Integration of Repeatomic and Cytogenetic Data on Satellite DNA for the Genome Analysis in the Genus Salvia (Lamiaceae)
Abstract
:1. Introduction
2. Results
2.1. Comparative Analyses of the Repetitive DNA Sequences
2.2. BLAST Results
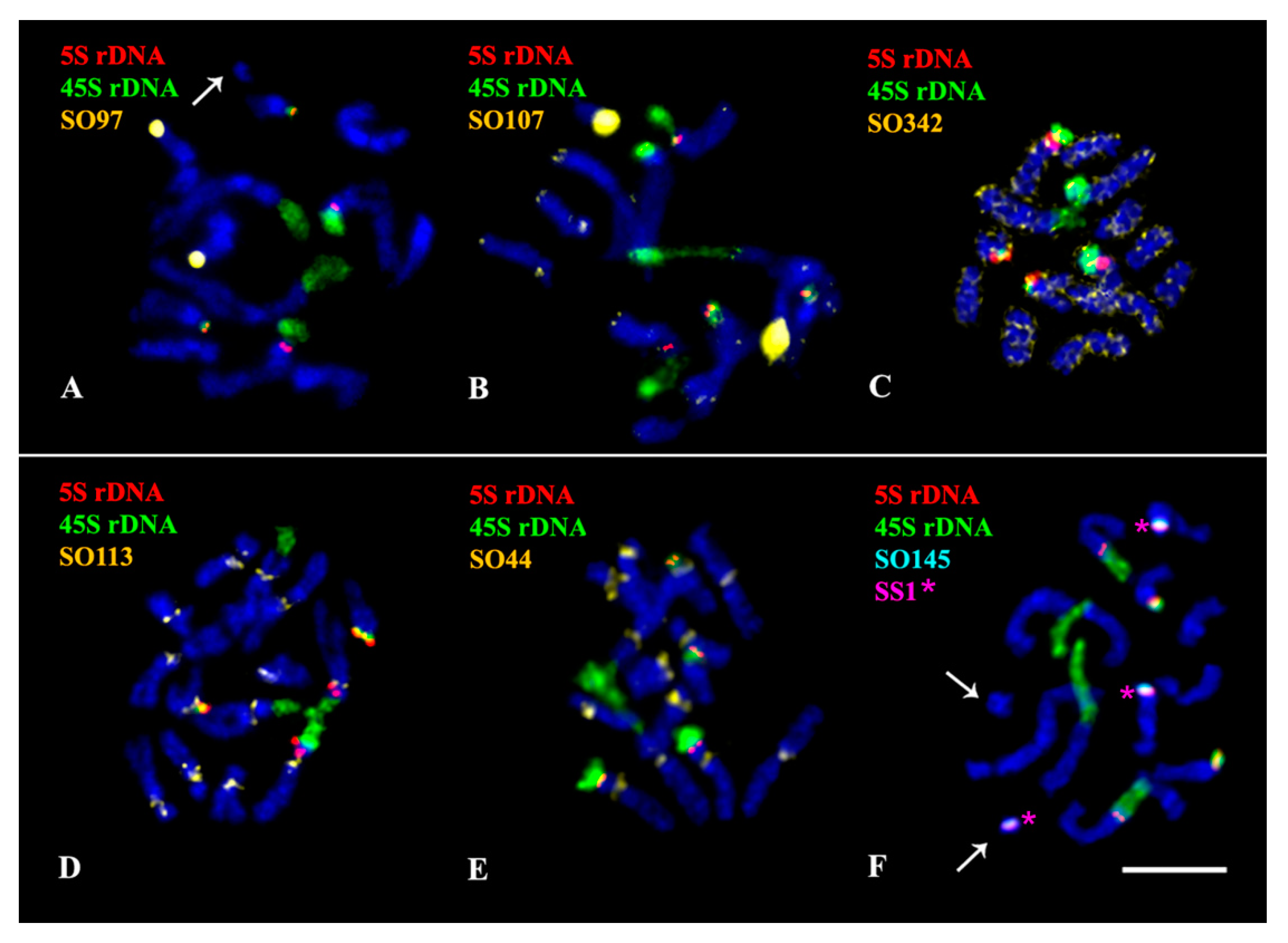
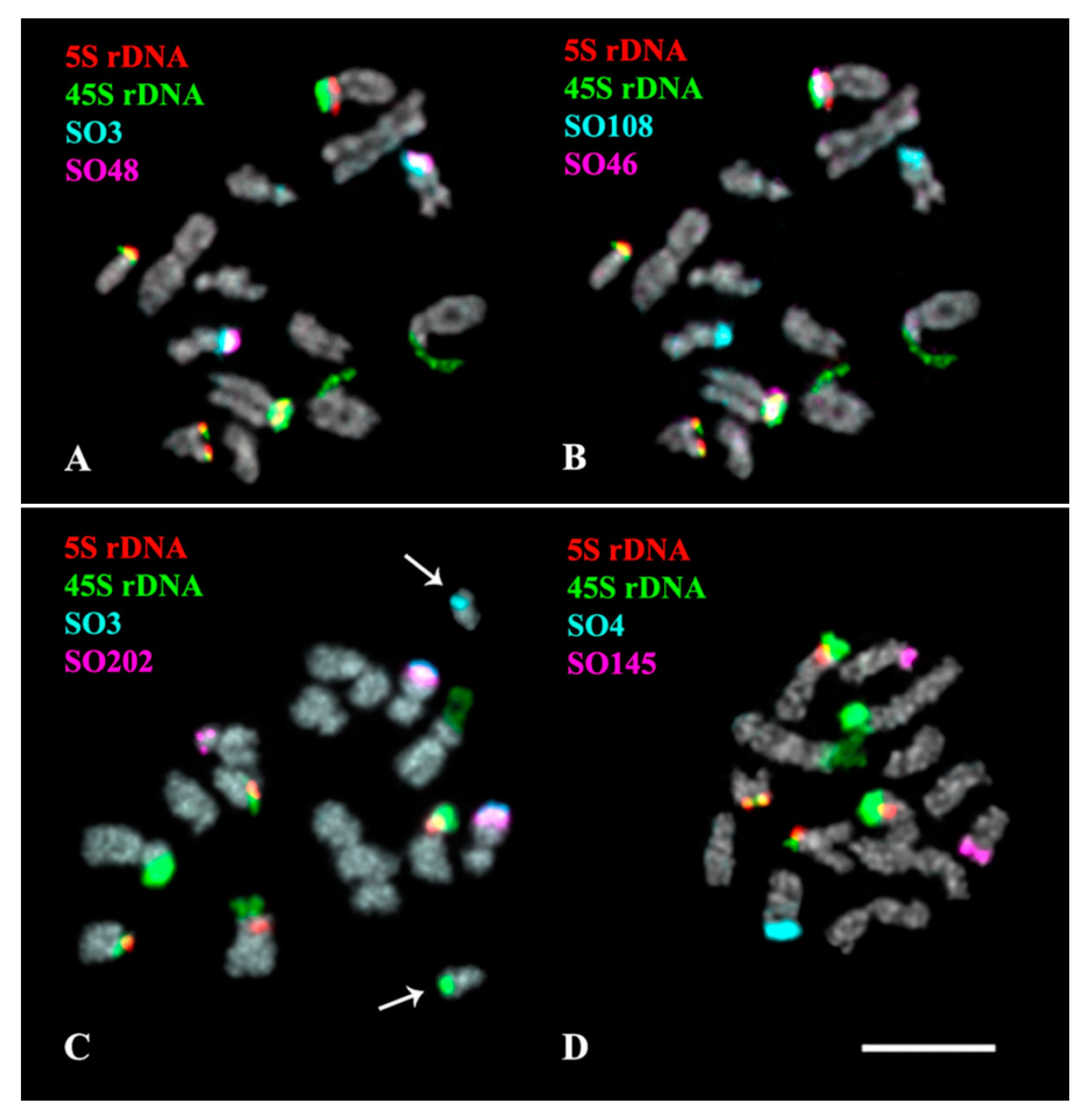
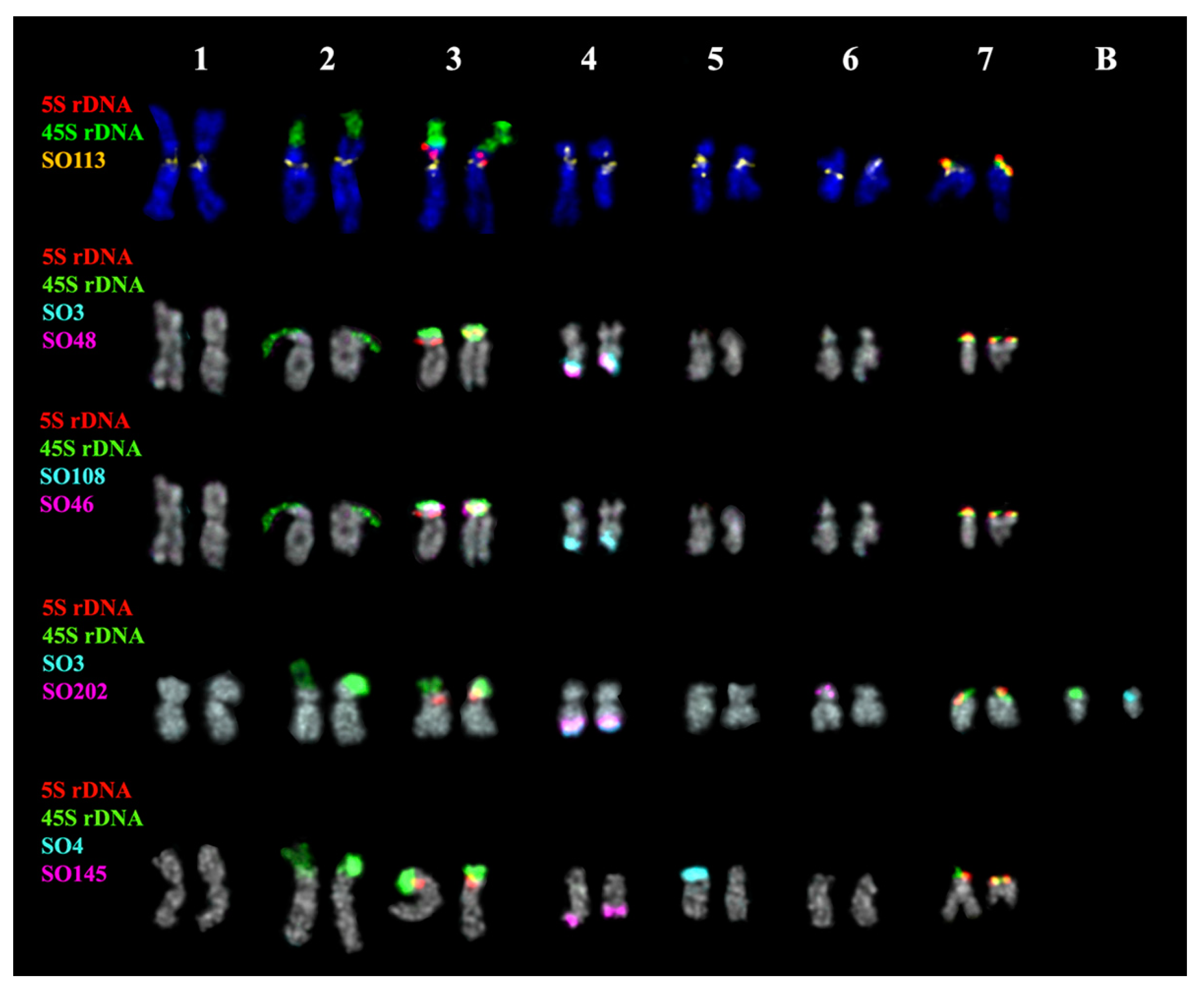
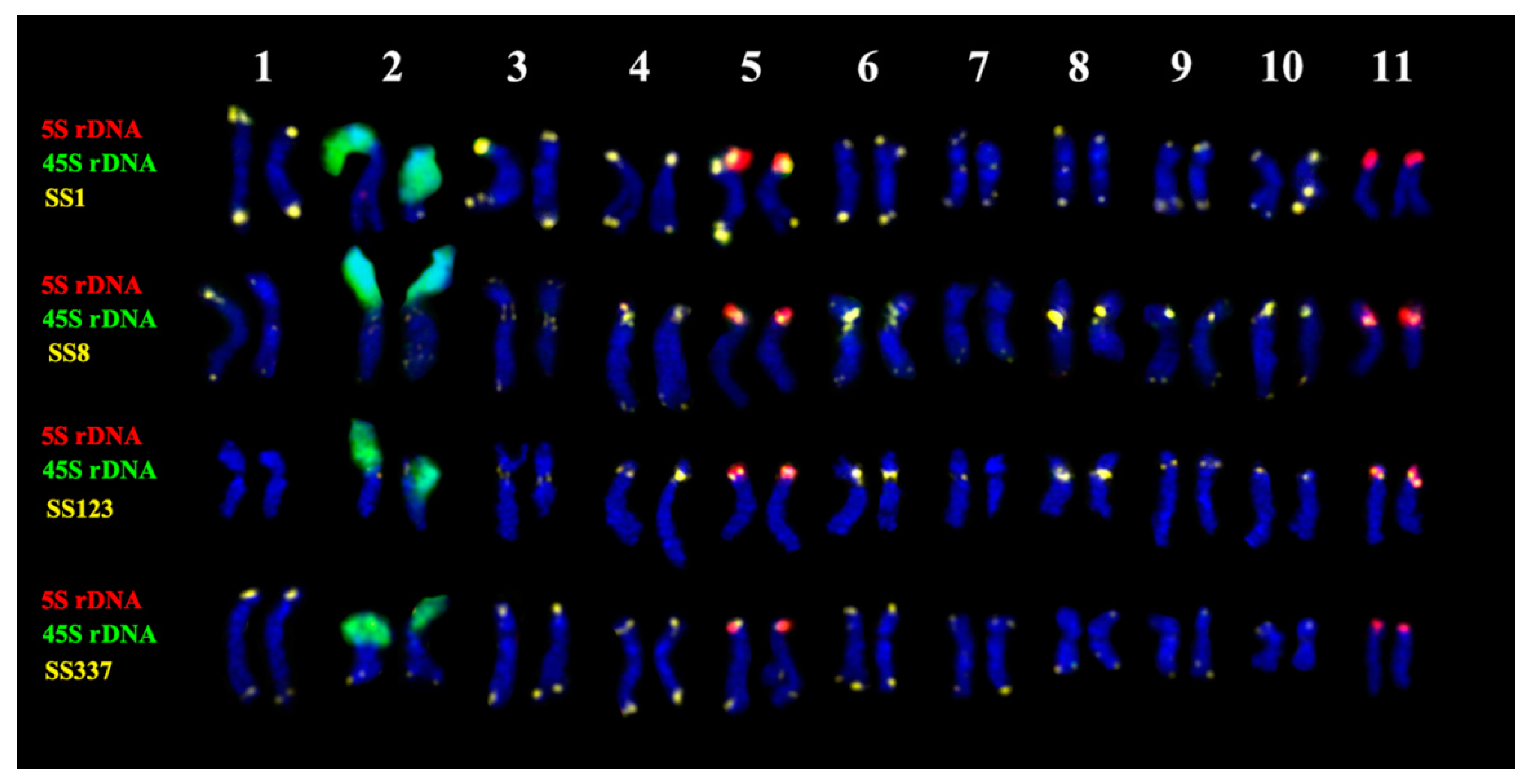
2.3. Chromosomal Structural Variations
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Genomic DNA Extraction and Sequencing
4.3. Sequence Analysis and Identification of DNA Repeats
4.4. Chromosome Spread Preparation
4.5. Fluorescence In Situ Hybridization
4.6. Chromosome Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pobedimova, E.G. Salvia L. In Flora of the USSR; Schischkin, B.K., Ed.; Akad. Scient. URSS: Moskva, Russia, 1954; Volume 21, pp. 244–363. [Google Scholar]
- Kintzios, S.E. Sage: The Genus Salvia, 1st ed.; CRC Press: London, UK, 2000; p. 20. [Google Scholar]
- Kriebel, R.; Drew, B.T.; Drummond, C.P.; González-Gallegos, J.G.; Celep, F.; Mahdjoub, M.M.; Rose, J.P.; Xiang, C.L.; Hu, G.X.; Walker, J.B.; et al. Tracking temporal shifts in area, biomes, and pollinators in the radiation of Salvia (sages) across continents: Leveraging anchored hybrid enrichment and targeted sequence data. Am. J. Bot. 2019, 106, 573–597. [Google Scholar] [PubMed]
