, 57, 487–497. [Google Scholar] [CrossRef]
- Colas, I.; Shaw, P.; Prieto, P.; Wanous, M.; Spielmeyer, W.; Mago, R.; Moore, G. Effective chromosome pairing requires chromatin remodeling at the onset of meiosis. Proc. Natl. Acad. Sci. USA 2008, 105, 6075–6080. [Google Scholar] [CrossRef]
- Barakate, A.; Higgins, J.D.; Vivera, S.; Stephens, J.; Perry, R.M.; Ramsay, L.; Colas, I.; Oakey, H.; Waugh, R.; Franklin, F.C.H.; et al. The Synaptonemal Complex Protein ZYP1 Is Required for Imposition of Meiotic Crossovers in Barley. Plant Cell 2014, 26, 729–740. [Google Scholar] [CrossRef]
- Voelkel-Meiman, K.; Taylor, L.F.; Mukherjee, P.; Humphreys, N.; Tsubouchi, H.; MacQueen, A.J. SUMO Localizes to the Central Element of Synaptonemal Complex and Is Required for the Full Synapsis of Meiotic Chromosomes in Budding Yeast. PLoS Genet. 2013, 9, e1003837. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S. Comparative Genome Organization in Plants: From Sequence and Markers to Chromatin and Chromosomes. Plant Cell 2000, 12, 617–636. [Google Scholar] [CrossRef]
- El-Twab, M.H.A. Physical mapping of the 45S rDNA on the chromosomes of Triticum turgidum and T. aestivum using fluorescence in situ hybridization for chromosome ancestors. Arab. J. Biotech. 2007, 10, 69–80. [Google Scholar]
- Lloyd, A.; Ranoux, M.; Vautrin, S.; Glover, N.M.; Fourment, J.; Charif, D.; Choulet, F.; Lassalle, G.; Marande, W.; Tran Ngoc Danh, J.; et al. Meiotic gene evolution: Can you teach a new dog new tricks? Mol. Biol. Evol. 2014, 31, 1724–1727. [Google Scholar] [CrossRef] [PubMed]
- Crismani, W.; Girard, C.; Froger, N.; Pradillo, M.; Santos, J.L.; Chelysheva, L.; Copenhaver, G.P.; Horlow, C.; Mercier, R. FANCM limits meiotic crossovers. Science 2012, 336, 1588–1590. [Google Scholar] [CrossRef]
- Emmanuel, E.; Yehuda, E.; Melamed-Bessudo, C.; Avivi-Ragolsky, N.; Levy, A.A. The role of AtMSH2 in homologous recombination in Arabidopsis thaliana. EMBO Rep. 2006, 7, 100–105. [Google Scholar] [CrossRef]
- Tam, S.M.; Hays, J.B.; Chetelat, R.T. Effects of suppressing the DNA mismatch repair system on homeologous recombination in tomato. Theor. Appl. Genet. 2011, 123, 1445–1458. [Google Scholar] [CrossRef]
