Molecular Cloning and Expression Analysis of the Cryptochrome Gene CiPlant-CRY1 in Antarctic Ice Alga Chlamydomonas sp. ICE-L
Abstract
1. Introduction
2. Results
2.1. Bioinformatics Analysis of CiPlant-CRY1
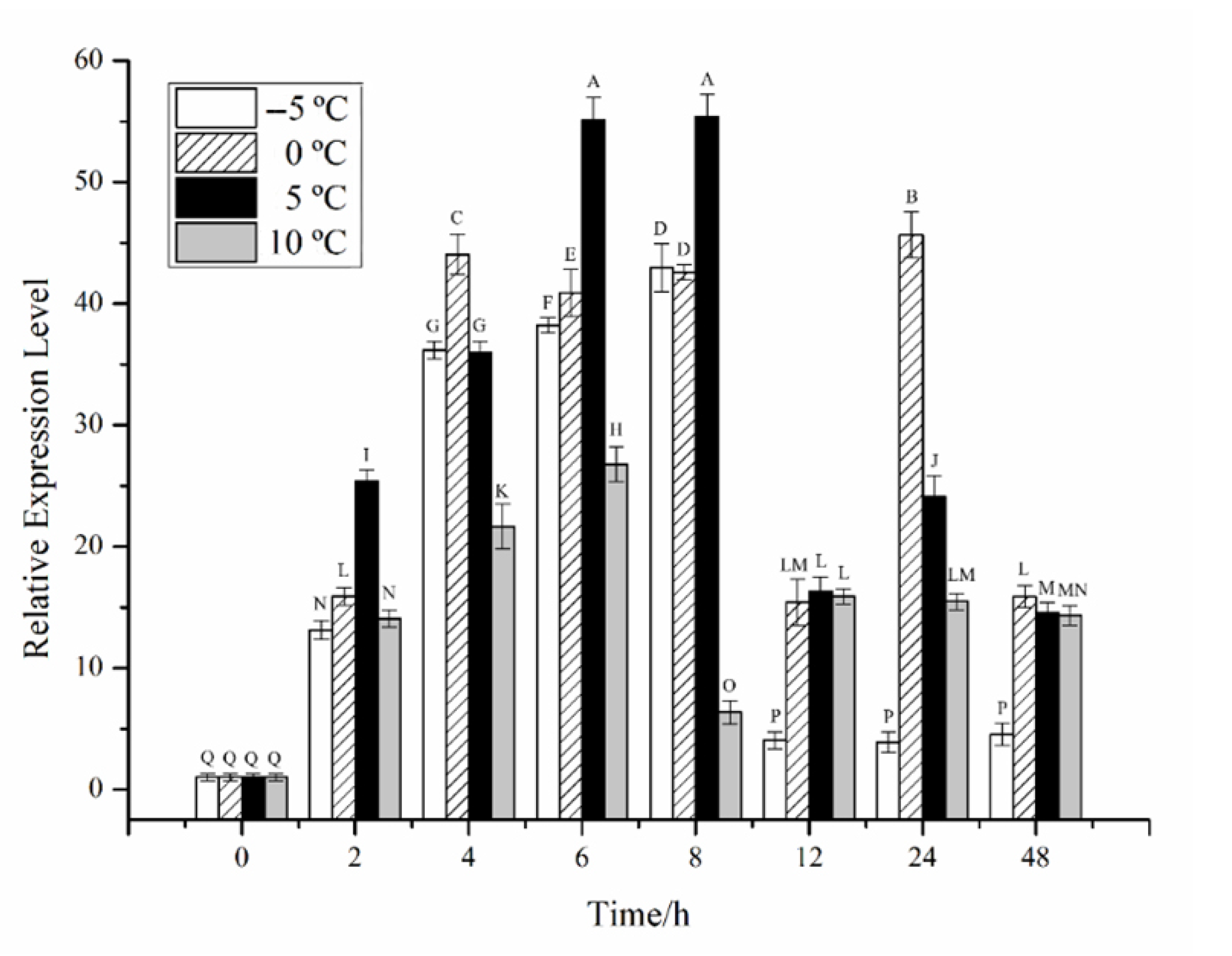
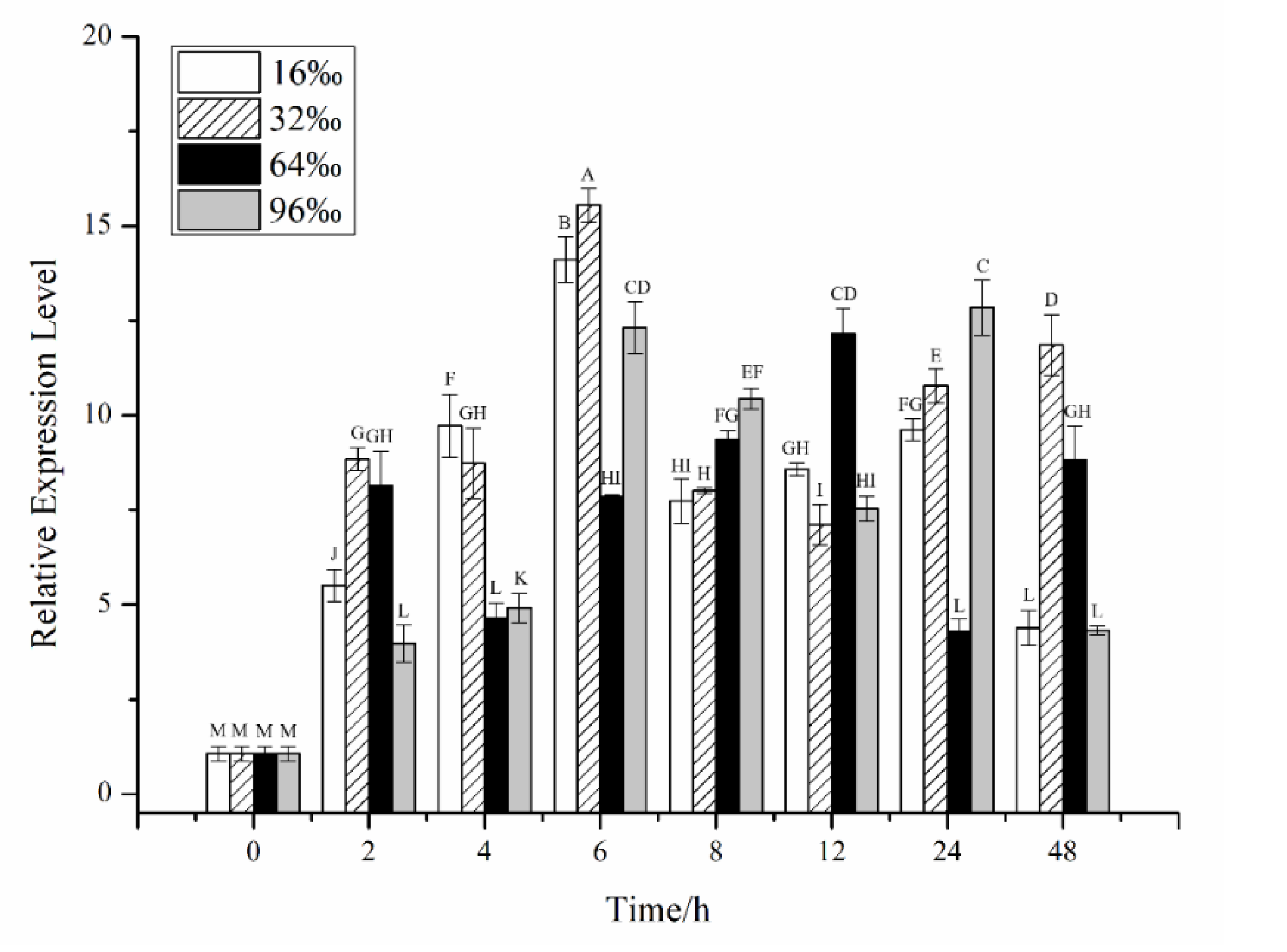
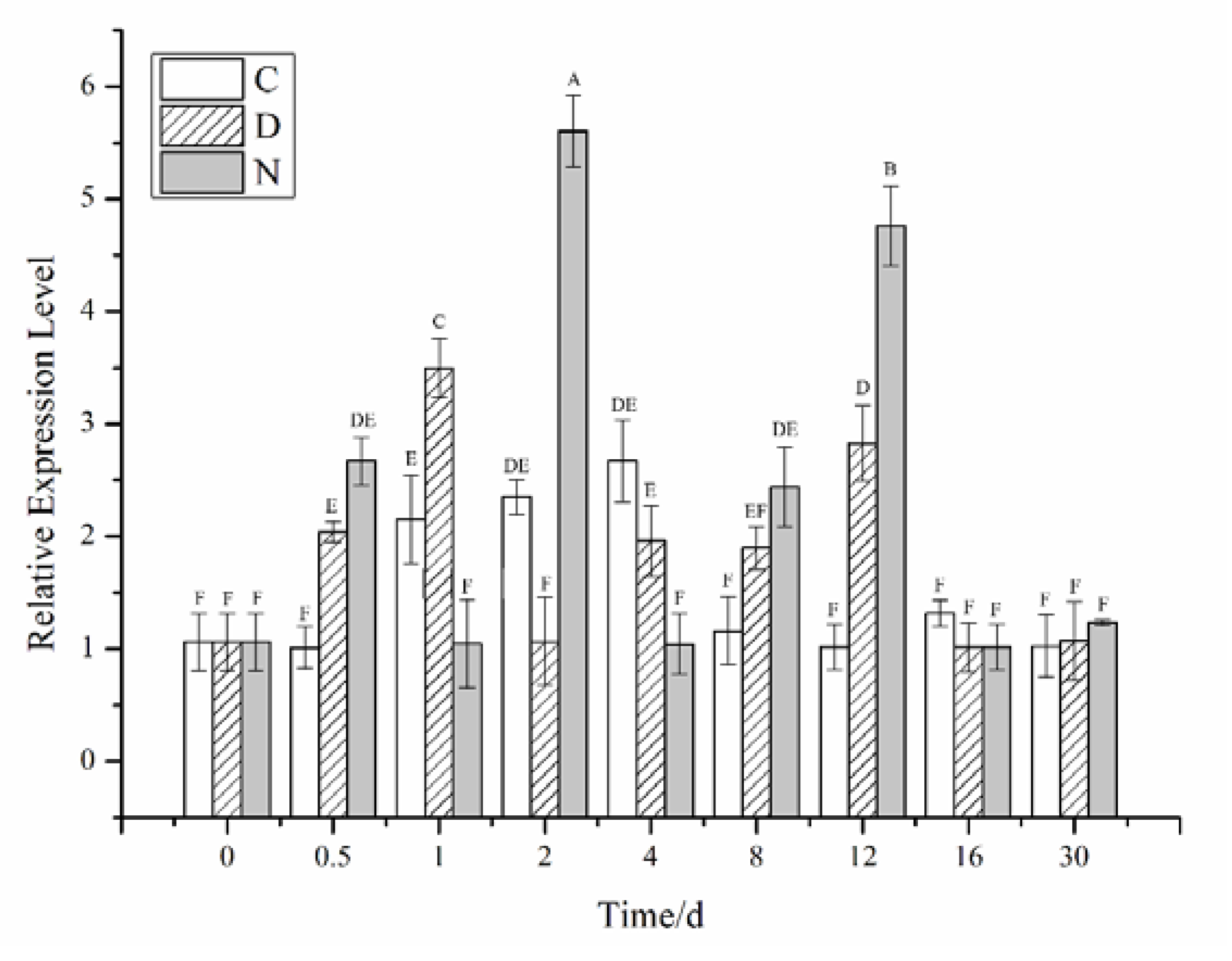
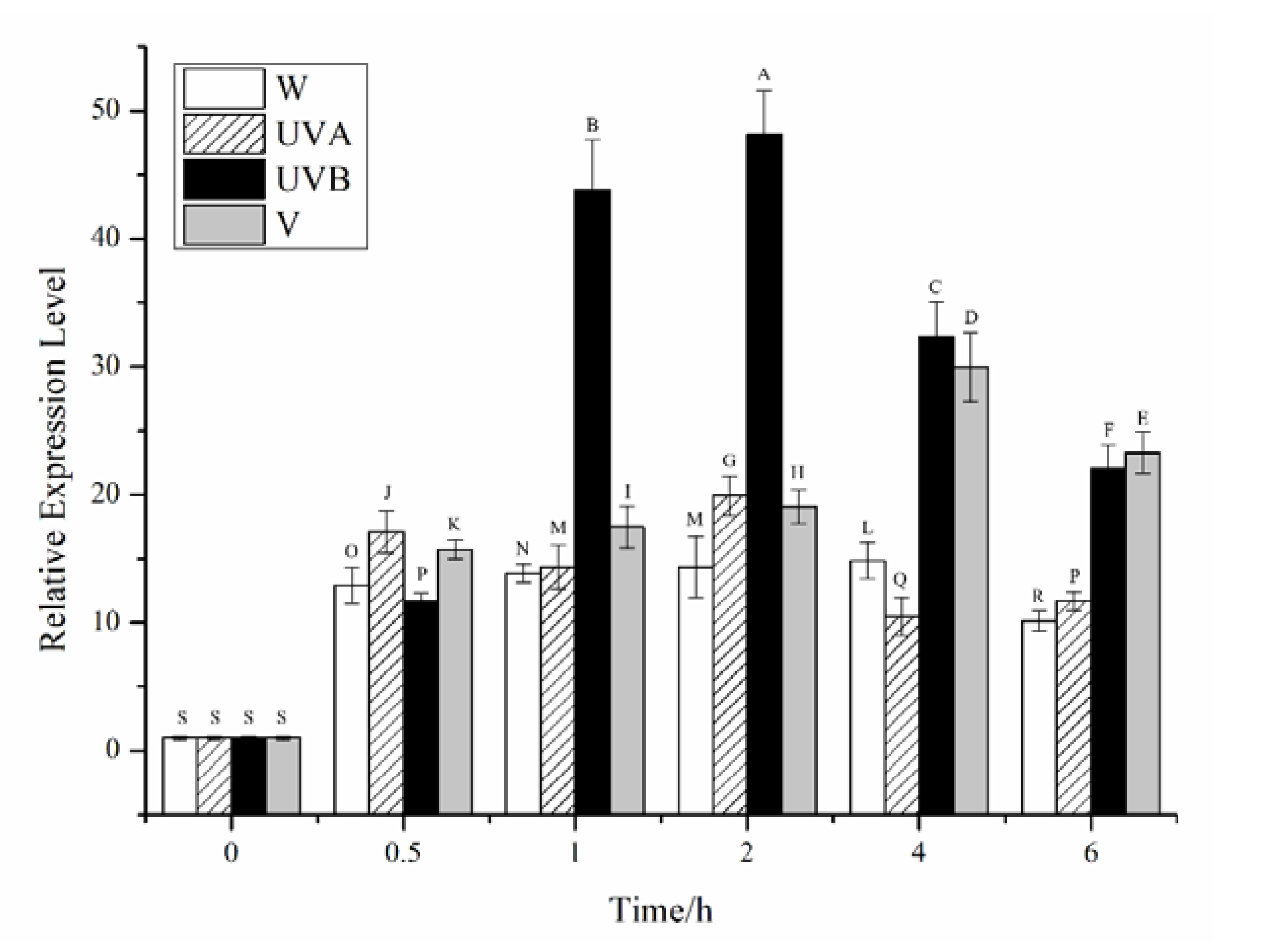
2.2. Effects of Different Stress Conditions on the Expression of CiPlant-CRY1
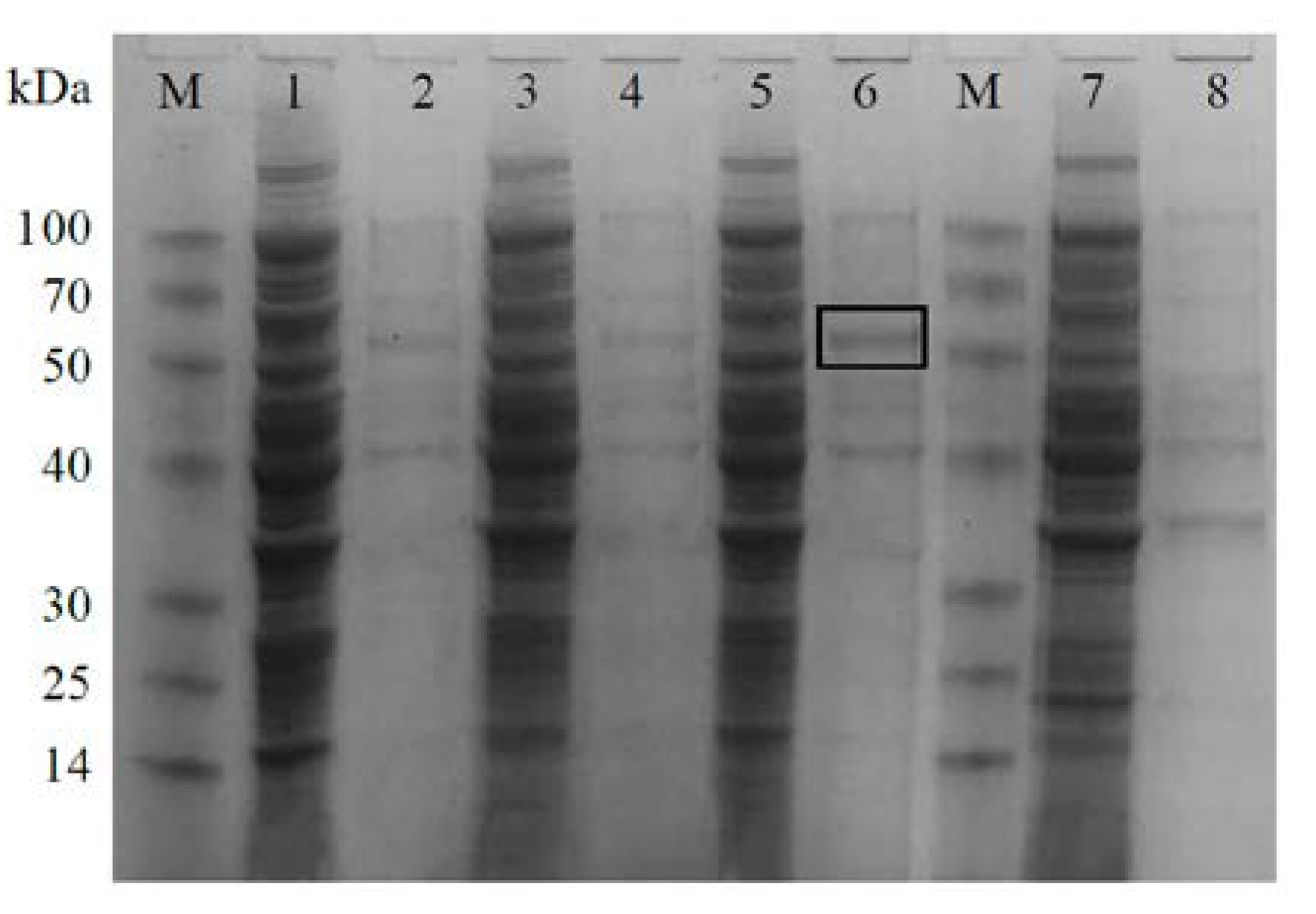
2.3. Protein Expression
3. Discussion
4. Materials and Methods
4.1. Algal Culture
4.2. RNA Extraction and cDNA Synthesis
4.3. Cloning and Sequencing of CRY Complete ORF
4.4. Bioinformatic Analysis
4.5. Analysis of CiPlant-CRY1 mRNA Expression by Quantitative Real-Time PCR (qRT-PCR)
4.6. Protein Expression
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, C.T.; Todo, T. The cryptochromes. Genome. Biol. 2005, 6, 220. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, L.; Guan, Z.Y.; Wang, Q.; Yan, X.H.; Wang, J.; Wang, Z.Z.; Cao, J.B.; Zhang, D.L.; Gong, X.; Yin, P. Structural insights into the photoactivation of Arabidopsis CRY2. Nat. Plants 2020, 6, 1432–1438. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, L.; Oldemeyer, S.; Kottke, T. Time-Resolved Infrared Spectroscopy on Plant Cryptochrome-Relevance of Proton Transfer and ATP Binding for Signaling. J. Phys. Chem. A 2018, 122, 140–147. [Google Scholar] [CrossRef]
