Advances in the Genetic Basis and Molecular Mechanism of Lesion Mimic Formation in Rice
Abstract
:1. Introduction
2. Discovery, Excavation, and Classification of Rice Lesion Mimic Mutants
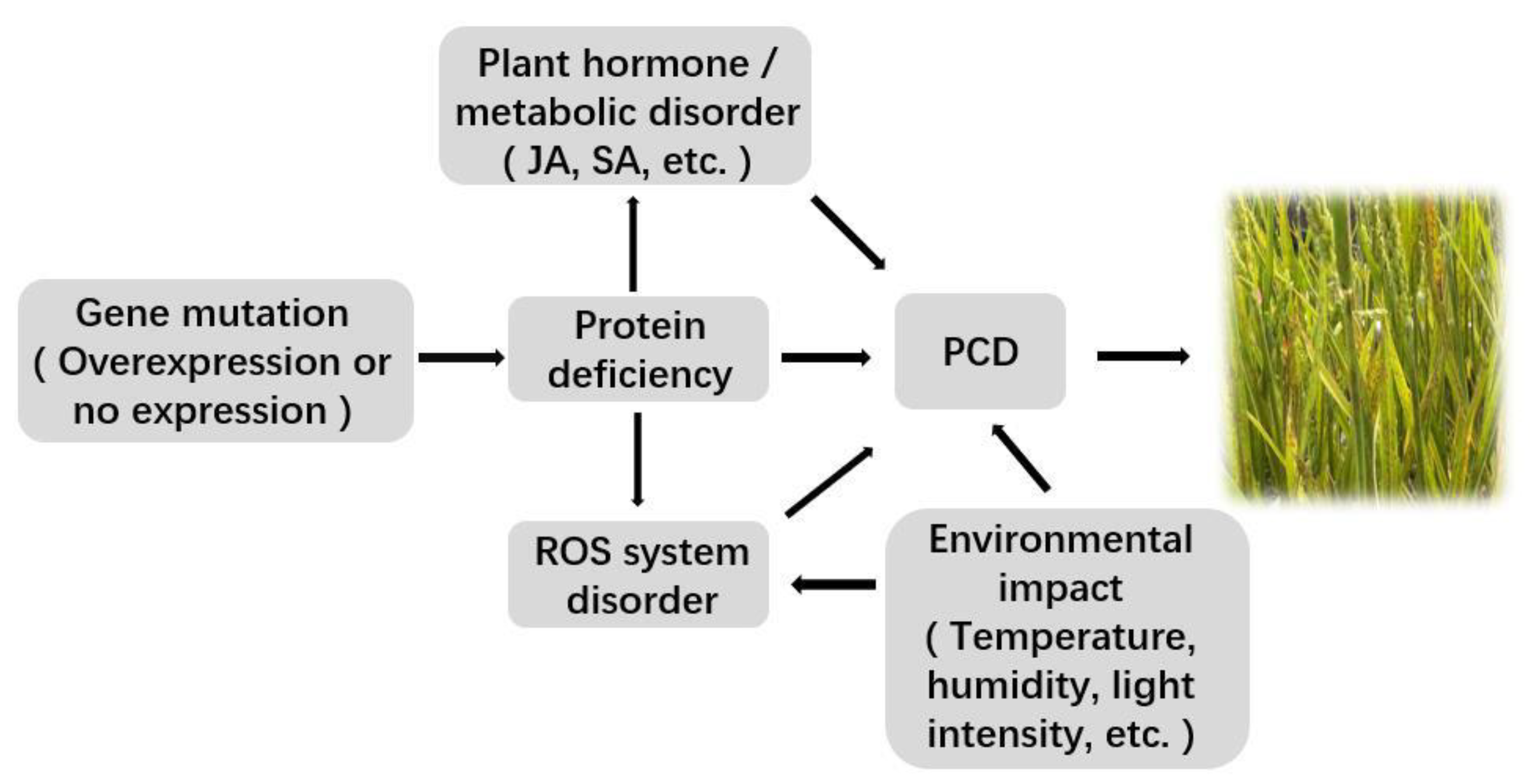
3. Mechanism of Rice Lesion Occurrence
3.1. Related Gene Mutation or Expression Change
3.2. Accumulation of ROS
3.3. Effects of Plant Hormones
3.4. Disorder of Plant Metabolic Pathways
3.5. Uncontrolled PCD
3.6. Influence of the External Environment
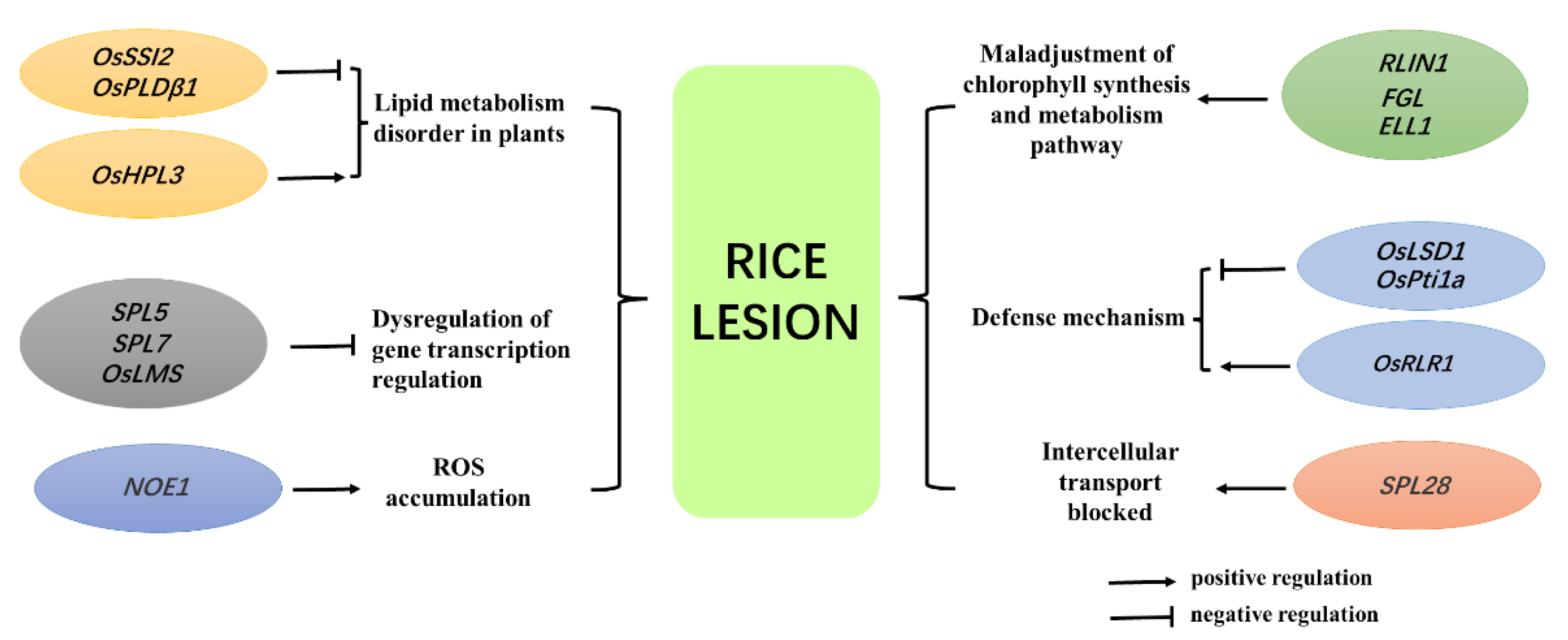
4. Identification, Cloning, and Functional Analysis of Rice Lesion Mimic Genes
4.1. Plant Lipid Metabolism Pathway
4.2. Gene Transcription Regulation Pathway
4.3. Chlorophyll Metabolism Pathway
4.4. Plant Defense Response Pathway
5. Disease Resistance of Rice Lesion Mimic Mutants
5.1. Lesion Mimic Genes Involved in Plant Hormone Regulation
5.2. Lesion Mimic Genes Involved in ROS Signaling Pathway
6. Research Prospect
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jiao, R.; Xu, N.; Hu, J.; Song, Z.L.; Hu, J.Q.; Rao, Y.C.; Wang, Y.X. Research progress on traits and molecular mechanisms of rice lesion mimic mutants. Chin. J. Rice Sci. 2018, 32, 285–295. [Google Scholar]
- Matin, M.N.; Pandeya, D.; Baek, K.H.; Dong, S.L.; Lee, J.H.; Kang, H.; Kang, S.G. Phenotypic and genotypic analysis of rice lesion mimic mutants. Plant Pathol. J. 2010, 26, 159–169. [Google Scholar] [CrossRef] [Green Version]
- Xia, C.Y.; Yan, B.Y.; Cai, Y.F.; Wang, H.H.; Gong, Y.H. Advances in programmed cell death in plants. Lett. Biotechnol. 2008, 19, 296–298. [Google Scholar]
- Huang, Q.N.; Yang, Y.; Shi, Y.F.; Chen, J.; Wu, J.L. Research progress on spot leaf variation in rice. Chin. J. Rice Sci. 2010, 24, 108–115. [Google Scholar]
- McGrann, G.R.; Steed, A.; Burt, C.; Paul, N.; Brown, J.K. Differential effects of lesion mimic mutants in barley on disease development by facultative pathogens. J. Exp. Bot. 2015, 66, 3417–3428. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Dietrich, R.A.; Richberg, M.H. Death Don’t Have No Mercy: Cell Death Programs in Plant-Microbe Interactions. Plant Cell 1996, 8, 1793–1807. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Liu, G.; Wang, L.; Zhou, Z.; Leung, H.; Wang, G.L.; He, C. Physical mapping of a rice lesion mimic gene, Spl1, to a 70-kb segment of rice chromosome 12. Mol. Genet. Genom. 2004, 272, 108–115. [Google Scholar] [CrossRef]
- Yamanouchi, U.; Yano, M.; Lin, H.; Ashikari, M.; Yamada, K. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. USA 2002, 99, 7530–7535. [Google Scholar] [CrossRef] [Green Version]
