LiDAR Platform for Acquisition of 3D Plant Phenotyping Database
Abstract
:1. Introduction
2. Materials and Methods
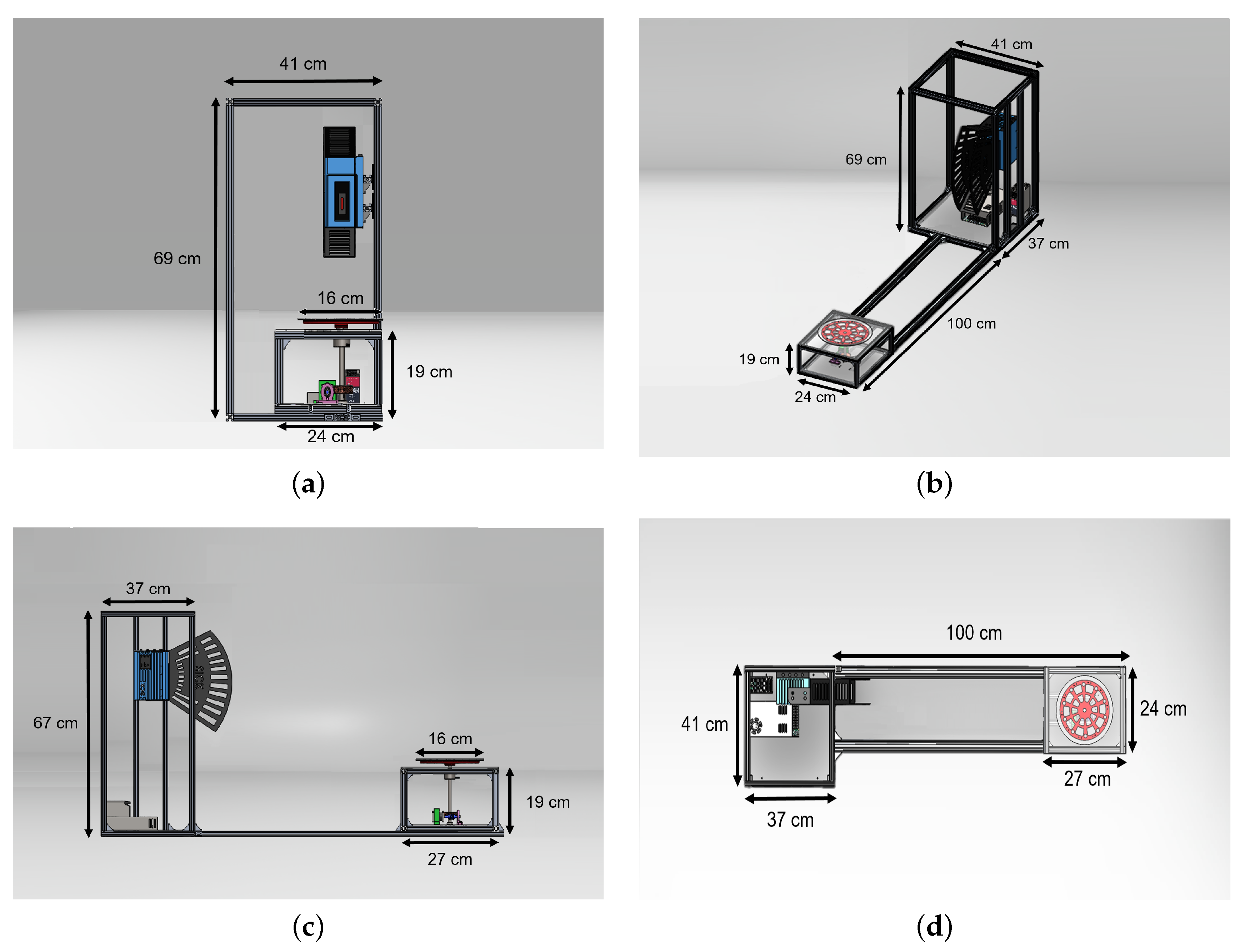
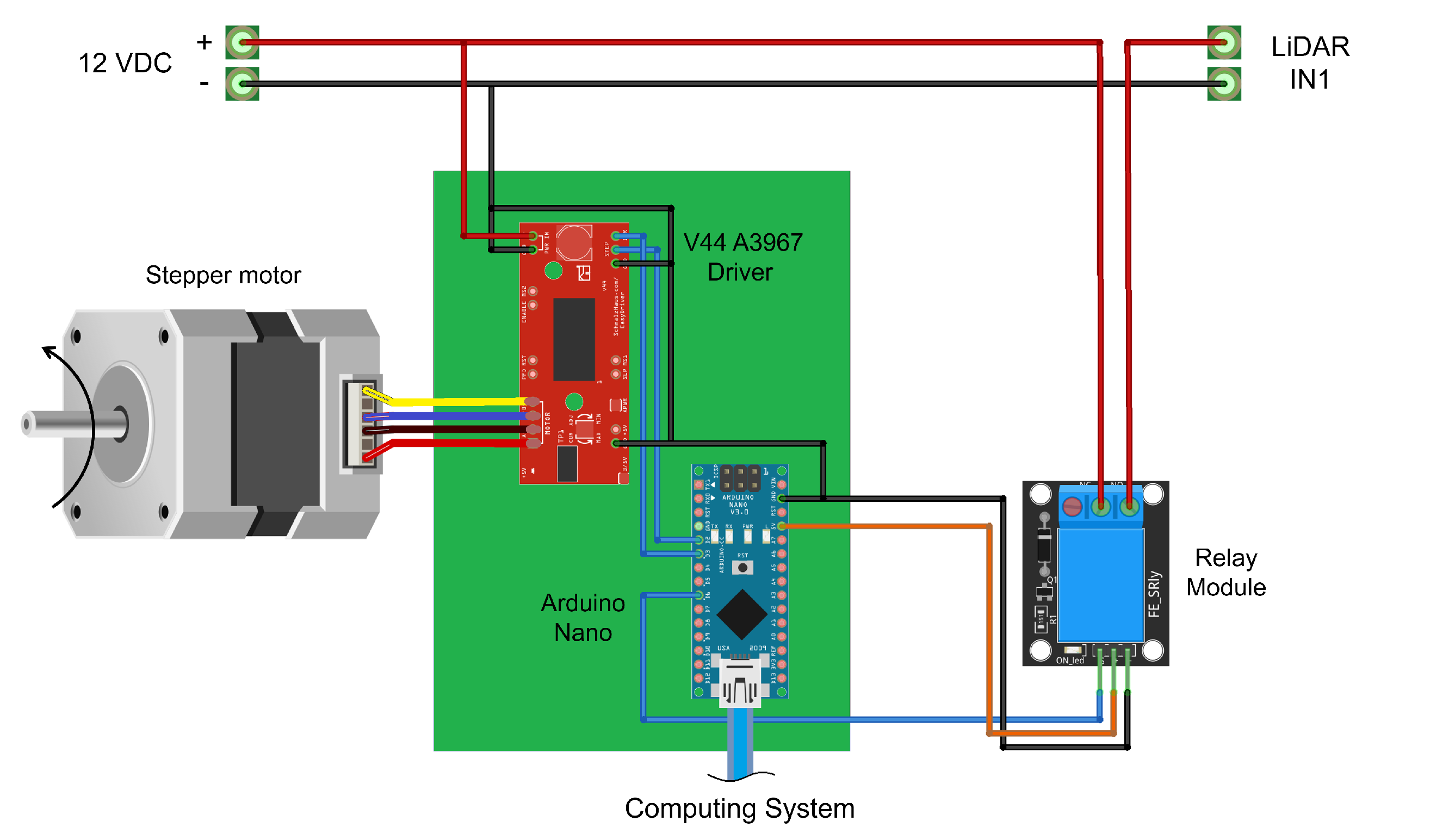