- Chaves, I.; Pokorny, R.; Byrdin, M.; Hoang, N.; Ritz, T.; Brettel, K.; Essen, L.O.; van der Horst, G.T.; Batschauer, A.; Ahmad, M. The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011, 62, 335–364. [Google Scholar] [CrossRef] [PubMed]
- Nohr, D.; Franz, S.; Rodriguez, R.; Paulus, B.; Essen, L.O.; Weber, S.; Schleicher, E. Extended Electron-Transfer in Animal Cryptochromes Mediated by a Tetrad of Aromatic Amino Acids. Biophys. J. 2016, 111, 301–311. [Google Scholar] [CrossRef]
- Shao, K.; Zhang, X.; Li, X.; Hao, Y.H.; Huang, X.W.; Ma, M.L.; Zhang, M.H.; Yu, F.; Liu, H.T.; Zhang, P. The oligomeric structures of plant cryptochromes. Nat. Struct. Mol. Biol. 2020, 27, 480–488. [Google Scholar] [CrossRef]
- Kottke, T.; Oldemeyer, S.; Wenzel, S.; Zou, Y.; Mittag, M. Cryptochrome photoreceptors in green algae: Unexpected versatility of mechanisms and functions. J. Plant Physiol. 2017, 217, 4–14. [Google Scholar] [CrossRef]
- König, S.; Juhas, M.; Jäger, S.; Kottke, T.; Büchel, C. The cryptochrome-photolyase protein family in diatoms. J. Plant Physiol. 2017, 217, 15–19. [Google Scholar] [CrossRef]