- Hoang, T.V.; Vo, K.T.X.; Rahman, M.M.; Choi, S.H.; Jeon, J.S. Heat stress transcription factor OsSPL7 plays a critical role in reactive oxygen species balance and stress responses in rice. Plant Sci. 2019, 289, 110273. [Google Scholar] [CrossRef]
- Bai, J.T.; Zhu, X.D.; Wang, Q.; Zhang, J.; Chen, H.Q.; Dong, G.J.; Zhu, L.; Zheng, H.K.; Xie, Q.J.; Nian, J.Q.; et al. Rice TUTOU1 encodes a suppressor of camp receptor-like protein that is important for actin organization and panicle development. Plant Physiol. 2015, 169, 1179. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Chen, Z.J.; Tian, D.G.; Yang, G.K.; Yang, S.H.; Liu, H.Q.; Chen, S.B.; Wang, F. Identification and gene mapping of a lesion mimic and senescence mutant lms1 in Rice. J. Fujian Agric. Sci. 2014, 29, 29–34. [Google Scholar]
- Han, X.Y.; Yang, Y.; Yu, C.L.; Zhang, W.H.; Ye, S.H.; Chen, B.; Chen, C.; Cheng, Y.; Yan, C.Q.; Chen, J.P. Proteomics study of an enhanced disease-resistant mutant of rice. Chin. J. Rice Sci. 2014, 28, 559–569. [Google Scholar]
- Wu, C.; Bordeos, A.; Madamba, M.R.; Baraoidan, M.; Ramos, M.; Wang, G.L.; Leach, J.E.; Leung, H. Rice lesion mimic mutants with enhanced resistance to diseases. Mol. Genet. Genom. 2008, 279, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Zhu, X.D.; Wang, L.Y.; Zhang, L.H.; Xue, Q.Z.; He, Z.H. Physiological and genetic analysis of rice lesion mimic mutants. J. Plant Physiol. Mol. Biol. 2004, 30, 331–338. [Google Scholar]
- Wang, Z.H.; Jia, Y.L. Induction and preliminary analysis of rice lesion mutant lmm1. J. Nucl. Agric. Sci. 2006, 20, 255–258. [Google Scholar]
- Tang, J.; Zhu, X.; Wang, Y.; Liu, L.; Xu, B.; Li, F.; Fang, J.; Chu, C. Semi-dominant mutations in the CC-NB-LRR-type R gene, NLS1, lead to constitutive activation of defense responses in rice. Plant J. 2011, 66, 996–1007. [Google Scholar] [CrossRef]
- Mori, M.; Tomita, C.; Sugimoto, K.; Hasegawa, M.; Hayashi, N.; Dubouzet, J.G.; Ochiai, H.; Sekimoto, H.; Hirochika, H.; Kikuchi, S. Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol. Biol. 2007, 63, 847–860. [Google Scholar] [CrossRef]
- Shirano, Y.; Kachroo, P.; Shah, J.; Klessig, D.F. A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 2002, 14, 3149–3162. [Google Scholar] [CrossRef]
- Danon, A.; Miersch, O.; Felix, G.; Camp, R.G.; Apei, K. Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J. 2005, 41, 68–80. [Google Scholar] [CrossRef]