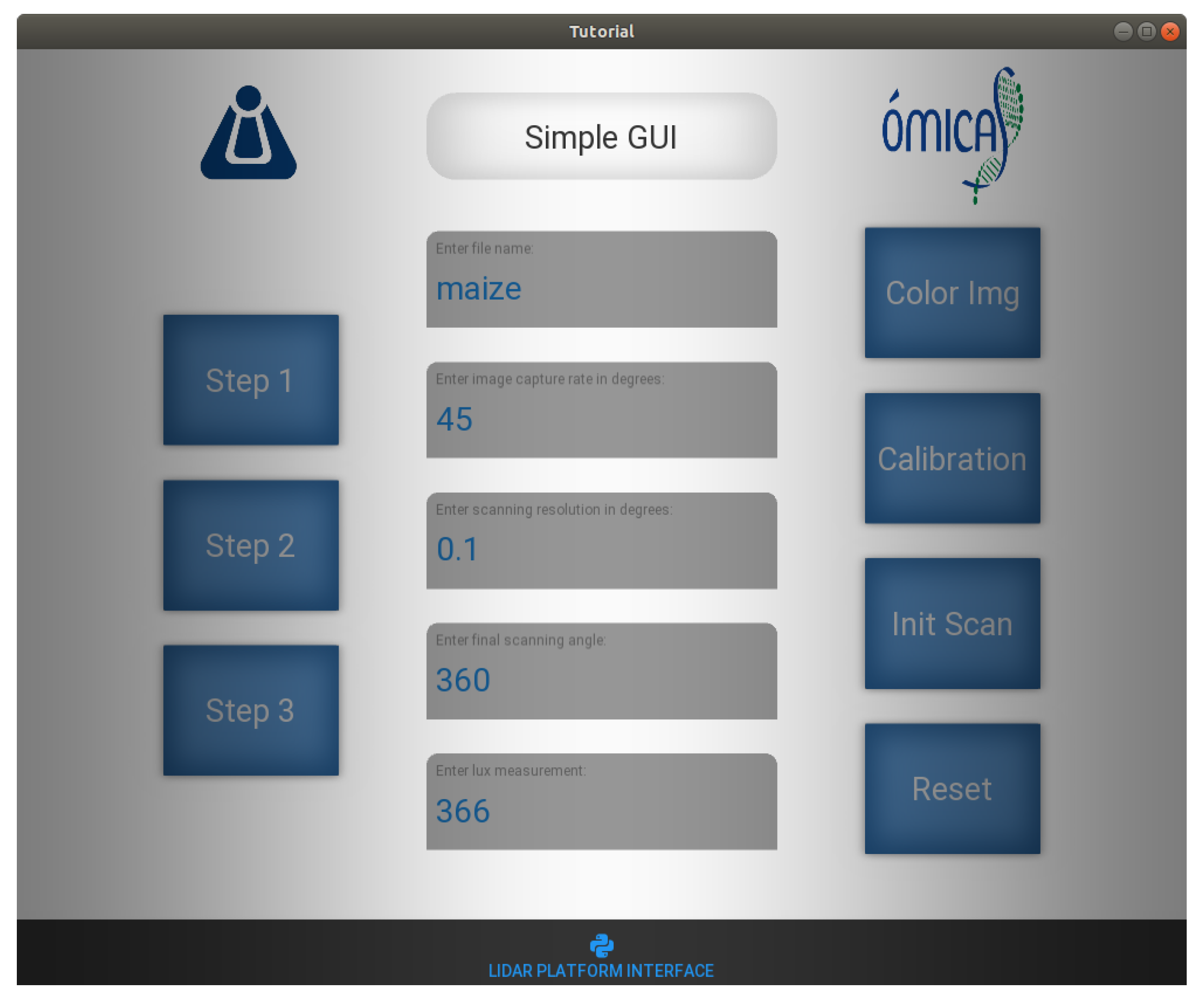
2.1. Platform LiDAR
| Algorithm 1: 3D model generation with the proposed LiDAR scanning platform. |
 |
2.2. Application