- Himmelbaur, W.; Stibal, E. Entwicklungsrichtungen in der blutenregion der gattung Salvia L. I–III. Biol. Gen. 1933, 8, 449–474. [Google Scholar]
- Claßen-Bockhoff, R.; Wester, P.; Tweraser, E. The staminal lever mechanism in Salvia L. (Lamiaceae)—A review. Plant Biol. 2003, 5, 33–41. [Google Scholar]
- Walker, J.B.; Sytsma, K.J.; Treutlein, J.; Wink, M. Salvia (Lamiaceae) is not monophyletic: Implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am. J. Bot. 2004, 91, 1115–1125. [Google Scholar] [PubMed]
- Will, M.; Claßen-Bockhoff, R. Time to split Salvia sl (Lamiaceae)–new insights from Old World Salvia phylogeny. Mol. Phylogenetics Evol. 2017, 109, 33–58. [Google Scholar]
- Walker, J.B.; Sytsma, K.J. Staminal evolution in the genus Salvia (Lamiaceae): Molecular phylogenetic evidence for multiple origins of the staminal lever. Ann. Bot. 2007, 100, 375–391. [Google Scholar]
- Drew, B.T.; González-Gallegos, J.G.; Xiang, C.L.; Walked, J.B.; Sytsma, K.J. Salvia united: The greatest good for the greatest number. Taxon 2017, 66, 133–145. [Google Scholar]