- Jin, B.; Zhou, X.; Jiang, B.; Gu, Z.; Zhang, P.; Qian, Q.; Chen, X.; Ma, B. Transcriptome profiling of the spl5 mutant reveals that SPL5 has a negative role in the biosynthesis of serotonin for rice disease resistance. Rice 2015, 8, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, A.; Wang, Y.; Tang, J.; Xue, P.; Li, C.; Liu, L.; Hu, B.; Yang, F.; Loake, G.J.; Chu, C. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 2011, 158, 451–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, X. SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1998, 1, 316–323. [Google Scholar] [CrossRef]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14 (Suppl. 1), S153–S164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Liu, H.; Yuan, B.; Li, X.; Xu, C.; Wang, S. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ. 2011, 34, 179–191. [Google Scholar] [CrossRef]
- Jiang, C.J.; Shimono, M.; Maeda, S.; Inoue, H.; Mori, M.; Hasegawa, M.; Sugano, S.; Takatsuji, H. Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol. Plant-Microbe Interact. 2009, 22, 820–829. [Google Scholar] [CrossRef] [Green Version]
- Tu, R.; Wang, H.; Liu, Q.; Wang, D.; Zhou, X.; Xu, P.; Zhang, Y.; Wu, W.; Chen, D.; Cao, L. Characterization and genetic analysis of the oshpl3 rice lesion mimic mutant showing spontaneous cell death and enhanced bacterial blight resistance. Plant Physiol. Biochem. 2020, 154, 94–104. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Hong, X.; Hu, D.; Liu, C.; Yang, J.; Li, Y.; Huang, Y.; Feng, Y.; Gong, H. Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J. Exp. Bot. 2015, 66, 973–987. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Rahman, M.L.; Cho, S.; Kim, Y.; Koh, H.; Yoo, S.; Paek, N. The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J. 2013, 74, 122–133. [Google Scholar] [CrossRef]
- Qiao, Y.; Jiang, W.; Lee, J.; Park, B.S.; Choi, M.S.; Piao, R.; Woo, M.O.; Roh, J.H.; Han, L.; Paek, N.C. SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 2010, 185, 258–274. [Google Scholar] [CrossRef]
- Broker, L.E.; Kruyt, F.A.; Giaccone, G. Cell Death Independent of Caspases: A Review. Clin. Cancer Res. 2005, 11, 3155–3162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, L.R.; Qu, S.; Bordeos, A.; Yang, C.; Baraoidan, M.; Yan, H.; Xie, Q.; Nahm, B.H.; Leung, H.; Wang, G.L. Spotted leaf 11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 2004, 16, 2795–2808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Li, j.; Ma, L.; Xue, Y.; Yin, P.; Xiao, J.; Wang, S. Pathogen-inducible OsMPKK10.2-OsMPK6 cascade phosphorylates the Raf-like kinase OsEDR1 and inhibits its scaffold function to promote rice disease resistance. Mol. Plant 2021, 14, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Manosalva, P.M.; Bruce, M.; Leach, J.E. Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance. Plant J. 2011, 68, 777–787. [Google Scholar] [CrossRef]