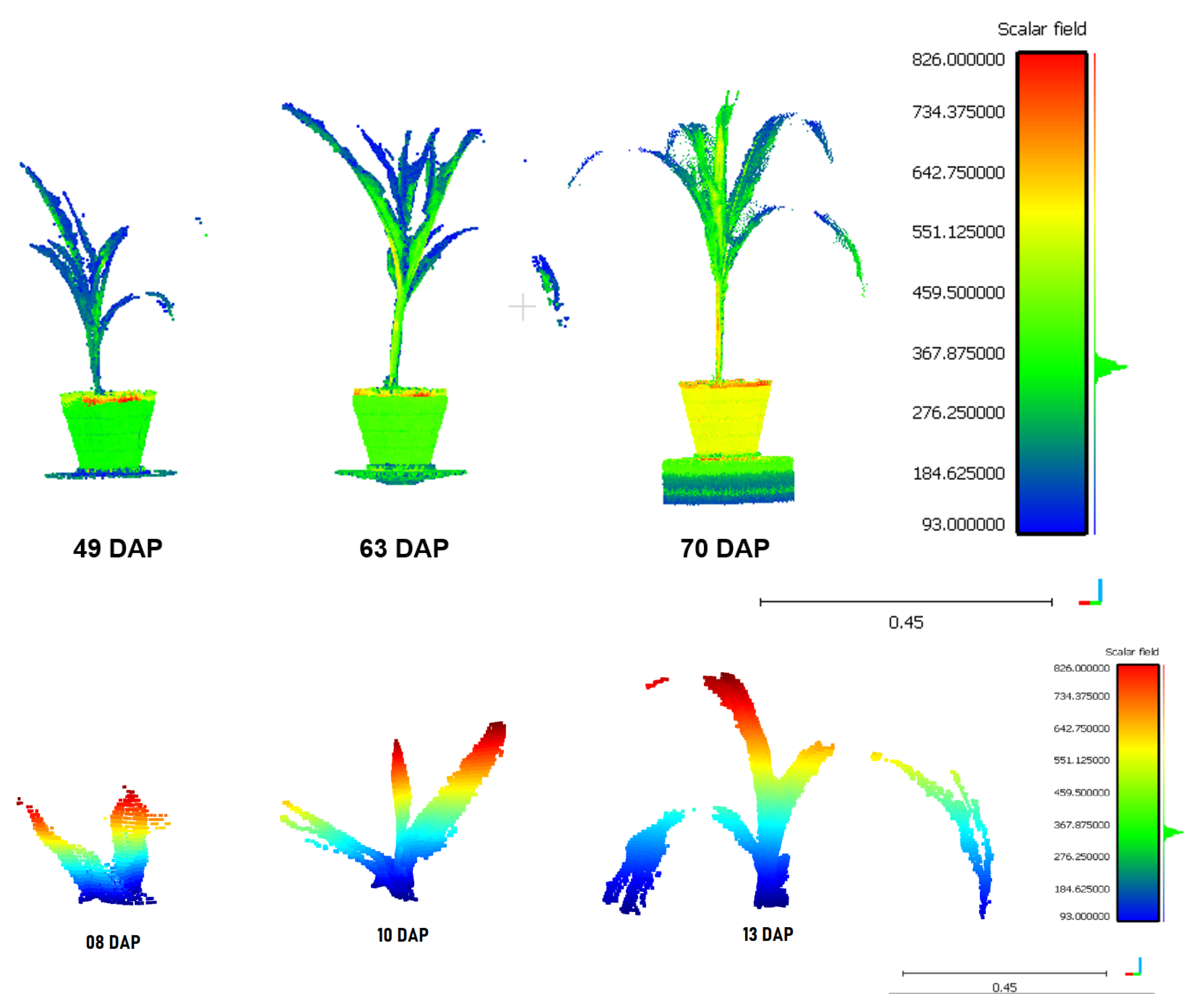
- Seedling height h:To determine the height of the plant, the point in the cloud with the highest value on the Z-axis is required.