- Kempanna, C.; Riley, R. Relationships between the genetic effects of deficiencies for chromosome III and V on meiotic pairing in Triticum aestivum. Nature 1962, 195, 1270–1273. [Google Scholar] [CrossRef]
- Girard, C.; Crismani, W.; Froger, N.; Mazel, J.; Lemhemdi, A.; Horlow, C.; Mercier, R. FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 2014, 42, 9087–9095. [Google Scholar] [CrossRef] [PubMed]
- Girard, C.; Chelysheva, L.; Choinard, S.; Froger, N.; Macaisne, N.; Lemhemdi, A.; Mazel, J.; Crismani, W.; Mercier, R. AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 2015, 11, e1005369. [Google Scholar] [CrossRef]
- Séguéla-Arnaud, M.; Crismani, W.; Larchevêque, C.; Mazel, J.; Froger, N.; Choinard, S.; Lemhemdi, A.; Macaisne, N.; van Leene, J.; Gevaert, K.; et al. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl. Acad. Sci. USA 2015, 112, 4713–4718. [Google Scholar] [CrossRef]
- Séguéla-Arnaud, M.; Choinard, S.; Larchevêque, C.; Girard, C.; Froger, N.; Crismani, W.; Mercier, R. RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 2017, 45, 1860–1871. [Google Scholar] [CrossRef]
- Fernandes, J.B.; Séguéla-Arnaud, M.; Larchevêque, C.; Lloyd, A.H.; Mercier, R. Unleashing meiotic crossovers in hybrid plants. Proc. Natl. Acad. Sci. USA 2018, 115, 2431–2436. [Google Scholar] [CrossRef]
- Mieulet, D.; Aubert, G.; Bres, C.; Klein, A.; Droc, G.; Vieille, E.; Rond-Coissieux, C.; Snachez, M.; Dalmais, M.; Mauxion, J.-P.; et al. Unleashing meiotic crossovers in crops. Nat. Plant 2018, 4, 1010–1016. [Google Scholar] [CrossRef]
- Bassi, F.M.; Kumar, A.; Zhang, Q.; Paux, E.; Huttner, E.; Kilian, A.; Dizon, R.; Feuillet, C.; Xu, S.S.; Kianian, S.F. Radiation hybrid QTL mapping of Tdes2 involved in the first meiotic division of wheat. Theor. Appl. Genet. 2013, 126, 1977–1990. [Google Scholar] [CrossRef]
- Devos, K.; Sorrells, M.; Anderson, J.; Miller, T.E.; Reader, S.M.; Lukaszewski, A.J.; Dubcovsky, J.; Sharp, P.J.; Faris, J.; Gale, M.D. Chromosome aberrations in wheat nullisomic-tetrasomic and ditelosomic lines. Cereal Res. Commun. 1999, 27, 231–239. [Google Scholar] [CrossRef][Green Version]