- Hu, G.X.; Takano, A.; Drew, B.T.; Liu, E.D.; Soltis, D.E.; Soltis, P.S.; Peng, H.; Xiang, C.L. Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Ann. Bot. 2018, 122, 649–668. [Google Scholar]
- Wu, H.; Ma, P.F.; Li, H.T.; Hu, G.X.; Li, D.Z. Comparative plastomic analysis and insights into the phylogeny of Salvia (Lamiaceae). Plant Divers. 2021, 43, 15–26. [Google Scholar]
- Kriebel, R.; Drew, B.T.; González-Gallegos, J.G.; Celep, F.; Heeg, L.; Mahdjoub, M.M.; Sytsma, K.J. Pollinator shifts, contingent evolution, and evolutionary constraint drive floral disparity in Salvia (Lamiaceae): Evidence from morphometrics and phylogenetic comparative methods. Evolution 2020, 74, 1335–1355. [Google Scholar]
- Rose, J.P.; Kriebel, R.; Kahan, L.; DiNicola, A.; González-Gallegos, J.G.; Celep, F.; Lemmon, E.M.; Lemmon, A.R.; Sytsma, K.J.; Drew, B.T. Sage insights into the phylogeny of Salvia: Dealing with sources of discordance within and across genomes. Front Plant Sci. 2021, 12, 767478. [Google Scholar] [PubMed]
- The Chromosome Counts Database (CCDB). Available online: http://ccdb.tau.ac.il/Angiosperms/Lamiaceae/Salvia (accessed on 25 July 2022).