- Total Volume v:To calculate the volume of the plant, a voxelization of the points with a distance of 25 cm is performed. Then the voxel count is denoted in the (9) equation as the summation of the V parameter is performed and multiplied by the distance value used. The distance value was calculated experimentally.
- Classification of organs:In order to separate the organs of the seedling, a classification is made with respect to the stem and leaves. The database is split in two, taking the first days of shoot and seedling up to the third leaf as one database and the rest as another. In each database, 60% of the point clouds are used for training a Random Forest classifier, 20% for tuning the classifier parameters, and the rest for model validation.Once this result has been obtained, filtering of leaf segments that were considered stems is carried out. To do this, a virtual ring is used that goes up from the base of the plant to the highest point [31]. The radius of the ring was 0.0015 and was estimated and validated experimentally.
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- United Nations Department of Economic and Social Affairs Population Division. Available online: https://n9.cl/vbs5ri (accessed on 6 October 2021).
- Li, L.; Zhang, Q.; Huang, D. A Review of Imaging Techniques for Plant Phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guo, R.; Li, M.; Chen, Y.; Li, G. A review of computer vision technologies for plant phenotyping. Comput. Electron. Agric. 2020, 176, 105672. [Google Scholar] [CrossRef]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef]
- Gao, M.; Yang, F.; Wei, H.; Liu, X. Individual Maize Location and Height Estimation in Field from UAV-Borne LiDAR and RGB Images. Remote Sens. 2022, 14, 2292. [Google Scholar] [CrossRef]
- Chen, Q.; Gao, T.; Zhu, J.; Wu, F.; Li, X.; Lu, D.; Yu, F. Individual Tree Segmentation and Tree Height Estimation Using Leaf-Off and Leaf-On UAV-LiDAR Data in Dense Deciduous Forests. Remote Sens. 2022, 14, 2787. [Google Scholar] [CrossRef]
- Gyawali, A.; Aalto, M.; Peuhkurinen, J.; Villikka, M.; Ranta, T. Comparison of Individual Tree Height Estimated from LiDAR and Digital Aerial Photogrammetry in Young Forests. Sustainability 2022, 14, 3720. [Google Scholar] [CrossRef]
- Wang, Y.; Wen, W.; Wu, S.; Wang, C.; Yu, Z.; Guo, X.; Zhao, C. Maize Plant Phenotyping: Comparing 3D Laser Scanning, Multi-View Stereo Reconstruction, and 3D Digitizing Estimates. Remote Sens. 2018, 11, 63. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, C.; Wu, D.; Qiao, F.; Li, W.; Duan, L.; Wang, K.; Xiao, Y.; Chen, G.; Liu, Q.; et al. High-Throughput Phenotyping and QTL Mapping Reveals the Genetic Architecture of Maize Plant Growth. Plant Physiol. 2017, 173, 1554–1564. [Google Scholar] [CrossRef]
- Cabrera-Bosquet, L.; Fournier, C.; Brichet, N.; Welcker, C.; Suard, B.; Tardieu, F. High-throughput estimation of incident light, light interception and radiation-use efficiency of thousands of plants in a phenotyping platform. New Phytol. 2016, 212, 269–281. [Google Scholar] [CrossRef]
- Guo, Q.; Wu, F.; Pang, S.; Zhao, X.; Chen, L.; Liu, J.; Xue, B.; Xu, G.; Li, L.; Jing, H.; et al. Crop 3D—A LiDAR based platform for 3D high-throughput crop phenotyping. Sci. China Life Sci. 2018, 61, 328–339. [Google Scholar] [CrossRef]
- Young, S.N.; Kayacan, E.; Peschel, J.M. Design and field evaluation of a ground robot for high-throughput phenotyping of energy sorghum. Precis. Agric. 2019, 20, 697–722. [Google Scholar] [CrossRef]
- Leotta, M.J.; Vandergon, A.; Taubin, G. Interactive 3D Scanning Without Tracking. In Proceedings of the XX Brazilian Symposium on Computer Graphics and Image Processing (SIBGRAPI 2007), Minas Gerais, Brazil, 7–10 October 2007; pp. 205–212. [Google Scholar] [CrossRef]