- Dvořák, J. Genetic variability in Aegilops speltoides affecting homoeologouis pairing in wheat. Can. J. Genet. Cytol. 1972, 14, 371–380. [Google Scholar] [CrossRef]
- Dvorak, J.; Deal, K.R.; Luo, M.-C. Discovery and Mapping of Wheat Ph1 Suppressors. Genetics 2006, 174, 17–27. [Google Scholar] [CrossRef]
- Beck, M.; Komis, G.; Muller, J.; Menzel, D.; Samaj, J. Arabidopsis Homologs of Nucleus- and Phragmoplast-Localized Kinase 2 and 3 and Mitogen-Activated Protein Kinase 4 Are Essential for Microtubule Organization. Plant Cell 2010, 22, 755–771. [Google Scholar] [CrossRef]
- Zeng, Q.; Chen, J.-G.; Ellis, B.E. AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J. 2011, 67, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Alou, A.H.; Jean, M.; Domingue, O.; Belzile, F.J. Structure and expression of AtPMS1, the Arabidopsis ortholog of the yeast DNA repair gene PMS1. Plant Sci. 2004, 167, 447–456. [Google Scholar] [CrossRef]
- Franklin, F.C.H.; Higgins, J.D.; Sanchez-Moran, E.; Armstrong, S.J.; Osman, K.E.; Jackson, N.; Jones, G.H. Control of meiotic recombination in Arabidopsis: Role of the MutL and MutS homologues. Biochem. Soc. Trans. 2006, 34, 542–544. [Google Scholar] [CrossRef]
- Rogacheva, M.V.; Manhart, C.M.; Chen, C.; Guarne, A.; Surtees, J.; Alani, E. Mlh1-Mlh3, a Meiotic Crossover and DNA Mismatch Repair Factor, Is a Msh2-Msh3-stimulated Endonuclease. J. Biol. Chem. 2014, 289, 5664–5673. [Google Scholar] [CrossRef]
- Dion, É.; Li, L.; Jean, M.; Belzile, F. An Arabidopsis MLH1 mutant exhibits reproductive defects and reveals a dual role for this gene in mitotic recombination. Plant J. 2007, 51, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Martín, A.C.; Shaw, P.; Phillips, D.; Reader, S.; Moore, G. Licensing MLH1 sites for crossover during meiosis. Nat. Commun. 2014, 5, 4580. [Google Scholar] [CrossRef]
- Sutani, T.; Yuasa, T.; Tomonaga, T.; Dohmae, N.; Takio, K.; Yanagida, M. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999, 13, 2271–2283. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, N.U.; Stronghill, P.E.; Dengler, R.E.; Hasenkampf, C.A.; Riggs, C.D. Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 2003, 130, 3283–3295. [Google Scholar] [CrossRef]