- Delestaing, N. Contribution a!’etude cytologique du genre Salvia. Rev. Cytol. Bioi. Veg. 1954, 15, 195–236. [Google Scholar]
- Ranjbar, M.; Pakatchi, A.; Babataheri, Z.J. Chromosome number evolution, biogeography and phylogenetic relationships in Salvia (Lamiaceae). Webbia J. Plant Taxon Geogr. 2015, 70, 293–312. [Google Scholar]
- Haque, M.S.; Ghoshal, K. Karyotypes and chromosome morphology in the genus Salvia Linn. Cytologia 1980, 45, 627–640. [Google Scholar]
- Haque, M.S. Chromosome numbers in the genus Salvia LIN. Proc. Indian Natl. Sci. Acad. B. 1981, 47, 419–426. [Google Scholar]
- Sudarmono; Okada, H. Genetic Differentiations among the Populations of Salvia japonica (Lamiaceae) and Its Related Species. HAYATI J. Biosci. 2008, 15, 18–26. [Google Scholar]
- Eroğlu, H.E.; Martin, E.; Kahraman, A.; Aslan, E.G. The new chromosomal data and karyotypic variations in genus Salvia L. (Lamiaceae): Dysploidy, polyploidy and symmetrical karyotypes. Caryologia 2021, 74, 21–28. [Google Scholar]
- Alberto, C.M.; Sanso, A.M.; Xifreda, C.C. Chromosomal studies in species of Salvia (Lamiaceae) from Argentina. Bot. J. Linn. Soc. 2003, 141, 483–490. [Google Scholar]
- Masoud, S.; Alijanpoo, B.; Khayyami, M. Karyotype analysis in some Salvia species (Lamiaceae) of Iran. Caryologia 2010, 63, 405–410. [Google Scholar]
- Martin, E.; Celep, F.; Eroğlu, H.E. Comparative chromosomal features and new karyological data in Salvia: B-chromosomes, polyploidy, dysploidy and symmetric karyotypes. Braz. J. Bot. 2022, 45, 625–634. [Google Scholar]
- Tropicos.org. Missouri Botanical Garden. IPCN Chromosome Reports. Available online: http://legacy.tropicos.org/NameSearch.aspx?projectid=9 (accessed on 25 July 2022).
- Özdemir, C.; Şenel, G. The morphological, anatomical and karyological properties of Salvia sclarea L. Turk J Bot. 1999, 23, 7–18. [Google Scholar]
- Martin, E.; Ozlem, C.; Kahraman, A.; Celep, F.; Dogan, M. A cytomorphological study in some taxa of the genus Salvia L. (Lamiaceae). Caryologia 2011, 64, 272–287. [Google Scholar]
- Hu, G.X.; Xiang, C.L.; Liu, E.D.; Dong, H.J.; Funamoto, T. Karyotypic study of eighteen taxa of Salvia (Lamiaceae) from China. Caryologia 2016, 69, 50–57. [Google Scholar]
- Song, Z.; Lin, C.; Xing, P.; Fen, Y.; Jin, H.; Zhou, C.; Gu, Y.Q.; Wang, J.; Li, X. A high-quality reference genome sequence of Salvia miltiorrhiza provides insights into tanshinone synthesis in its red rhizomes. Plant Genome 2020, 13, e20041. [Google Scholar]
- Liu, Q.; Li, X.; Zhou, X.; Li, M.; Zhang, F.; Schwarzacher, T.; Heslop-Harrison, J.S. The repetitive DNA landscape in Avena (Poaceae): Chromosome and genome evolution defined by major repeat classes in whole-genome sequence reads. BMC Plant Biol. 2019, 19, 226. [Google Scholar]
- Zagorski, D.; Hartmann, M.; Bertrand, Y.J.; Paštová, L.; Slavíková, R.; Josefiová, J.; Fehrer, J. Characterization and dynamics of repeatomes in closely related species of Hieracium (Asteraceae) and their synthetic and apomictic hybrids. Front Plant Sci. 2020, 11, 591053. [Google Scholar]
- Dogan, M.; Pouch, M.; Mandáková, T.; Hloušková, P.; Guo, X.; Winter, P.; Chumová, Z.; Van Niekerk, A.; Mummenhoff, K.; Al-Shehbaz, I.; et al. Evolution of tandem repeats is mirroring post-polyploid cladogenesis in Heliophila (Brassicaceae). Front Plant Sci. 2021, 11, 607893. [Google Scholar]
- Yurkevich, O.Y.; Samatadze, T.E.; Selyutina, I.Y.; Suprun, N.A.; Suslina, S.N.; Zoshchuk, S.A.; Amosova, A.V.; Muravenko, O.V. Integration of genomic and cytogenetic data on tandem DNAs for analyzing the genome diversity within the genus Hedysarum L. (Fabaceae). Front Plant Sci. 2022, 29, 865958. [Google Scholar]
- Božin, B.; Lakić, N.; Čonić, B.S.; Kladar, N.; Orčić, D.; Mimica-Dukić, N. Antioxidant and antimicrobial properties of a new chemotype of woodland sage (Salvia nemorosa L. subsp. nemorosa, Lamiaceae) essential oil. Biol. Serbica 2012, 34, 51–60. [Google Scholar]
- Bochkarev, N.I.; Zelentsov, S.V.; Shuvaeva, T.P.; Borodkina, A.P. State of taxonomy, morphology and breeding of clary sage (review). Oil Crops 2014, 1, 165–177. [Google Scholar]
- Badiee, P.; Nasirzadeh, A.R.; Motaffaf, M. Comparison of Salvia officinalis L. essential oil and antifungal agents against candida species. J. Pharm. Technol. Drug Res. 2012, 1, 7. [Google Scholar] [CrossRef]
- Wong, J.; Chiang, Y.F.; Shih, Y.H.; Chiu, C.H.; Chen, H.Y.; Shieh, T.M.; Wang, K.L.; Huang, T.C.; Hong, Y.H.; Hsia, S.M. Salvia sclarea l. Essential oil extract and its antioxidative phytochemical sclareol inhibit oxytocin-induced uterine hypercontraction dysmenorrhea model by inhibiting the ca2+–mlck–mlc20 signaling cascade: An ex vivo and in vivo study. Antioxidants 2020, 9, 991. [Google Scholar] [CrossRef] [PubMed]