- Wang, L.; Pei, Z.; Tian, Y.; He, C. OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol. Plant-Microbe Interact. 2005, 18, 375–384. [Google Scholar] [CrossRef] [Green Version]
- Xia, S.S.; Cui, Y.; Li, F.F.; Tan, J.; Ling, Y.H. Phenotypic identification and gene mapping of premature senescence mutant lmps1 in rice. Acta Agron. Sin. 2019, 45, 46–54. [Google Scholar] [CrossRef]
- Wang, J.J.; Zhang, L.X.; Wang, L.Y.; Zhang, L.H.; Zhu, C.N.; He, Z.H.; Jin, Q.S.; Fan, H.H.; Xin, Y. Induced responses of rice Lesion Resembling Disease mutants to light and temperature. Sci. Agric. Sin. 2010, 43, 2039–2044. [Google Scholar]
- Yuan, Y.; Zhong, S.; Li, Q.; Zhu, Z.; Lou, Y.; Wang, L.; Wang, J.; Wang, M.; Li, Q.; Yang, D.; et al. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 2007, 5, 313–324. [Google Scholar] [CrossRef]
- Takahashi, A.; Agrawal, G.K.; Yamazaki, M.; Onosato, K.; Miyao, A.; Kawasaki, T.; Shimamoto, K.; Hirochika, H. Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell 2007, 19, 2940–2951. [Google Scholar] [CrossRef] [Green Version]
- Park, C.J.; Peng, Y.; Chen, X.; Dardick, C.; Ruan, D.; Bart, R.; Canlas, P.E.; Ronald, P.C. Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PLoS Biol. 2008, 6, e231. [Google Scholar]
- Yamaguchi, T.; Kuroda, M.; Yamakawa, H.; Ashizawa, H.; Hirayae, K.; Kurimoto, L.; Shinya, T.; Shibuya, N. Suppression of a phospholipase D gene, OsPLDβ1, activates defense responses and increases disease resistance in rice. Plant Physiol. 2009, 150, 308–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, T.; Maisonneuve, S.; Isshiki, M.; Mizutani, M.; Chen, L.; Wong, H.L.; Kawasaki, T.; Shimamoto, K. Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 2010, 285, 11308–11313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Peng, Y.; Zhang, Q.; Xia, S.; Ruan, B.; Xu, Q.; Yu, X.; Zhou, T.; Liu, H.; Zeng, D.; et al. Disruption of EARLY LESION LEAF 1, encoding a cytochrome P450 monooxygenase, induces ROS accumulation and cell death in rice. Plant J. 2021, 105, 942–956. [Google Scholar] [CrossRef]
- Sun, C.; Liu, L.; Tang, J.; Lin, A.; Zhang, F.; Fang, J.; Zhang, G.; Chu, C. RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J. Genet. Genom. 2011, 38, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hao, L.; Pan, J.; Zheng, X.; Jiang, G.; Yang, J.; Gu, Z.; Qian, Q.; Zhai, W.; Ma, B. SPL5, a cell death and defense-related gene, encodes a putative splicing factor 3b subunit 3 (SF3b3) in rice. Mol. Breed. 2012, 30, 939–949. [Google Scholar] [CrossRef]