- Quan, L.; Wang, J.; Tan, P.; Yuan, L. Image-based modeling by joint segmentation. Int. J. Comput. Vis. 2007, 75, 135–150. [Google Scholar] [CrossRef]
- Pollefeys, M.; Koch, R.; Vergauwen, M.; Van Gool, L. An automatic method for acquiring 3D models from photographs: Applications to an archaeological site. In Proceedings of the ISPRS International Workshop on Photogrammetric Measurements, Object Modeling and Documentation in Architecture and Industry, Thessaloniki, Greece, 7–9 July 1999. [Google Scholar]
- Leiva, F.; Vallenback, P.; Ekblad, T.; Johansson, E.; Chawade, A. Phenocave: An Automated, Standalone, and Affordable Phenotyping System for Controlled Growth Conditions. Plants 2021, 10, 1817. [Google Scholar] [CrossRef] [PubMed]
- Murcia, H.F.; Tilaguy, S.; Ouazaa, S. Development of a Low-Cost System for 3D Orchard Mapping Integrating UGV and LiDAR. Plants 2021, 10, 2804. [Google Scholar] [CrossRef]
- Murcia, H.; Sanabria, D.; Méndez, D.; Forero, M.G. A Comparative Study of 3D Plant Modeling Systems Based on Low-Cost 2D LiDAR and Kinect. In Proceedings of the Mexican Conference on Pattern Recognition, Mexico City, Mexico, 23–26 June 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 272–281. [Google Scholar] [CrossRef]
- Brichet, N.; Fournier, C.; Turc, O.; Strauss, O.; Artzet, S.; Pradal, C.; Welcker, C.; Tardieu, F.; Cabrera-Bosquet, L. A robot-assisted imaging pipeline for tracking the growths of maize ear and silks in a high-throughput phenotyping platform. Plant Methods 2017, 13, 96. [Google Scholar] [CrossRef]
- Reiser, D.; Vázquez-Arellano, M.; Paraforos, D.S.; Garrido-Izard, M.; Griepentrog, H.W. Iterative individual plant clustering in maize with assembled 2D LiDAR data. Comput. Ind. 2018, 99, 42–52. [Google Scholar] [CrossRef]
- Vázquez-Arellano, M.; Reiser, D.; Paraforos, D.S.; Garrido-Izard, M.; Burce, M.E.C.; Griepentrog, H.W. 3-D reconstruction of maize plants using a time-of-flight camera. Comput. Electron. Agric. 2018, 145, 235–247. [Google Scholar] [CrossRef]
- Vázquez-Arellano, M.; Paraforos, D.S.; Reiser, D.; Garrido-Izard, M.; Griepentrog, H.W. Determination of stem position and height of reconstructed maize plants using a time-of-flight camera. Comput. Electron. Agric. 2018, 154, 276–288. [Google Scholar] [CrossRef]
- Bao, Y.; Tang, L.; Srinivasan, S.; Schnable, P.S. Field-based architectural traits characterisation of maize plant using time-of-flight 3D imaging. Biosyst. Eng. 2019, 178, 86–101. [Google Scholar] [CrossRef]
- Qiu, Q.; Sun, N.; Bai, H.; Wang, N.; Fan, Z.; Wang, Y.; Meng, Z.; Li, B.; Cong, Y. Field-Based High-Throughput Phenotyping for Maize Plant Using 3D LiDAR Point Cloud Generated With a “Phenomobile”. Front. Plant Sci. 2019, 10, 554. [Google Scholar] [CrossRef]
- McCormick, R.F.; Truong, S.K.; Mullet, J.E. 3D sorghum reconstructions from depth images identify QTL regulating shoot architecture. Plant Physiol. 2016, 172, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Paulus, S.; Schumann, H.; Kuhlmann, H.; Léon, J. High-precision laser scanning system for capturing 3D plant architecture and analysing growth of cereal plants. Biosyst. Eng. 2014, 121, 1–11. [Google Scholar] [CrossRef]
- Thapa, S.; Zhu, F.; Walia, H.; Yu, H.; Ge, Y. A Novel LiDAR-Based Instrument for High-Throughput, 3D Measurement of Morphological Traits in Maize and Sorghum. Sensors 2018, 18, 1187. [Google Scholar] [CrossRef]
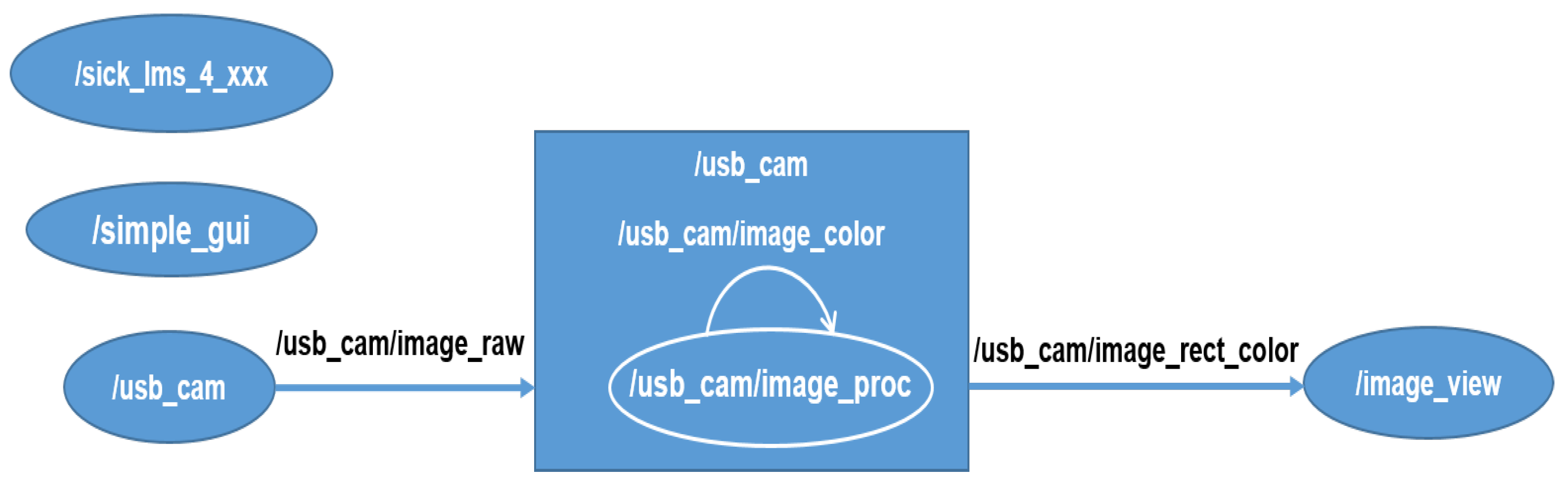
- Lehning, M.; SICK. sick_scan. Available online: https://github.com/SICKAG/sick_scan (accessed on 6 October 2021).