- Lopresti, A.L. Salvia (Sage): A review of its potential cognitive-enhancing and protective effects. Drugs RD 2017, 17, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Jug-Dujaković, M.; Ristić, M.; Pljevljakušić, D.; Dajić-Stevanović, Z.; Liber, Z.; Hančević, K.; Radić, T.; Šatović, Z. High diversity of indigenous populations of dalmatian sage (Salvia officinalis L.) in essential-oil composition. Chem. Biodivers. 2012, 9, 2309–2323. [Google Scholar] [CrossRef]
- Afonso, A.F.; Pereira, O.R.; Fernandes, Â.; Calhelha, R.C.; Silva, A.M.; Ferreira, I.C.; Cardoso, S.M. Phytochemical composition and bioactive effects of Salvia africana, Salvia officinalis ‘Icterina’and Salvia mexicana aqueous extracts. Molecules 2019, 24, 4327. [Google Scholar] [CrossRef]
- Hamidpour, M.; Hamidpour, R.; Hamidpour, S.; Shahlari, M. Chemistry, pharmacology, and medicinal property of sage (Salvia) to prevent and cure illnesses such as obesity, diabetes, depression, dementia, lupus, autism, heart disease, and cancer. J. Tradit. Complementary Med. 2014, 4, 82–88. [Google Scholar] [CrossRef]
- Aćimović, M.G.; Cvetković, M.T.; Stanković Jeremić, J.M.; Pezo, L.L.; Varga, A.O.; Čabarkapa, I.S.; Kiprovski, B. Biological activity and profiling of Salvia sclarea essential oil obtained by steam and hydrodistillation extraction methods via chemometrics tools. Flavour Fragr. J. 2022, 37, 20–32. [Google Scholar] [CrossRef]
- Olarte, A.; Mantri, N.; Nugent, G.; Wohlmuth, H.; Li, C.G.; Xue, C.; Pang, E. A gDNA microarray for genotyping Salvia species. Mol. Biotechnol. 2013, 54, 770–783. [Google Scholar] [CrossRef]
- Martin, E.; Altınordu, F.; Celep, F.; Kahraman, A.; Doğan, M. Karyomorphological studies in seven taxa of the genus Salvia (Lamiaceae) in Turkey. Caryologia 2015, 68, 13–18. [Google Scholar] [CrossRef]
- Tarinejad, A.R.; Mirshekari, B. Study on Variation of Karyotypes between and within Species of Salvia officinalis L., Stachys byzantine L. and Dracocephalum molarica L. Acta Hortic. 2010, 853, 39–46. [Google Scholar] [CrossRef]
- Hao, D.C.; Xiao, P.G. Genomics and evolution in traditional medicinal plants: Road to a healthier life. Evol. Bioinform. 2015, 11, EBO-S31326. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Osbourn, A.; Kontogianni, V.G.; Liu, L.W.; Jordán, M.J. Temporal transcriptome changes induced by methyl jasmonate in Salvia sclarea. Gene 2015, 558, 41–53. [Google Scholar]
- Vautrin, S.; Song, C.; Zhu, Y.J.; Berges, H.; Sun, C.; Chen, S.L. The first insight into the Salvia (Lamiaceae) genome via BAC library construction and high-throughput sequencing of target BAC clones. Pak. J. Bot. 2015, 47, 1347–1357. [Google Scholar]
- Radosavljević, I.; Jakse, J.; Javornik, B.; Satovic, Z.; Liber, Z. New microsatellite markers for Salvia officinalis (Lamiaceae) and cross-amplification in closely related species. Am J Bot. 2011, 98, e316–e318. [Google Scholar] [CrossRef] [PubMed]
- McCann, J.; Macas, J.; Novák, P.; Stuessy, T.F.; Villasenor, J.L.; Weiss-Schneweiss, H. Differential genome size and repetitive DNA evolution in diploid species of Melampodium sect Melampodium (Asteraceae). Front. Plant Sci. 2020, 11, 362. [Google Scholar] [CrossRef] [PubMed]
- Zwyrtková, J.; Němečková, A.; Čížková, J.; Holušová, K.; Kapustová, V.; Svačina, R.; Kopecký, D.; Till, B.; Doležel, J.; Hřibová, E. Comparative analyses of DNA repeats and identification of a novel Fesreba centromeric element in fescues and ryegrasses. BMC Plant Biol. 2020, 20, 280. [Google Scholar] [CrossRef] [PubMed]
- Waring, M.; Britten, R.J. Nucleotide sequence repetition: A rapidly reassociating fraction of mouse DNA. Science 1966, 154, 791–794. [Google Scholar] [CrossRef]
- Britten, R.J.; Kohne, D.E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 1968, 161, 529–540. [Google Scholar] [CrossRef]
- SanMiguel, P.; Bennetzen, J.L. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotranposons. Ann. Bot. 1998, 82, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Bennetzen, J.L.; Wang, H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef]
- Meštrovic, N.; Mravinac, B.; Pavlek, M.; Vojvoda-Zeljko, T.; Šatovi’c, E.; Plohl, M. Structural and functional liaisons between transposable elements and satellite DNAs. Chromosome Res. 2015, 23, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, D.J. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989, 5, 103–107. [Google Scholar] [CrossRef]