- Liu, X.; Li, F.; Tang, J.; Wang, W.; Zhang, F.; Wang, G.; Chu, J.; Yan, C.; Wang, T.; Chu, C.; et al. Activation of the Jasmonic Acid Pathway by Depletion of the Hydroperoxide Lyase OsHPL3 Reveals Crosstalk between the HPL and AOS Branches of the Oxylipin Pathway in Rice. PLoS ONE 2012, 7, e50089. [Google Scholar] [CrossRef]
- Undan, J.R.; Tamiru, M.; Abe, A.; Yoshida, K.; Kosugi, S.; Takagi, H.; Yoshida, K.; Kanzaki, H.; Saitoh, H.; Fekih, R.; et al. Mutation in OsLMS, a gene encoding a protein with two double-stranded RNA binding motifs, causes lesion mimic phenotype and early senescence in rice (Oryza sativa L.). Genes Genet. Syst. 2012, 87, 169–179. [Google Scholar] [CrossRef] [Green Version]
- Vega-Sánchez, M.E.; Verhertbruggen, Y.; Christensen, U.; Chen, X.; Sharma, V.; Varanasi, P.; Jobling, S.A.; Talbot, M.; White, R.G.; Joo, M.; et al. Loss of Cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 2012, 159, 56–69. [Google Scholar] [CrossRef] [Green Version]
- Jiao, B.B.; Wang, J.J.; Zhu, X.D.; Zeng, L.J.; Li, Q.; He, Z.H. A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol. Plant 2012, 5, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Fekih, R.; Tamiru, M.; Kanzaki, H.; Abe, A.; Yoshida, K.; Kanzaki, E.; Saitoh, H.; Takagi, H.; Natsume, S.; Undan, J.R.; et al. The rice (Oryza sativa L.) LESION MIMIC RESEMBLING, which encodes an AAA-type ATPase, is implicated in defense response. Mol. Genet. Genom. 2015, 290, 611–622. [Google Scholar] [CrossRef]
- You, Q.; Zhai, K.; Yang, D.; Yang, W.; Wu, J.; Liu, J.; Pan, W.; Wang, J.; Zhu, A.; Jian, Y.; et al. An E3 Ubiquitin Ligase-BAG Protein Module Controls Plant Innate Immunity and Broad-Spectrum Disease Resistance. Cell Host Microbe 2016, 20, 758–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Gong, P.; Li, K.; Huang, F.; Cheng, F.; Pan, G. A single cytosine deletion in the OsPLS1 gene encoding vacuolar-type H+-ATPase subunit A1 leads to premature leaf senescence and seed dormancy in rice. J. Exp. Bot. 2016, 67, 2761–2776. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Wang, Y.; Liu, L.L.; Wang, C.; Gan, T.; Zhang, Z.; Wang, Y.; Wang, D.; Niu, M.; Long, W.; et al. Isolation and characterization of a spotted leaf 32 mutant with early leaf senescence and enhanced defense response in rice. Sci. Rep. 2017, 7, 41846. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Lei, C.; Wang, J.; Ma, J.; Tang, S.; Wang, C.; Zhao, K.; Tian, P.; Zhang, H.; Qi, C.; et al. SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J. Exp. Bot. 2017, 68, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ning, Y.; Zhang, Y.; Yu, N.; Zhao, C.; Zhan, X.; Wu, W.; Chen, D.; Wei, X.; Wang, G.L.; et al. OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell 2017, 29, 345–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, P.; Fan, S.; Deng, L.; Zhong, G.; Zhang, S.; Li, M.; Chen, W.; Wang, G.; Tu, B.; Wang, Y.; et al. LML1, encoding a conserved eukaryotic release factor 1 protein, regulates cell death and pathogen resistance by forming a conserved complex with SPL33 in rice. Plant Cell Physiol. 2018, 59, 887–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, J.; Bai, P.; Ning, Y.; Wang, J.; Shi, X.; Xiong, Y.; Zhang, K.; He, F.; Zhang, C.; Wang, R.; et al. The Monocot-Specific Receptor-like Kinase SDS2 controls cell death and immunity in rice. Cell Host Microbe 2018, 23, 498–510.e5. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Wang, Y.; Ma, X.; Meng, L.; Jing, R.; Wang, F.; Wang, S.; Cheng, Z.; Zhang, X.; Jiang, L.; et al. Disruption of gene SPL35, encoding a novel CUE domain-containing protein, leads to cell death and enhanced disease response in rice. Plant Biotechnol. J. 2019, 17, 1679–1693. [Google Scholar] [CrossRef] [Green Version]
- Sathe, A.P.; Su, X.; Chen, Z.; Chen, T.; Wei, X.; Tang, S.; Zhang, X.B.; Wu, J.L. Identification and characterization of a spotted-leaf mutant spl40 with enhanced bacterial blight resistance in rice. Rice 2019, 12, 68. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Q.; Zhang, Y.; Chen, Y.; Yu, N.; Cao, Y.; Zhan, X.; Cheng, S.; Cao, L. LMM24 Encodes Receptor-Like Cytoplasmic Kinase 109, which regulates cell death and defense responses in rice. Int. J. Mol. Sci. 2019, 20, 3243. [Google Scholar] [CrossRef] [Green Version]
- Du, D.; Zhang, C.; Xing, Y.; Lu, X.; Cai, L.; Yun, H.; Zhang, Q.; Zhang, Y.; Chen, X.; Liu, M.; et al. The CC-NB-LRR OsRLR1 mediates rice disease resistance through interaction with OsWRKY19. Plant Biotechnol. J. 2021, 19, 1052–1064. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Fujiwara, M.; Hamada, S.; Shimamoto, K.; Nomura, Y.; Nakagami, H.; Takahashi, A.; Hirochika, H. Plasma membrane localization is essential for Oryza sativa Pto-interacting protein 1a-mediated negative regulation of immune signaling in rice. Plant Physiol. 2014, 166, 327–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Wang, X.; Shu, X.; Wu, D. Research progress on signal pathways and disease resistance of plant lesion mutants. J. Nucl. Agric. Sci. 2009, 23, 631–638. [Google Scholar]