- Duanmu, D.; Rockwell, N.C.; Lagarias, J.C. Algal light sensing and photoacclimation in aquatic environments. Plant Cell Environ. 2017, 40, 2558–2570. [Google Scholar] [CrossRef]
- Liu, C.L.; Wang, X.L.; Wang, X.N.; Sun, C.J. Acclimation of Antarctic Chlamydomonas to the sea-ice environment: A transcriptomic analysis. Extremophiles 2016, 20, 437–450. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, Z.; He, Y.Y.; Liu, L.N.; Qu, C.F.; Miao, J.L. Molecular Cloning and Expression of a Cryptochrome Gene CiCRY-DASH1 from the Antarctic microalga Chlamydomonas sp. ICE-L. Mol. Biotechnol. 2020, 62, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.N.; Dieckmann, G.S. Antarctic Sea ice-a habitat for extremophiles. Science 2002, 295, 641–644. [Google Scholar] [CrossRef] [PubMed]
- Petrou, K.; Kranz, S.A.; Trimborn, S.; Hassler, C.S.; Ameijeiras, S.B.; Sackett, O.; Ralph, P.J.; Davidson, A.T. Southern Ocean phytoplankton physiology in a changing climate. J. Plant Physiol. 2016, 203, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Essen, L.O.; Franz, S.; Banerjee, A. Structural and evolutionary aspects of algal blue light receptors of the cryptochrome and aureochrome type. J. Plant Physiol. 