- Smith, S.J.; Osman, K.; Franklin, F.C.H. The condensin complexes play distinct roles to ensure normal chromosome morphogenesis during meiotic division in Arabidopsis. Plant J. 2014, 80, 255–268. [Google Scholar] [CrossRef]
- Shen, Y.; Tang, D.; Wang, K.; Wang, M.; Huang, J.; Luo, W.; Luo, Q.; Hong, L.; Li, M.; Cheng, Z. ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J. Cell Sci. 2012, 125, 2581–2591. [Google Scholar] [CrossRef]
- Chelysheva, L.; Gendrot, G.; Vezon, D.; Doutriaux, M.P.; Mercier, R.; Grelon, M. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 2007, 3, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Yang, X.; Ellis, J.L.; Fisher, N.M.; Makaroff, C.A. The Arabidopsis SYN3 cohesin protein is important for early meiotic events. Plant J. 2012, 71, 147–160. [Google Scholar] [CrossRef]
- Hartung, F.; Suer, S.; Bergmann, T.; Puchta, H. The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 2006, 34, 4438–4448. [Google Scholar] [CrossRef]
- Ramirez-Gonzalez, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; van Ex, F.; Pasha, A.; et al. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef]
- Sears, E.R. Nullisomic-Tetrasomic Combinations in Hexaploid Wheat. In Chromosome Manipulations and Plant Genetics; Riley, R., Lewis, K.R., Eds.; Springer US: Boston, MA, USA, 1966; pp. 29–45. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Slater, G.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef]
- Colas, I.; Macaulay, M.; Higgins, J.D.; Phillips, D.; Barakate, A.; Posch, M.; Armstrong, S.J.; Franklin, F.C.H.; Halpin, C.; Waugh, R.; et al. A spontaneous mutation in MutL-Homolog 3 (HvMLH3) affects synapsis and crossover resolution in the barley desynaptic mutant des10. New Phytol. 2016, 212, 693–707. [Google Scholar] [CrossRef]
- Darrier, B.; Arrieta, M.; Mittmann, S.U.; Sourdille, P.; Ramsay, L.; Waugh, R.; Colas, I. Following the Formation of Synaptonemal Complex Formation in Wheat and Barley by High-Resolution Microscopy. Methods Mol. Biol. 2020, 2061, 207–215. [Google Scholar] [CrossRef]