- Yoshioka, K.; Kachroo, P.; Tsui, F.; Sharma, S.B.; Shan, J.; Klessig, D.F. Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 2001, 26, 447–459. [Google Scholar] [CrossRef] [Green Version]
- Feng, B.H.; Yang, Y.; Shi, Y.F.; Shen, H.C.; Wang, H.M.; Huang, Q.N.; Xu, X.; Lü, X.G.; Wu, J.L. Characterization and genetic analysis of a novel rice spotted-leaf mutant HM47 with broad-spectrum resistance to Xanthomonas oryzae pv. oryzae. Chin. Bull. Bot. 2013, 55, 473–483. [Google Scholar] [CrossRef]
- Li, X.; Yang, D.L.; Sun, L.; Li, Q.; Mao, B.; He, Z. The systemic acquired resistance regulator OsNPR1 attenuates growth by repressing auxin signaling through promoting IAA-Amido synthase expression. Plant Physiol. 2016, 172, 546–558. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Wu, J.; Zheng, X.; Zhang, S.; Sun, X.; Qiu, Q.; Lu, T. Gene knockout study reveals that cytosolic ascorbate peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 2013, 8, e57472. [Google Scholar] [CrossRef]
- Tian, M.; Rao, L.B.; Li, J.Y. Active oxygen species in plant cells and their physiological functions. Plant Physiol. Commun. 2005, 41, 235–241. [Google Scholar]
- Torres, M.A.; Dang, J.L.; Jones, J. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Dat, J.F.; Lopezdelgado, H.; Foyer, C.H.; Scott, I.M. Effects of Salicylic Acid on Oxidative Stress and Thermotolerance in Tobacco. J. Plant Physiol. 2000, 156, 659–665. [Google Scholar] [CrossRef]
- Chamnongpol, S.; Willekens, H.; Moeder, W.; Langebartels, C.; Sandermann, H.; Montagu, M.V.; Inze, D.; Camp, W.V. Defense activation and enhanced pathogens tolerance induced by H2O2 in transgenic tobacco. Proc. Natl. Acad. Sci. USA 1998, 95, 5818–5823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Mutant Gene | Mutant Type | Accession Number | Protein Function Analysis | Reference |
|---|---|---|---|---|
| SPL7 | RILM | Os05g05304 | Heat shock transcription factor | [9] |
| SPL11 | RILM | Os12g05700 | E3 ubiquitin ligase | [32] |
| LSD1 | VILM | Os08g01595 | C2C2 zinc finger protein | [35] |
| OsNPR1(NH1) | VILM | Os01g01943 | Transcriptional coactivators | [38] |
| SPL18(OsAT1) | WLLM | Os10g01956 | Acyltransferase | [18] |
| OsPti1a | VILM | Os05g01358 | Rice protein kinase | [39] |
| XB15 | VILM | Os03g60650 | Protein phosphatase | [40] |
| OsACDR1 (OsEDR1,SPL3) | RILM | Os03g01601 | Mitogen-activated protein kinase | [33] |
| OsSSI2 | RILM | Os01g09199 | Fatty acid dehydrogenase | [26] |