- Makałowski, W.; Gotea, V.; Pande, A.; Makałowska, I. Transposable elements: Classification, identification, and their use as a tool for comparative genomics. In Evolutionary Genomics. Methods in Molecular Biology; Anisimova, M., Ed.; Humana: New York, NY, USA, 2019; Volume 1910, pp. 170–270. [Google Scholar]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Neumann, P.; Novák, P.; Hoštáková, N.; Macas, J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA 2019, 10, 1. [Google Scholar] [CrossRef]
- Vitte, C.; Panaud, O. LTR retrotransposons and flowering plant genome size: Emergence of the increase/decrease model. Cytogenet. Genome Res. 2005, 110, 91–107. [Google Scholar] [CrossRef]
- Baucom, R.; Estill, J.; Chaparro, C.; Upshaw, N.; Jogi, A.; Deragon, J.-M.; Westerman, R.P.; SanMiguel, P.J.; Bennetzen, J.L. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 2009, 5, e1000732. [Google Scholar] [CrossRef]
- Macas, J.; Novák, P.; Pellicer, J.; ˇCížková, J.; Koblížková, A.; Neumann, P.; Fuková, I.; Doležel, J.; Kelly, L.J.; Leitch, I.J. In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS ONE 2015, 10, e0143424. [Google Scholar]
- Zhang, Q.-J.; Gao, L.-I. Rapid and recent evolution of LTR retrotransposons drives rice genome evolution during the speciation of AA-genome Oryza species. G3 2017, 7, 1875–1885. [Google Scholar] [CrossRef]
- Wang, D.; Zheng, Z.; Li, Y.; Hu, H.; Wang, Z.; Du, X. Which factors contribute most to genome size variation within angiosperms? Ecol. Evol. 2021, 11, 2660–2668. [Google Scholar] [CrossRef]
- Vitte, C.; Bennetzen, J.L. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA 2006, 103, 17638–17643. [Google Scholar] [CrossRef] [Green Version]
- Becher, H.; Powell, R.F.; Brown, M.R.; Metherell, C.; Pellicer, J.; Leitch, I.J.; Twyford, A.D. The nature of intraspecific and interspecific genome size variation in taxonomically complex eyebrights. Ann. Bot. 2021, 128, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Maksimović, M.; Vidic, D.; Miloš, M.; Šolić, M.E.; Abadžić, S.; Siljak-Yakovlev, S. Effect of the environmental conditions on essential oil profile in two Dinaric Salvia species: S. brachyodon Vandas and S. officinalis L. Biochem. Syst. Ecol. 2007, 35, 473–478. [Google Scholar] [CrossRef]
- Siljak-Yakovlev, S.; Pustahija, F.; Šolic, E.M.; Bogunić, F.; Muratović, E.; Bašić, N.; Catrice, O.; Brown, S.C. Towards a genome size and chromosome number database of Balkan flora: C-values in 343 taxa with novel values for 242. Adv. Sci. Lett. 2010, 3, 190–213. [Google Scholar] [CrossRef]
- Maynard, R.C.I.; Ruter, J.M. DNA Content estimation in the genus Salvia. J. Amer. Soc. Hort. Sci. 2022, 147, 123–134. [Google Scholar] [CrossRef]
- Jia, K.H.; Liu, H.; Zhang, R.G.; Xu, J.; Zhou, S.S.; Jiao, S.Q.; Yan, X.M.; Tian, X.C.; Shi, T.L.; Luo, H.; et al. Chromosome-scale assembly and evolution of the tetraploid Salvia splendens (Lamiaceae) genome. Hortic. Res. 2021, 8, 177. [Google Scholar] [CrossRef]
- Stapley, J.; Santure, A.W.; Dennis, S.R. Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol. Ecol. 2015, 24, 2241–2252. [Google Scholar] [CrossRef]
- Mhiri, C.; Borges, F.; Grandbastien, M.A. Specificities and dynamics of transposable elements in land plants. Biology 2022, 11, 488. [Google Scholar] [CrossRef]
- McStay, B. Nucleolar organizer regions: Genomic ‘dark matter’ requiring illumination. Genes Dev. 2016, 30, 1598–1610. [Google Scholar] [CrossRef]
- Mehrotra, S.; Goyal, V. Repetitive sequences in plant nuclear DNA: Types, distribution, evolution and function. Genom. Proteom. Bioinform. 2014, 12, 164–171. [Google Scholar] [CrossRef]
- Kubis, S.; Schmidt, T.; Heslop-Harrison, J.S. Repetitive DNA elements as a major component of plant genomes. Ann. Bot. 1998, 82, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Hemleben, V.; Kovaˇrík, A.; Torres-Ruiz, R.A.; Volkov, R.A.; Beridze, T. Plant highly repeated satellite DNA: Molecular evolution, distribution and use for identification of hybrids. Syst. Biodivers. 2007, 5, 277–289. [Google Scholar] [CrossRef]
- Macas, J.; Mészáros, T.; Nouzová, M. PlantSat: A Specialized Database for Plant Satellite Repeats. Bioinformatics 2002, 18, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Raina, S.N. Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet. Genome Res. 2005, 109, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Meštrovic, N.; Mravinac, B. Satellite DNA evolution. In Repetitive DNA; Garrido-Ramos, M.A., Ed.; Karger: Granada, Spain, 2012; pp. 126–152. [Google Scholar]
- Plohl, M.; Petrovi’c, V.; Luchetti, A.; Ricci, A.; Šatovi’c, E.; Passamonti, M.; Mantovani, B. Long-term conservation vs high sequence divergence: The case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity 2010, 104, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA in Plants: More than Just Rubbish. Cytogenet. Genome Res. 2015, 146, 153–170. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef]