2017, 217, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lin, C.T. Mechanisms of Cryptochrome-Mediated Photoresponses in Plants. Annu Rev Plant Biol. 2020, 71, 103–129. [Google Scholar] [CrossRef]
- Kondoh, M.; Shiraishi, C.; Müller, P.; Ahmad, M.; Hitomi, K.; Getzoff, E.D.; Terazima, M. Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome. J. Mol. Biol. 2011, 413, 128–137. [Google Scholar] [CrossRef]
- Pooam, M.; Dixon, N.; Hilvert, M.; Misko, P.; Waters, K.; Jourdan, N.; Drahy, S.; Mills, S.; Engle, D.; Link, J.; et al. Effect of temperature on the Arabidopsis cryptochrome photocycle. Physiol. Plant 2021, 172, 1653–1661. [Google Scholar] [CrossRef]
- D’Amico-Damião, V.; Carvalho, R.F. Cryptochrome-Related Abiotic Stress Responses in Plants. Front. Plant Sci. 2018, 9, 1897. [Google Scholar] [CrossRef]
- Serrano-Bueno, G.; Romero-Campero, F.J.; Lucas-Reina, E.; Romero, J.M.; Valverde, F. Evolution of photoperiod sensing in plants and algae. Curr. Opin. Plant Biol. 2017, 37, 10–17. [Google Scholar] [CrossRef]
- Petroutsos, D.; Tokutsu, R.; Maruyama, S.; Flori, S.; Greiner, A.; Magneschi, L.; Cusant, L.; Kottke, T.; Mittag, M.; Hegemann, P.; et al. A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 2016, 537, 563–566. [Google Scholar] [CrossRef]