| OsPLDβ1 | VILM | Os10g38060 | Phospholipase | [41] |
| SPL28 | RILM | Os01g07036 | Subunits of grid-related receptor protein complexes | [30] |
| OsSL(ELL1) | RILM | Os12g02680 | Cytochrome P450 monooxygenase | [42,43] |
| RLIN1 | WLLM | Os04g06108 | Porphyrin III oxidase | [44] |
| GF14e | WLLM | Os02g05803 | 14-3-3 protein | [34] |
| NOE1 | RILM | Os03g03910 | Catalase | [35] |
| NLS1 | RILM | Os11g14380 | CC-NB-LRR type R protein | [17] |
| SPL5 | RILM | Os07g02037 | Splicing factor 3b subunit | [45] |
| OsHPL3 | VILM | Os02g01102 | Hydroperoxide lyase | [28,46] |
| OsLMS | WLLM | Os02g06390 | RNA binding protein | [47] |
| CslF6 | VILM | Os08g01605 | Cellulose-like synthase F | [48] |
| RLS1 | VILM | Os02g10900 | NB-ARM domain protein | [49] |
| FGL | VILM | Os10g35370 | OsPORB protein, involved in cytochrome synthesis | [29] |
| SPL29 | WLLM | Os08g02069 | Acetylglucosamine pyrophosphatase | [27] |
| LMR | RILM | Os06g01300 | Adenosine triphosphatase | [50] |
| EBR1 | VILM | Os05g19970 | E3 ubiquitin ligase in RING domain | [51] |
| OsPLS1 | RILM | Os06g45120 | Vacuolar proton ATPase subunit | [52] |
| SPL32 | VILM | Os07g06584 | Ferroxin-dependent glutamate synthase | [53] |
| SPL33 | WLLM | Os01g01166 | eEF1A-like protein | [54] |
| OsCUL3a | VILM | Os02g07460 | Cullin protein | [55] |
| LML1 | WLLM | Os04g06599 | Eukaryotic Release Factor 1 Protein | [56] |
| SDS | VILM | Os01g57480 | SD-1 receptor-like kinase | [57] |
| SPL35 | WLLM | Os03g02050 | CUE domain protein | [58] |
| SPL40 | WLLM | Os05g03120 | Ribosome structural components | [59] |
| LMM24 | VILM | Os03g24930 | receptor-like cytoplasmic kinase | [60] |
| OsRLR1 | RILM | Os10g07978 | CC-NB-LRR protein | [61] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, J.; Fang, Y.; Xue, D. Advances in the Genetic Basis and Molecular Mechanism of Lesion Mimic Formation in Rice. Plants 2022, 11, 2169. https://doi.org/10.3390/plants11162169
Yan J, Fang Y, Xue D. Advances in the Genetic Basis and Molecular Mechanism of Lesion Mimic Formation in Rice. Plants. 2022; 11(16):2169. https://doi.org/10.3390/plants11162169
Chicago/Turabian StyleYan, Jiajie, Yunxia Fang, and Dawei Xue. 2022. "Advances in the Genetic Basis and Molecular Mechanism of Lesion Mimic Formation in Rice" Plants 11, no. 16: 2169. https://doi.org/10.3390/plants11162169
APA StyleYan, J., Fang, Y., & Xue, D. (2022). Advances in the Genetic Basis and Molecular Mechanism of Lesion Mimic Formation in Rice. Plants, 11(16), 2169. https://doi.org/10.3390/plants11162169