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA Evolution: Old Ideas, New Approaches. Curr. Opin. Genet. Dev. 2018, 49, 70–78. [Google Scholar] [CrossRef]
- Ugarkovic, D. Functional elements residing within satellite DNAs. EMBO Rep. 2005, 6, 1035–1039. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S. Comparative Genome Organization in Plants: From Sequence and Markers to Chromatin and Chromosomes. Plant Cell 2000, 12, 617–636. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S. Planning for remodelling: Nuclear architecture, chromatin and chromosomes. Trends Plant Sci. 2003, 8, 195–197. [Google Scholar] [CrossRef]
- Amosova, A.V.; Bolsheva, N.L.; Samatadze, T.E.; Twardovska, M.O.; Zoshchuk, S.A.; Andreev, I.O.; Badaeva, E.D.; Kunakh, V.A.; Muravenko, O.V. Molecular cytogenetic analysis of Deschampsia antarctica Desv. (Poaceae), Maritime Antarctic. PLoS ONE 2015, 10, e0138878. [Google Scholar] [CrossRef] [PubMed]
- Samoluk, S.S.; Robledo, G.; Bertioli, D.; Seijo, J.G. Evolutionary dynamics of an at-rich satellite DNA and its contribution to karyotype differentiation in wild diploid Arachis species. Mol. Genet. Genom. 2017, 292, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Afzal-Rafii, Z. Étude cytotaxonomique et phylogénetique de quelques Salvia de la region méditerranéenne: Groupe du Salvia officinalis L. Bull. La Soc. Bot. Fr. 1976, 123, 515–527. [Google Scholar] [CrossRef]
- Love, A.; Love, D. Arctic polypoloidy. Proc Genet. Soc Can 1957, 2, 23–27. [Google Scholar]
- Dhar, M.K.; Kour, J.; Kaul, S. Origin, behaviour, and transmission of B chromosome with special reference to Plantago lagopus. Genes 2019, 10, 152. [Google Scholar] [CrossRef]
- Houben, A.; Banaei-Moghaddam, A.M.; Klemme, S.; Timmis, J.N. Evolution and biology of supernumerary B chromosomes. Cell. Mol. Life Sci. 2014, 71, 467–478. [Google Scholar] [CrossRef]
- D’Ambrosio, U.; Alonso-Lifante, M.P.; Barros, K.; Kovaøík, A.; Mas de Xaxars, G.; Garcia, S. B-chrom: A database on B-chromosomes of plants, animals and fungi. New Phytol. 2017, 216, 635–642. [Google Scholar] [CrossRef]
- Belyayev, A.; Raskina, O. Chromosome evolution in marginal populations of Aegilops speltoides: Causes and consequences. Ann. Bot. 2013, 111, 531–538. [Google Scholar] [CrossRef]
- Houben, A.; Thompson, N.; Ahne, R.; Leach, C.R.; Verlin, D.; Timmis, J.N. A monophyletic origin of the B chromosomes of Brachycome dichromosomatica (Asteraceae). Plant Syst. Evol. 1999, 219, 127–135. [Google Scholar]
- Huang, W.; Du, Y.; Zhao, X.; Jin, W. B chromosome contains active genes and impacts the transcription of A chromosomes in maize (Zea mays L. ) BMC Plant Biol. 2016, 16, 88. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Chikida, N.N.; Belousova, M.K.; Ruban, A.S.; Surzhikov, S.A.; Zoshchuk, S.A. A new insight on the evolution of polyploid Aegilops species from the complex Crassa: Molecular-cytogenetic analysis. Plant Syst. Evol. 2021, 307, 3. [Google Scholar] [CrossRef]
- de Assis, R.; Baba, V.Y.; Cintra, L.A.; Gonçalves, L.S.A.; Rodrigues, R.; Vanzela, A.L.L. Genome relationships and LTR-retrotransposon diversity in three cultivated Capsicum L. (Solanaceae) species. BMC Genom. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, A.; Hufnagel, B.; Soriano, A.; Péret, B. The Highly Repeat-Diverse (Peri) Centromeres of White Lupin (Lupinus albus L.). Front. Plant Sci. 2022, 13, 862079. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, X.; Zhou, J.; Yu, L.A.; Li, S.; Zhang, Y.; Qin, R.; Gao, W.; Deng, C. Genome-wide analysis of transposable elements and satellite DNAs in Spinacia species to shed light on their roles in sex chromosome evolution. Front. Plant Sci. 2021, 11, 575462. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.C.; Waminal, N.E.; Kim, H.H. In silico mining and FISH mapping of a chromosome-specific satellite DNA in Capsicum annuum L. Genes Genom. 2019, 41, 1001–1006. [Google Scholar] [CrossRef]
- Amosova, A.; Ghukasyan, L.; Yurkevich, O.; Bolsheva, N.; Samatadze, T.; Zoshchuk, S.; Muravenko, O. Cytogenomics of Deschampsia P. Beauv (Poaceae) species based on sequence analyses and FISH mapping of CON/COM satellite DNA families. Plants 2021, 10, 1105. [Google Scholar] [CrossRef]
- Orooji, F.; Mirzaghaderi, G.; Kuo, Y.T.; Fuchs, J. Variation in the Number and Position of rDNA Loci Contributes to the Diversification and Speciation in Nigella (Ranunculaceae). Front. Plant Sci. 2022, 1874. [Google Scholar] [CrossRef]