- Villafani, Y.; Yang, H.W.; Park, Y.I. Color Sensing and Signal Transmission Diversity of Cyanobacterial Phytochromes and Cyanobacteriochromes. Mol. Cells 2020, 43, 509–516. [Google Scholar] [PubMed]
- Beel, B.; Prager, K.; Spexard, M.; Sasso, S.; Weiss, D.; Müller, N.; Heinnickel, M.; Dewez, D.; Ikoma, D.; Grossman, A.R.; et al. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell 2012, 24, 2992–3008. [Google Scholar] [CrossRef] [PubMed]
- Müller, N.; Wenzel, S.; Zou, Y.; Künzel, S.; Sasso, S.; Weiß, D.; Prager, K.; Grossman, A.; Kottke, T.; Mittag, M. A Plant Cryptochrome Controls Key Features of the Chlamydomonas Circadian Clock and Its Life Cycle. Plant Physiol. 2017, 174, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009, 60, 357–377. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Q.; Nguyen, P.; Lin, C.T. Cryptochrome-mediated light responses in plants. Enzymes 2014, 35, 167–189. [Google Scholar] [PubMed]
- Fortunato, A.E.; Annunziata, R.; Jaubert, M.; Bouly, J.P.; Falciatore, A. Dealing with light: The widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J. Plant Physiol. 2015, 172, 42–54. [Google Scholar] [CrossRef]