- Pitzer, B.; Toris, R. usb_cam. Available online: https://github.com/ros-drivers/usb_cam (accessed on 6 October 2021).
- Balta, H.; Velagic, J.; Bosschaerts, W.; De Cubber, G.; Siciliano, B. Fast statistical outlier removal based method for large 3D point clouds of outdoor environments. IFAC-PapersOnLine 2018, 51, 348–353. [Google Scholar] [CrossRef]
- Gelard, W.; Devy, M.; Herbulot, A.; Burger, P. Model-based segmentation of 3D point clouds for phenotyping sunflower plants. In Proceedings of the 12 International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications, Porto, Portugal, 27 February 2017. [Google Scholar]
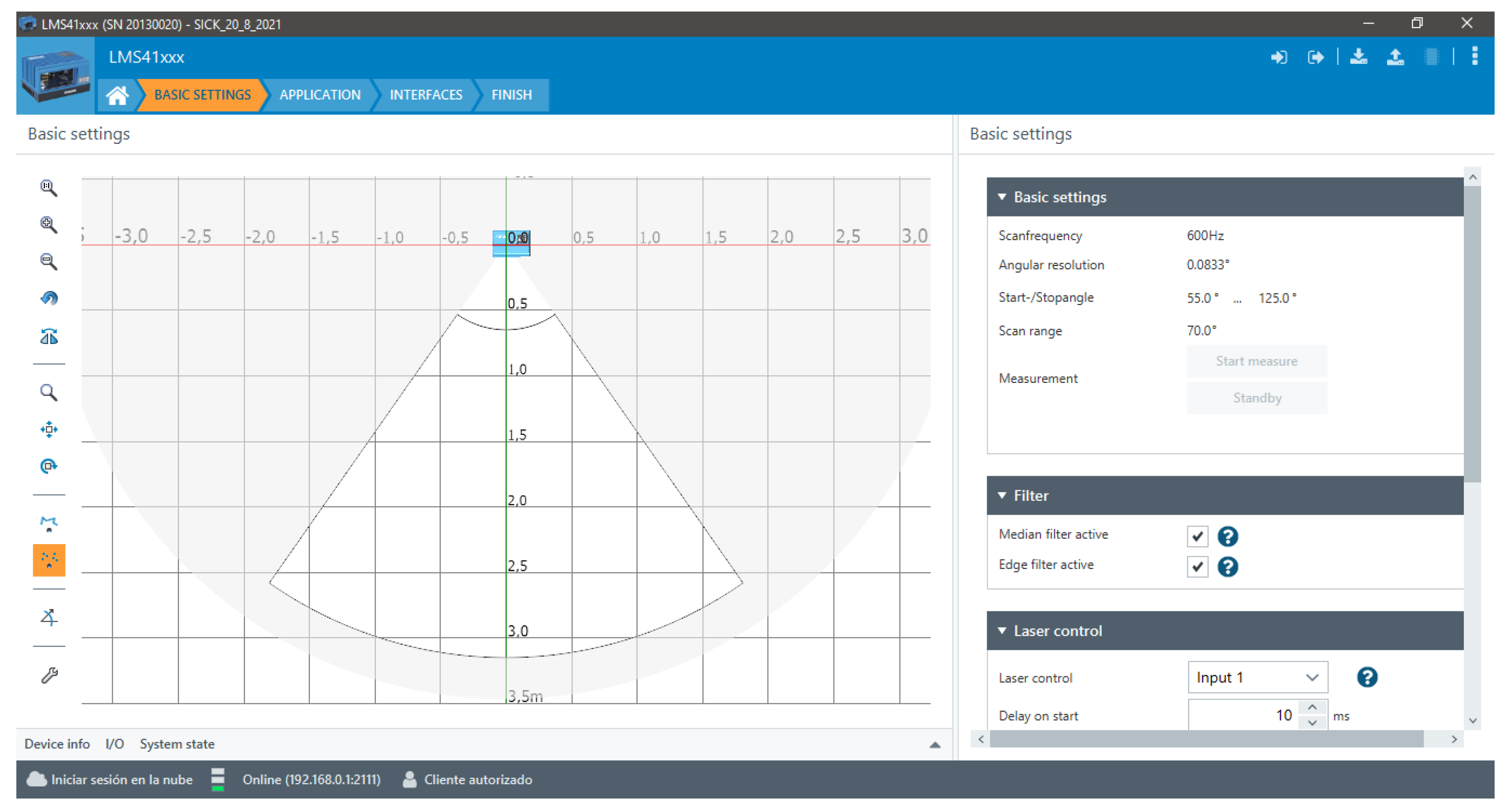


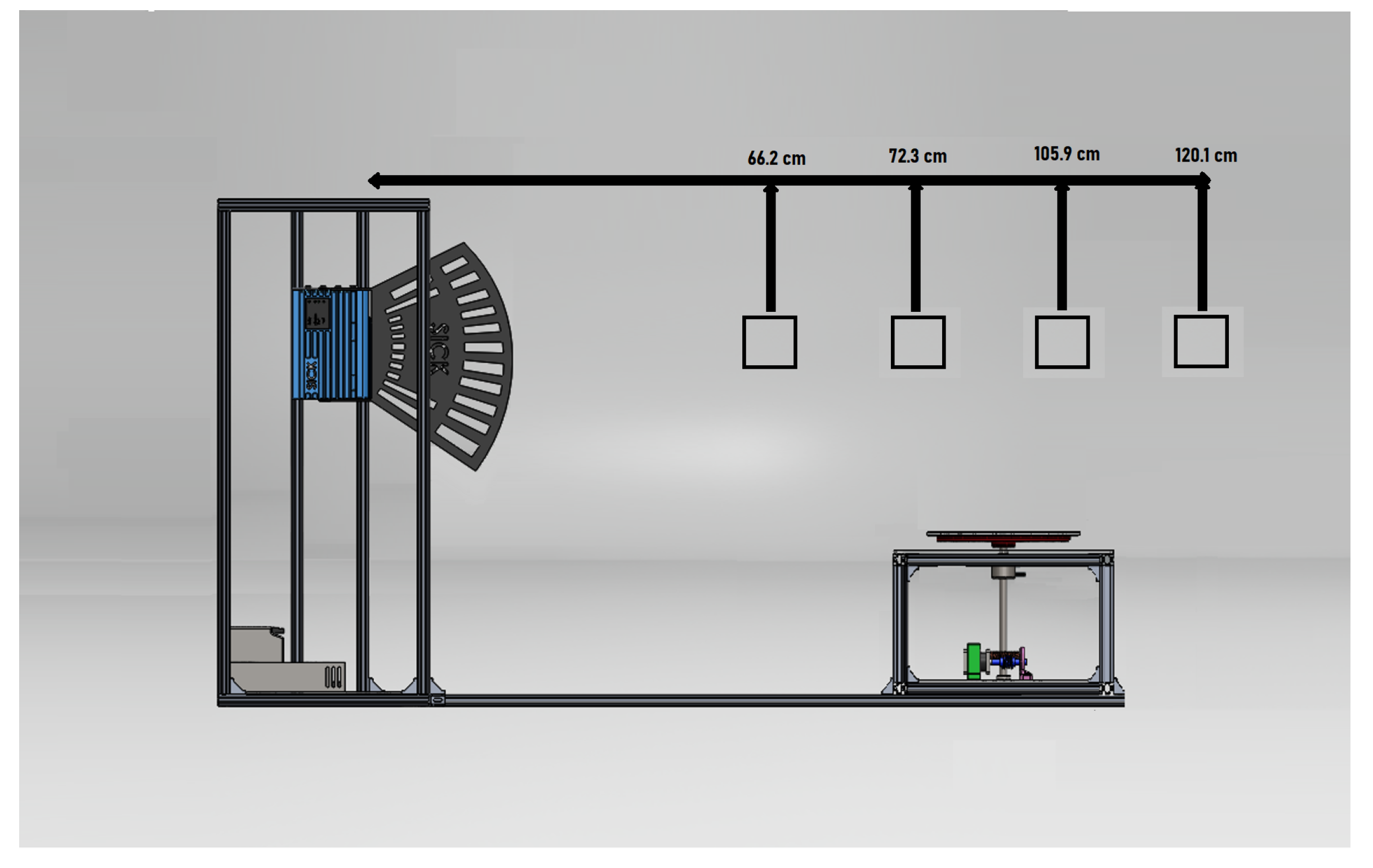

| Feature | LMS4121R-13000 |
|---|---|
| Application | Indoor |
| Reading field | Front |
| Light source | Visible red light |
| Laser class | 2 (IEC 60825-1:2014, EN 60825-1:2014) |
| Aperture angle | 70° |
| Scanning frequency | 600 Hz |
| Angular resolution | 0.0833° |
| Working range | 70 cm … 300 cm |
| Ground Truth | Point Cloud | Error Calculation | |||||
|---|---|---|---|---|---|---|---|
| Coord. | ID | [cm] | [cm] | Indiv. Error [%] | Prom. Error [%] | ||
| X | U1 | 5.5 | 5.4383 | 1.1218 | 1.6773 | 0.0134 | 0.0002 |
| U2 | 5.5 | 5.5573 | 1.0418 | 0.1056 | 0.0111 | ||
| D1 | 5.5 | 5.3808 | 2.1673 | 0.0709 | 0.0050 | ||
| D2 | 5.5 | 5.3692 | 2.3782 | 0.0825 | 0.0068 | ||
| Y | U3 | 5.5 | 5.5431 | 0.7836 | 2.1418 | 0.0914 | 0.0084 |
| U4 | 5.5 | 5.6141 | 2.0745 | 0.1624 | 0.0264 | ||
| D3 | 5.5 | 5.3308 | 3.0764 | 0.1209 | 0.0146 | ||
| D4 | 5.5 | 5.3552 | 2.6327 | 0.0965 | 0.0093 | ||
| Z | L1 | 5.5 | 5.4363 | 1.1582 | 1.6755 | 0.0154 | 0.0002 |