- Vondrak, T.; Ávila Robledillo, L.; Novák, P.; Koblížková, A.; Neumann, P.; Macas, J. Characterization of repeat arrays in ultra-long nanopore reads reveals frequent origin of satellite DNA from retrotransposon-derived tandem repeats. Plant J. 2020, 101, 484–500. [Google Scholar] [CrossRef]
- Miklos, G.L.G.; Gill, A.C. Nucleotide sequences of highly repeated DNAs; compilation and comments. Genet. Res. 1982, 39, 1–30. [Google Scholar] [CrossRef]
- Langdon, T.; Seago, C.; Jones, R.N.; Ougham, H.; Thomas, H.; Forster, J.W.; Jenkins, G. De novo evolution of satellite DNA on the rye B chromosome. Genetics 2000, 154, 869–884. [Google Scholar] [CrossRef]
- Rosato, M.; Kovařík, A.; Garilleti, R.; Rosselló, J.A. Conserved organisation of 45S rDNA sites and rDNA gene copy number among major clades of early land plants. PLoS ONE 2016, 11, e0162544. [Google Scholar] [CrossRef] [PubMed]
- Roa, F.; Guerra, M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol. Biol. 2012, 12, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A galaxybased web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Robledillo, L.A.; Koblizkova, A.; Vrbova, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acid Res. 2017, 45, e111. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71-4. [Google Scholar] [CrossRef]
- Muravenko, O.V.; Amosova, A.V.; Samatadze, T.E.; Popov, K.V.; Poletaev, A.I.; Zelenin, A.V. 9-Aminoacridine: An efficient reagent to improve human and plant chromosome banding patterns and to standardize chromosome image analysis. Cytom. Part A J. Int. Soc. Anal. Cytol. 2003, 51, 52–57. [Google Scholar] [CrossRef]
- Gerlach, W.L.; Bedbrook, J.R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979, 7, 1869–1885. [Google Scholar] [CrossRef]
- Gerlach, W.L.; Dyer, T.A. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980, 8, 4851–4855. [Google Scholar] [CrossRef]
- Muravenko, O.V.; Yurkevich, O.Y.; Bolsheva, N.L.; Samatadze, T.E.; Nosova, I.V.; Zelenina, D.A.; Volkov, A.A.; Popov, K.V.; Zelenin, A.V. Comparison of genomes of eight species of sections Linum and Adenolinum from the genus Linum based on chromosome banding, molecular markers and RAPD analysis. Genetica 2009, 135, 245–255. [Google Scholar] [CrossRef]
- Amosova, A.V.; Yurkevich, O.Y.; Bolsheva, N.L.; Samatadze, T.E.; Zoshchuk, S.A.; Muravenko, O.V. Repeatome Analyses and Satellite DNA Chromosome Patterns in Deschampsia sukatschewii, D. cespitosa, and D. antarctica (Poaceae). Genes 2022, 13, 762. [Google Scholar] [CrossRef]
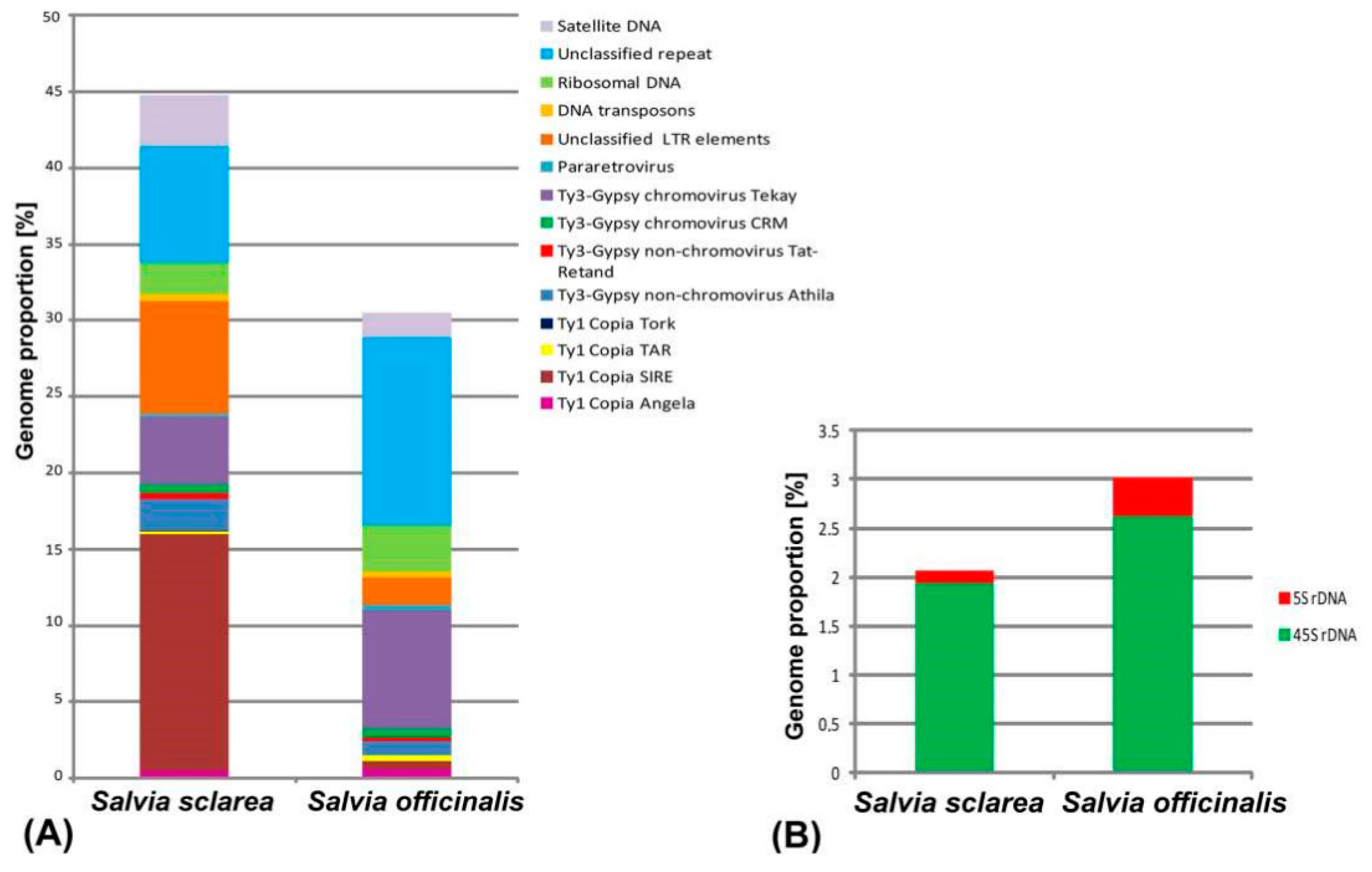


| Repeat Name | Genome Proportion (%) | |
|---|---|---|
| Salvia sclarea | Salvia officinalis | |
| Retrotransposons (Class I) | 31.33 | 13.2 |
| Ty1-Copia | 16.28 | 1.53 |
| Angela | 0.48 | 0.62 |
| Bianca | 0.03 | - |
| Ikeros | 0.03 | 0.02 |
| SIRE | 15,49 | 0.51 |
| TAR | 0.22 | 0.36 |
| Tork | 0.03 | 0.02 |
| Ty3-Gypsy | 7.47 | 9.54 |
| Non-chromovirus Athila | 2.05 | 0.92 |
| Non-chromovirus Tat-Retand | 0.4 | 0.22 |
| Chromovirus CRM | 0.61 | 0.61 |
| Chromovirus Tekay | 4.41 | 7.79 |
| Pararetrovirus | 0.2 | 0.32 |
| Unclassified LTR elements | 7.38 | 1.81 |
| Transposons (Class II) | 0.45 | 0.34 |
| EnSpm_CACTA MuDR_Mutator | 0.25 - | 0.3 0.04 |
| PIF_Harbinger | 0.15 | - |
| Helitron | 0.05 | - |
| Ribosomal DNA | 2.07 | 3.01 |
| 45S rDNA | 1.94 | 2.62 |
| 5S rDNA | 0.13 | 0.39 |
| Unclassified repeats | 7.62 | 12.6 |
| Satellite DNA | 3.4 | 1.63 |
| Organelle | 13.78 | 15.25 |
| Putative satellites | 3 high confident 1 low confident | 8 high confident 4 low confident |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muravenko, O.V.; Yurkevich, O.Y.; Kalnyuk, J.V.; Samatadze, T.E.; Zoshchuk, S.A.; Amosova, A.V. Integration of Repeatomic and Cytogenetic Data on Satellite DNA for the Genome Analysis in the Genus Salvia (Lamiaceae). Plants 2022, 11, 2244. https://doi.org/10.3390/plants11172244
Muravenko OV, Yurkevich OY, Kalnyuk JV, Samatadze TE, Zoshchuk SA, Amosova AV. Integration of Repeatomic and Cytogenetic Data on Satellite DNA for the Genome Analysis in the Genus Salvia (Lamiaceae). Plants. 2022; 11(17):2244. https://doi.org/10.3390/plants11172244
Chicago/Turabian StyleMuravenko, Olga V., Olga Yu. Yurkevich, Julia V. Kalnyuk, Tatiana E. Samatadze, Svyatoslav A. Zoshchuk, and Alexandra V. Amosova. 2022. "Integration of Repeatomic and Cytogenetic Data on Satellite DNA for the Genome Analysis in the Genus Salvia (Lamiaceae)" Plants 11, no. 17: 2244. https://doi.org/10.3390/plants11172244
APA StyleMuravenko, O. V., Yurkevich, O. Y., Kalnyuk, J. V., Samatadze, T. E., Zoshchuk, S. A., & Amosova, A. V. (2022). Integration of Repeatomic and Cytogenetic Data on Satellite DNA for the Genome Analysis in the Genus Salvia (Lamiaceae). Plants, 11(17), 2244. https://doi.org/10.3390/plants11172244