- Boden, S.A.; Shadiac, N.; Tucker, E.J.; Langridge, P.; Able, J.A. Expression and functional analysis of TaASY1 during meiosis of bread wheat (Triticum aestivum). BMC Mol. Biol. 2007, 8, 65. [Google Scholar] [CrossRef]
- Gerlach, W.L.; Bedbrook, J.R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979, 7, 1869–1885. [Google Scholar] [CrossRef]
- Aragon-Alcaide, L.; Beven, A.; Moore, G.; Shaw, P. The use of vibratome sections of cereal spikelets to study anther development and meiosis. Plant J. 1998, 14, 503–508. [Google Scholar] [CrossRef]
- Higgins, J.D.; Perry, R.M.; Barakate, A.; Ramsay, L.; Waugh, R.; Halpin, C.; Armstrong, S.J.; Franklin, F.C.H. Spatiotemporal Asymmetry of the Meiotic Program Underlies the Predominantly Distal Distribution of Meiotic Crossovers in Barley. Plant Cell 2012, 24, 4096–4109. [Google Scholar] [CrossRef] [PubMed]
- Das, R.M.; Storey, K.G. Apical Abscission Alters Cell Polarity and Dismantles the Primary Cilium During Neurogenesis. Science 2014, 343, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Vonesch, C.; Unser, M. A Fast Thresholded Landweber Algorithm for Wavelet-Regularized Multidimensional Deconvolution. IEEE Trans. Image Process. 2008, 17, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saafeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
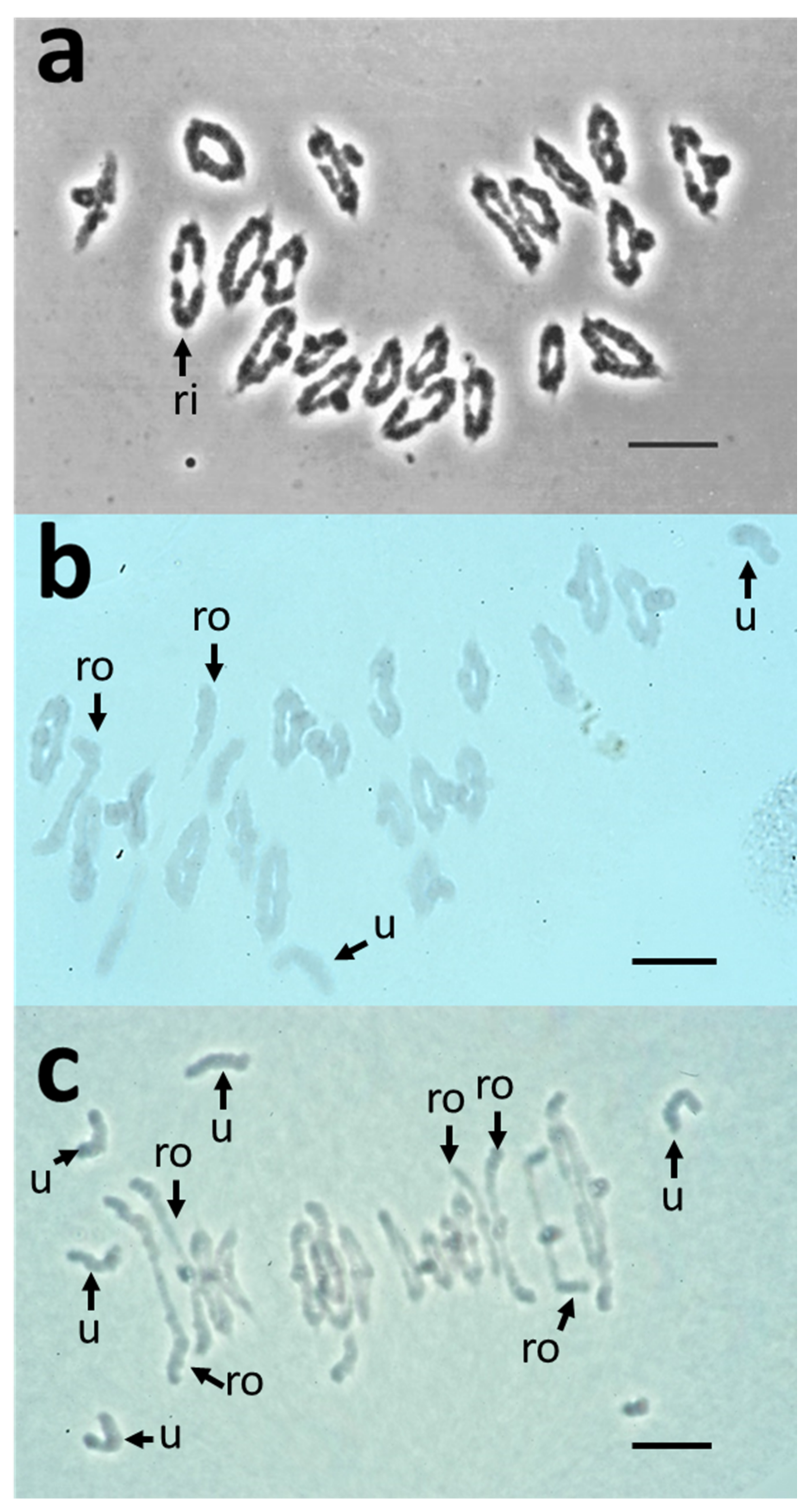

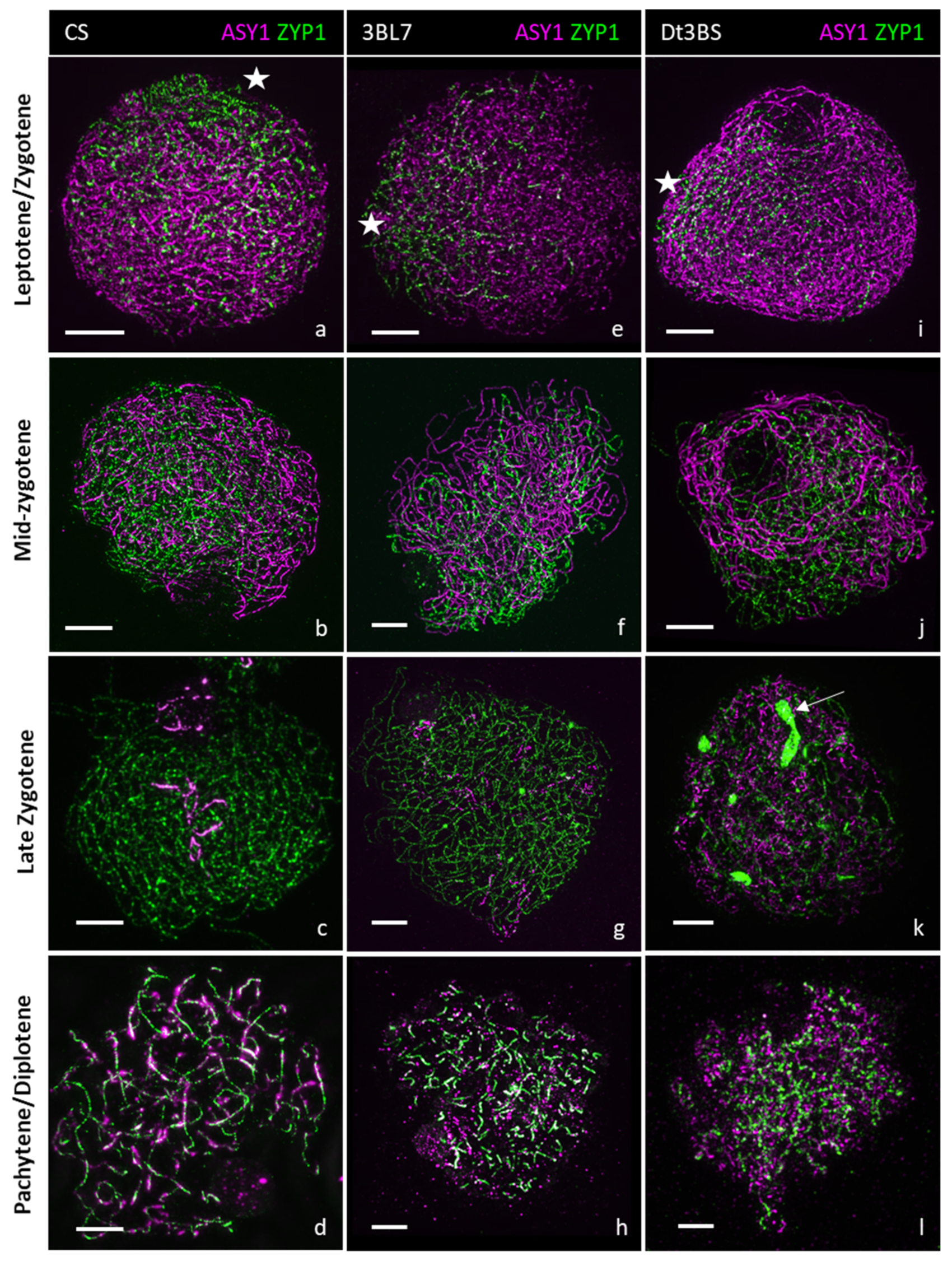

| Line | % Arm | 2n | Chiasmata | Univalents | Bivalents | Rods | Rings | Multivalents |
|---|---|---|---|---|---|---|---|---|
| CS | 100 | 42 | 40.98 ± 1.12 A,B | 0.04 ± 0.28 E | 20.98 ± 0.14 B | 1.02 ± 1.12 NA | 19.98 ± 1.12 A,B | 0 C |
| N3B | 0 | 40 | 35.22 ± 1.99 H,I,J | 0.44 ± 0.84 D,E | 19.78 ± 0.42 J,K,L | 4.34 ± 1.76 B,C | 15.44 ± 1.83 F,G,H | 0 C |
| N3AT3B | 100 | 42 | 38.44 ± 2.35 D,E,F,G | 0.04 ± 0.28 E | 20.42 ± 0.91 B,C,D,E,F | 2.34 ± 1.49 D | 18.08 ± 1.69 C,D,E | 0.28 ± 0.45 B |
| N3AT3D | 100 | 42 | 38.82 ± 2.09 C,D,E,F,G | 0.28 ± 0.70 D,E | 20.22 ± 0.95 D,E,F,G,H,I,J | 2.90 ± 1.93 C,D | 17.32 ± 1.96 D,E,F | 0.32 ± 0.47 A,B |
| N3BT3A | 0 | 42 | 36.74 ± 1.98 G,H,I,J | 0.88 ± 1.14 C,D | 19.46 ± 1.16 I,J,K,L | 4.34 ± 1.49 B,C | 15.12 ± 1.75 G,H | 0.56 ± 0.50 A |