- Hang, W.; Gujar, A.; Zhang, H.J.; Xu, W.X.; Zhao, C.C.; Zhu, X.L.; Xue, J.A.; Zhang, C.H.; Ji, C.L.; Qin, S.; et al. Cloning, expression, and characterization of a novel plant type cryptochrome gene from the green alga Haematococcus pluvialis. Protein. Expr Purif. 2020, 172, 105633. [Google Scholar] [CrossRef]
- Tissot, N.; Ulm, R. Cryptochrome-mediated blue-light signalling modulates UVR8 photoreceptor activity and contributes to UV-B tolerance in Arabidopsis. Nat. Commun. 2020, 11, 1323. [Google Scholar] [CrossRef]
- An, M.L.; Mou, S.L.; Zhang, X.W.; Ye, N.H.; Zheng, Z.; Cao, S.N.; Xu, D.; Fan, X.; Wang, Y.T.; Miao, J.L. Temperature regulates fatty acid desaturases at a transcriptional level and modulates the fatty acid profile in the Antarctic microalga Chlamydomonas sp. ICE-L. Bioresour. Technol. 2013, 134, 151–157. [Google Scholar] [CrossRef]
- Provasoli, L. Media and Prospects for the Cultivation of Marine Algae. In Culture and Collections of Algae, Proceedings of the U.S.-Japan Conference, Hakone, 12–15 September 1966; Watanabe, A., Hattori, R., Eds.; Japanese Society for Plant Physiology: Tokyo, Japan, 1968; pp. 63–75. [Google Scholar]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–4500. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Role | Name | Sequence (5′–3′) |
|---|---|---|
| cDNA | F1 | ATGCGCGTTGTGGCCGC |