| L2 | 5.5 | 5.4902 | 0.1782 | 0.0385 | 0.0015 | ||
| L3 | 5.5 | 5.3051 | 3.5436 | 0.1466 | 0.0215 | ||
| L4 | 5.5 | 5.6002 | 1.8218 | 0.1485 | 0.0220 | ||
| Accuracy [%] | 1.8315 | ||||||
| 5.4517 | 0.1271 | ||||||
| Abs. Error [cm] | 0.0310 | ||||||
| Campaign | Date | # Plants | Link |
|---|---|---|---|
| First | 7 July 2021 to 21 October 2021 | 21 | https://osf.io/fcgwk/ |
| Second | 21 October 2021 to 12 November 2021 | 40 | https://osf.io/x5cn9/ |
| Third | 2 February 2022 to 18 February 2022 | 45 | https://osf.io/5vykw/ |
| Fourth | 28 February 2022 to 23 March 2022 | 80 | https://osf.io/ks7my/ |
| Fifth | 28 March 2022 to 9 April 2022 | 55 | https://osf.io/tnhxy/ |
| Sixth | 25 April 2022 to 17 May 2022 | 80 | https://osf.io/h63jq/ |
| Seventh | 23 May 2022 to 14 June 2022 | 41 | https://osf.io/7uvm4/ |
| Maiz08 | |
|---|---|
| TAP (h) | Volume (cm) |
| 147.75 | 1.2068 |
| 167.91 | 1.5688 |
| 176.96 | 3.6859 |
| 192.02 | 7.3712 |
| 200.48 | 11.5586 |
| 297.30 | 19.7802 |
| 321.05 | 24.1338 |
| 345.16 | 38.5767 |
| 384.02 | 67.4274 |
| 432.32 | 100.5300 |
| 456.84 | 107.0390 |
| Name (Campaign 6) | Accuracy (%) | Name (Campaign 6) | Accuracy (%) |
|---|---|---|---|
| 01_01 | 93.44 | 05_11 | 93.34 |
| 01_02 | 89.16 | 06_01 | 98.92 |
| 01_03 | 97.97 | 06_02 | 87.41 |
| 01_04 | 89.03 | 06_03 | 94.10 |
| 01_05 | 78.71 | 06_04 | 87.31 |
| 01_06 | 93.69 | 06_05 | 98.02 |
| 01_07 | 83.61 | 06_06 | 86.13 |
| 01_08 | 91.26 | 06_07 | 89.54 |
| 01_09 | 95.12 | 06_08 | 82.57 |
| 01_10 | 92.23 | 06_09 | 78.65 |
| 01_11 | 91.63 | 06_10 | 79.32 |
| 02_04 | 90.34 | 06_11 | 91.80 |
| 02_05 | 88.81 | 09_01 | 90.91 |