| N3BT3D | 0 | 42 | 37.32 ± 1.91 F,G,H,I | 0.52 ± 0.89 D,E | 19.70 ± 0.91 G,H,I,J,K,L | 4.08 ± 1.91 B,C,D | 15.62 ± 2.18 F,G,H | 0.52 ± 0.50 A,B |
| N3DT3A | 100 | 42 | 39.94 ± 1.56 B,C,D | 0.08 ± 0.40 E | 20.28 ± 1.07 B,C,D,E,F,G,H | 1.92 ± 1.56 NA | 18.36 ± 1.75 C,D,E | 0.34 ± 0.52 A,B |
| N3DT3B | 100 | 42 | 39.82 ± 1.48 B,C,D | 0.08 ± 0.40 E | 20.24 ± 0.96 C,D,E,F,G,H,I | 1.98 ± 1.38 NA | 18.26 ± 1.58 C,D,E | 0.36 ± 0.48 A,B |
| Dt3BL | L | 40+2t | 39.44 ± 1.32 B,C,D | 0.08 ± 0.40 E | 20.96 ± 0.20 B | 2.48 ± 1.23 D | 18.48 ± 1.27 B,C,D,E | 0 C |
| Dt3BS | S | 40+2t | 25.84 ± 2.94 K | 6.16 ± 3.61 A | 17.92 ± 1.81 L | 10.00 ± 2.11 A | 7.92 ± 1.82 I | 0 C |
| F1CSx3BS | 100 | 41+t | 36.44 ± 1.89 G,H,I,J | 2.00 ± 1.47 A,B | 19.84 ± 0.99 F,G,H,I,J,K,L | 4.16 ± 1.77 B,C,D | 15.68 ± 1.89 F,G,H | 0.44 ± 0.51 A,B |
| F1CSx3BL | 100 | 41+t | 39.30 ± 1.09 C,D,E,F | 0.16 ± 0.55 D,E | 20.92 ± 0.27 B,C,D | 2.54 ± 1.11 D | 18.38 ± 1.07 C,D,E | 0 C |
| 3BS3 | 87 | 42 | 39.64 ± 1.32 B,C,D | 0.04 ± 0.28 E | 20.98 ± 0.14 B | 2.32 ± 1.33 D | 18.66 ± 1.32 B,C,D | 0 C |
| 3BS8 | 78 | 42 | 39.92 ± 1.63 B,C,D | 0.12 ± 0.48 E | 20.94 ± 0.24 B,C | 1.96 ± 1.58 NA | 18.98 ± 1.58 B,C,D | 0 C |
| 3BS7 | 75 | 42 | 40.40 ± 1.21 A,B,C | 0.12 ± 0.48 E | 20.94 ± 0.24 B,C | 1.48 ± 1.16 NA | 19.46 ± 1.16 A,B,C | 0 C |
| 3BS2 | 57 | 42 | 40.32 ± 1.20 A,B,C,D | 0.04 ± 0.28 E | 20.98 ± 0.14 B | 1.64 ± 1.16 NA | 19.34 ± 1.71 A,B,C | 0 C |
| 3BS4 | 55 | 42 | 39.78 ± 1.53 B,C,D | 0.04 ± 0.2 E8 | 20.98 ± 0.14 B | 2.18 ± 1.48 D | 18.80 ± 1.50 B,C,D | 0 C |
| 3BS1 | 33 | 42 | 40.20 ± 0.97 A,B,C,D | 0.04 ± 0.28 E | 20.98 ± 0.14 B | 1.76 ± 0.96 NA | 19.22 ± 0.95 A,B,C | 0 C |
| 3BS5 | 7 | 42 | 39.64 ± 1.50 B,C,D | 0.08 ± 0.57 E | 20.96 ± 0.28 B | 2.28 ± 1.34 D | 18.68 ± 1.39 B,C,D | 0 C |
| 3BL2 | 22 | 42 | 34.00 ± 2.48 I,J,K | 2.20 ± 1.91 B | 19.90 ± 0.95 F,G,H,I,J,K | 5.80 ± 1.84 A,B | 14.10 ± 1.92 H,I | 0 C |
| 3BL8 | 28 | 42 | 37.54 ± 1.84 E,F,G,H | 0.36 ± 0.78 D,E | 20.82 ± 0.39 B,C,D,E | 4.10 ± 1.69 B,C,D | 16.72 ± 1.73 E,F,G | 0 C |
| 3BL1 | 31 | 42 | 36.80 ± 2.16 G,H,I,J | 1.22 ± 1.11 B,C | 20.20 ± 0.83 E,F,G,H,I,J | 4.52 ± 1.95 B,C | 15.68 ± 2.03 F,G,H | 0.46 ± 0.50 A,B |