| R1 | CCACATCTTCTGTCGTTTCTGGGG | |
| qRT-PCR | F2 | GTCGAAGGATGTGCCAAGGG |
| R2 | GGTGCCATCACCAGTTCCTG | |
| 18S rRNA | F4 | ATGGAATAACACGATAGGACTCTGG |
| R4 | ACCTCTGACAATGAAATACGAATGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Zheng, Z.; Zhang, X.; Bao, Y.; Miao, J. Molecular Cloning and Expression Analysis of the Cryptochrome Gene CiPlant-CRY1 in Antarctic Ice Alga Chlamydomonas sp. ICE-L. Plants 2022, 11, 2213. https://doi.org/10.3390/plants11172213
Zhao Y, Zheng Z, Zhang X, Bao Y, Miao J. Molecular Cloning and Expression Analysis of the Cryptochrome Gene CiPlant-CRY1 in Antarctic Ice Alga Chlamydomonas sp. ICE-L. Plants. 2022; 11(17):2213. https://doi.org/10.3390/plants11172213
Chicago/Turabian StyleZhao, Yaoyao, Zhou Zheng, Xin Zhang, Yating Bao, and Jinlai Miao. 2022. "Molecular Cloning and Expression Analysis of the Cryptochrome Gene CiPlant-CRY1 in Antarctic Ice Alga Chlamydomonas sp. ICE-L" Plants 11, no. 17: 2213. https://doi.org/10.3390/plants11172213
APA StyleZhao, Y., Zheng, Z., Zhang, X., Bao, Y., & Miao, J. (2022). Molecular Cloning and Expression Analysis of the Cryptochrome Gene CiPlant-CRY1 in Antarctic Ice Alga Chlamydomonas sp. ICE-L. Plants, 11(17), 2213. https://doi.org/10.3390/plants11172213