| 02_06 | 86.30 | 09_02 | 96.38 |
| 04_01 | 95.75 | 09_03 | 95.66 |
| 04_02 | 94.59 | 09_04 | 90.42 |
| 04_03 | 91.96 | 09_05 | 93.55 |
| 04_04 | 96.02 | 09_06 | 94.53 |
| 04_05 | 84.09 | 09_07 | 89.51 |
| 04_06 | 97.00 | 09_08 | 94.05 |
| 04_07 | 95.52 | 09_09 | 95.33 |
| 04_08 | 89.24 | 09_10 | 90.12 |
| 04_09 | 98.79 | 09_11 | 94.18 |
| 04_10 | 92.25 | 09_12 | 94.64 |
| 04_11 | 90.64 | 12_01 | 90.00 |
| 04_12 | 87.11 | 12_02 | 87.63 |
| 05_01 | 79.91 | 12_03 | 81.86 |
| 05_02 | 88.58 | 12_04 | 90.02 |
| 05_03 | 90.22 | 12_05 | 89.71 |
| 05_04 | 91.42 | 12_06 | 91.28 |
| 05_05 | 93.28 | 12_07 | 86.62 |
| 05_06 | 95.22 | 12_08 | 85.76 |
| 05_07 | 86.77 | 12_09 | 90.09 |
| 05_08 | 94.34 | 12_10 | 90.77 |
| 05_10 | 96.21 | 12_11 | 96.06 |
| Average accuracy = 89.41% | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forero, M.G.; Murcia, H.F.; Méndez, D.; Betancourt-Lozano, J. LiDAR Platform for Acquisition of 3D Plant Phenotyping Database. Plants 2022, 11, 2199. https://doi.org/10.3390/plants11172199
Forero MG, Murcia HF, Méndez D, Betancourt-Lozano J. LiDAR Platform for Acquisition of 3D Plant Phenotyping Database. Plants. 2022; 11(17):2199. https://doi.org/10.3390/plants11172199
Chicago/Turabian StyleForero, Manuel G., Harold F. Murcia, Dehyro Méndez, and Juan Betancourt-Lozano. 2022. "LiDAR Platform for Acquisition of 3D Plant Phenotyping Database" Plants 11, no. 17: 2199. https://doi.org/10.3390/plants11172199
APA StyleForero, M. G., Murcia, H. F., Méndez, D., & Betancourt-Lozano, J. (2022). LiDAR Platform for Acquisition of 3D Plant Phenotyping Database. Plants, 11(17), 2199. https://doi.org/10.3390/plants11172199