| 3BL9 | 38 | 41 | 33.10 ± 2.38 J,K | 2.18 ± 1.51 A,B | 19.38 ± 0.83 K,L | 5.70 ± 1.82 A,B | 13.68 ± 2.00 H,I | 0.02 ± 0.14 C |
| 3BL10 | 50 | 42 | 35.90 ± 2.50 H,I,J | 1.04 ± 1.47 C,D | 20.44 ± 0.76 B,C,D,E,F,G | 5.04 ± 2.02 B | 15.40 ± 2.15 F,G,H | 0.02 ± 0.14 C |
| 3BL7 | 63 | 41 | 33.46 ± 2.57 J,K | 2.12 ± 1.22 A,B | 19.44 ± 0.61 K,L | 5.42 ± 2.13 A,B | 14.02 ± 2.28 G,H,I | 0 C |
| Arm | Deletion Bin | Estimated Size (Mb) | Number of Genes Mapped in Bins | Number of Genes Expressed | Percentage of Genes Expressed | Number of Genes on 3B Overexpressed |
|---|---|---|---|---|---|---|
| Short | 3BS3-0.87-1.00 | 4.46 | 93 | 28 | 30.11 | 7 |
| 3BS8-0.78-0.87 | 31.38 | 519 | 100 | 19.27 | 23 | |
| 3BS7-0.75-0.78 | 5.93 | 58 | 18 | 31.03 | 3 | |
| 3BS2-0.57-0.75 | 75.56 | 644 | 189 | 29.35 | 38 | |
| 3BS4-0.55-0.57 | 20.31 | 117 | 47 | 40.17 | 8 | |
| 3BS1-0.33-0.55 | 105.52 | 608 | 241 | 39.64 | 43 | |
| 3BS5-0.07-0.33 | 53.08 | 149 | 85 | 57.05 | 12 | |
| C-3BS5-0.07 | 48.04 | 67 | 38 | 56.72 | 11 | |
| Long | C-3BL2-0.22 | 90.60 | 416 | 223 | 53.61 | 40 |
| 3BL2-0.22-0.28 | 28.46 | 189 | 67 | 35.45 | 19 | |
| 3BL8-0.28-0.32 | 17.85 | 106 | 48 | 45.28 | 11 | |
| 3BL1-0.32-0.38 | 43.64 | 260 | 124 | 47.69 | 16 | |
| 3BL9-0.38-0.50 | 44.96 | 359 | 164 | 45.68 | 25 | |
| 3BL10-0.50-0.63 | 48.48 | 314 | 144 | 45.86 | 26 | |
| 3BL7-0.63-1.00 | 204.93 | 2161 | 583 | 26.98 | 115 | |
| Total Short Assigned | 344.28 | 2255 | 746 | 33.08 | 145 | |
| Total Long Assigned | 478.92 | 3805 | 1353 | 35.56 | 252 | |
| Total Assigned | 823.20 | 6060 | 2099 | 34.64 | 397 |
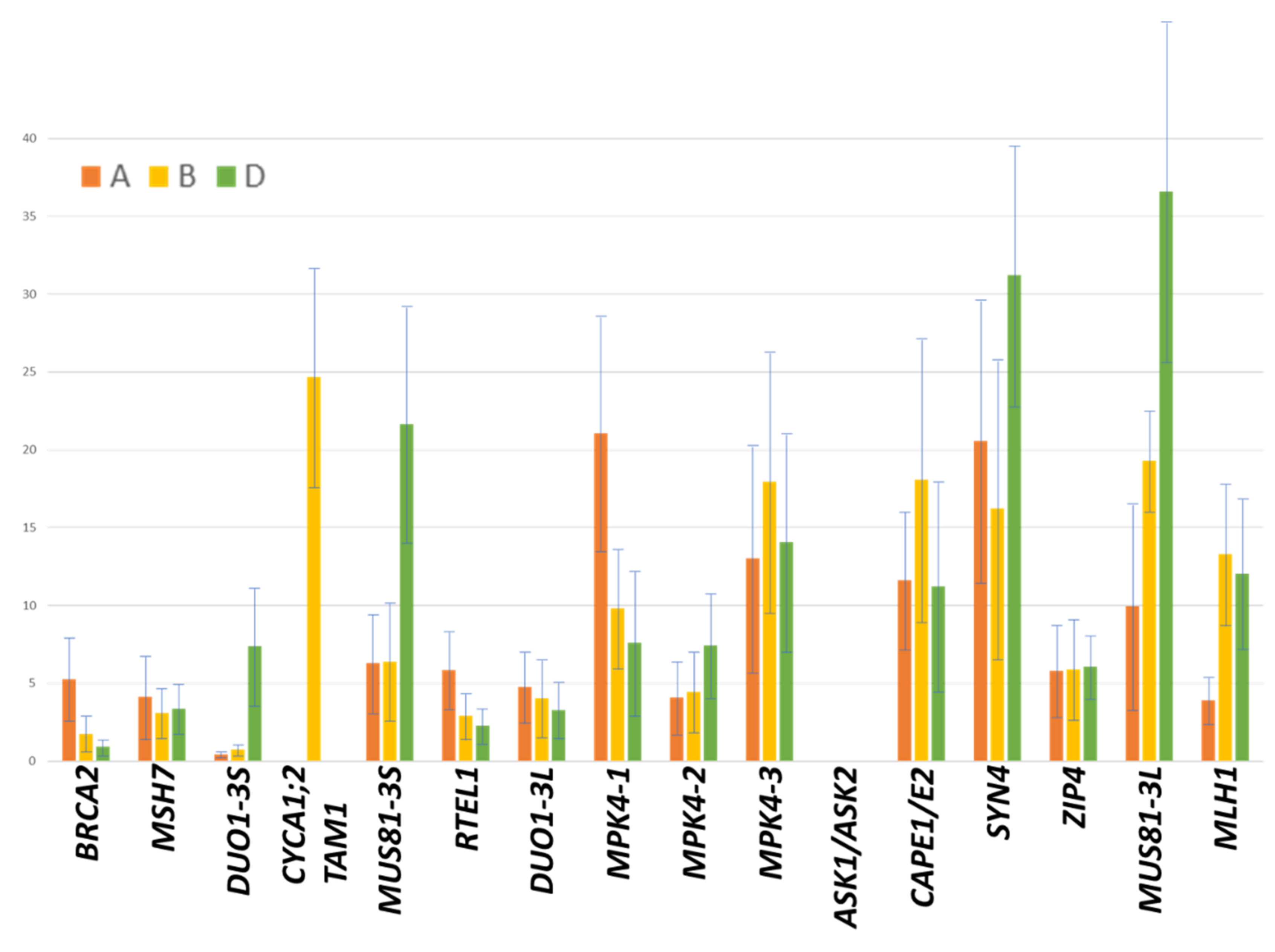
| Related | Deletion Bin | Gene ID | FPKM3A | FPKM3B | FPKM3D |
|---|---|---|---|---|---|
| BRCA2 | 3BS2-0.57-0.75 | TraesCS3B02G115500 | 5.26 ± 2.72 | 1.75 ± 0.99 | 0.93 ± 0.51 |
| MSH7 | 3BS4-0.55-0.57 | TraesCS3B02G136600 | 4.13 ± 2.54 | 3.08 ± 1.76 | 3.37 ± 1.61 |
| DUO1-3S | 3BS1-0.33-0.55 | TraesCS3B02G178200 | 0.45 ± 0.19 | 0.74 ± 0.39 | 7.37 ± 4.01 |
| CYCA1;2/TAM1 | 3BS1-0.33-0.55 | TraesCS3B02G183400 | 24.66 ± 7.16 | ||
| MUS81-3S | 3BS5-0.07-0.33 | TraesCS3B02G218300 | 6.29 ± 3.17 | 6.41 ± 3.93 | 21.67 ± 7.85 |
| RTEL1 | C-3BL2-0.22 | TraesCS3B02G242700 | 5.87 ± 2.53 | 2.92 ± 1.55 | 2.29 ± 1.26 |
| DUO1-3L | C-3BL2-0.22 | TraesCS3B02G254800 | 4.77 ± 2.45 | 4.07 ± 2.52 | 3.30 ± 1.77 |
| MPK4-1 | C-3BL2-0.22 | TraesCS3B02G256700 | 21.08 ± 7.62 | 9.83 ± 3.93 | 7.61 ± 4.64 |
| MPK4-2 | C-3BL2-0.22 | TraesCS3B02G260900 | 4.07 ± 2.39 | 4.47 ± 2.64 | 7.44 ± 3.32 |
| MPK4-3 | C-3BL2-0.22 | TraesCS3B02G270200 | 13.01 ± 7.36 | 17.93 ± 8.58 | 14.06 ± 7.12 |
| ASK1/ASK2 | 3BL1-0.32-0.38 | TraesCS3B02G308600 | |||
| CAP-E1/E2 | 3BL7-0.63-1.00 | TraesCS3B02G423800 | 11.63 ± 4.45 | 18.08 ± 9.11 | 11.23 ± 6.71 |
| SYN4 | 3BL7-0.63-1.00 | TraesCS3B02G429700 | 20.58 ± 9.22 | 16.23 ± 9.71 | 31.22 ± 8.59 |
| ZIP4 | 3BL7-0.63-1.00 | TraesCS3B02G434600 | 5.79 ± 2.94 | 5.91 ± 3.28 | 6.07 ± 2.13 |
| MUS81-3L | 3BL7-0.63-1.00 | TraesCS3B02G535000 | 9.96 ± 6.53 | 19.29 ± 3.30 | 36.59 ± 11.18 |
| MLH1* | 3BL7-0.63-1.00 | TraesCS3B02G564100 | 3.90 ± 1.89 | 13.29 ± 4.19 | 12.06 ± 4.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darrier, B.; Colas, I.; Rimbert, H.; Choulet, F.; Bazile, J.; Sortais, A.; Jenczewski, E.; Sourdille, P. Location and Identification on Chromosome 3B of Bread Wheat of Genes Affecting Chiasma Number. Plants 2022, 11, 2281. https://doi.org/10.3390/plants11172281
Darrier B, Colas I, Rimbert H, Choulet F, Bazile J, Sortais A, Jenczewski E, Sourdille P. Location and Identification on Chromosome 3B of Bread Wheat of Genes Affecting Chiasma Number. Plants. 2022; 11(17):2281. https://doi.org/10.3390/plants11172281
Chicago/Turabian StyleDarrier, Benoit, Isabelle Colas, Hélène Rimbert, Frédéric Choulet, Jeanne Bazile, Aurélien Sortais, Eric Jenczewski, and Pierre Sourdille. 2022. "Location and Identification on Chromosome 3B of Bread Wheat of Genes Affecting Chiasma Number" Plants 11, no. 17: 2281. https://doi.org/10.3390/plants11172281
APA StyleDarrier, B., Colas, I., Rimbert, H., Choulet, F., Bazile, J., Sortais, A., Jenczewski, E., & Sourdille, P. (2022). Location and Identification on Chromosome 3B of Bread Wheat of Genes Affecting Chiasma Number. Plants, 11(17), 2281. https://doi.org/10.3390/plants11172281








