Abstract
Plants subjected to stress need to respond rapidly and efficiently to acclimatize and survive. In this paper, we investigated a selected gene set potentially involved in early cell reprogramming in two rice genotypes with contrasting salinity tolerance (Pokkali tolerant and IR29 susceptible) in order to advance knowledge of early molecular mechanisms of rice in dealing with salt stress. Selected genes were evaluated in available transcriptomic data over a short period of 24 h and involved enzymes that avoid ROS formation (AOX, UCP and PTOX), impact ATP production (PFK, ADH and COX) or relate to the antioxidant system. Higher transcript accumulation of AOX (ROS balancing), PFK and ADH (alcohol fermentation) was detected in the tolerant genotype, while the sensitive genotype revealed higher UCP and PTOX transcript levels, indicating a predominant role for early transcription of AOX and fermentation in conferring salt stress tolerance to rice. Antioxidant gene analyses supported higher oxidative stress in IR29, with transcript increases of cytosolic CAT and SOD from all cell compartments (cytoplasm, peroxisome, chloroplast and mitochondria). In contrast, Pokkali increased mRNA levels from the AsA-GSH cycle as cytosolic/mitochondrial DHAR was involved in ascorbate recovery. In addition, these responses occurred from 2 h in IR29 and 10 h in Pokkali, indicating early but ineffective antioxidant activity in the susceptible genotype. Overall, our data suggest that AOX and ADH can play a critical role during early cell reprogramming for improving salt stress tolerance by efficiently controlling ROS formation in mitochondria. We discuss our results in relation to gene engineering and editing approaches to develop salinity-tolerant crops.
1. Introduction
Salinity is the major impediment shattering the productivity of cultivated land areas. Excessive salt accumulation causes severe ionic toxicity, increases soil compactness, reduces plants’ ability to acquire water and obstructs efficient transportation of nutrients, thus interfering with crop production and yields worldwide [1]. Rice (Oryza sativa), a model cereal crop, is a premier staple food, providing a large proportion of the human population’s food and income for billions across the globe. Rice is categorized as a typical glycophyte and is vulnerable to climate changes, thus harming food security [2,3]. However, abundant natural variability and various cultivated rice genotypes demonstrate contrasting responses to salt stress. Pokkali can withstand salinity, which is used as a positive control in screening salt-tolerant rice cultivars, while IR29 is considerably salt-sensitive and used as negative control [4]. Understanding the genes and mechanisms that regulate environmental stress in crops is critical for boosting agricultural yield and quality, safeguarding food security and even protecting important crops from extinction. Comparative analysis of stress-responding genes and their interconnected networks in rice genotypes with contrasting responses to salinity stress may lead to better comprehension of salinity-tolerating mechanisms and the identification of relevant genes for molecular breeding.
Tolerance of stresses is a complex phenomenon involving several particular gene loci with distinct regulation, molecular aspects and an array of interconnected mechanisms that maintain plant homeostasis upon exposure to hostile conditions [5]. In general, stress tolerance is linked to the maintenance of cellular redox homeostasis, regulating the levels of reactive oxygen species (ROS) required to initiate biological processes and function as signaling molecules to trigger plant defense responses [6]. Plants have different systems that act to regulate ROS formation by using alternative oxidase (AOX), uncoupling protein (UCP) and plastid terminal oxidase (PTOX) or ROS scavenging by enzymatic and non-enzymatic antioxidants.
The inner-facial mitochondrial membrane of plant cells harbors energy-dissipating alternative respiratory systems mediated by AOX. The AOX gene family in angiosperms is nucleus-encoded, composed of one to six members in two subfamilies (AOX1 and AOX2) linked to stress and housekeeping functions [7,8,9,10]. However, in monocots, the AOX2 subfamily is restricted only to some species of the Alismatales order [10], while most studies show that monocots have four or five AOX1 genes [9]. In rice, four AOX1 genes (AOX1a, 1c, 1d and 1e) have been found [9,11]; AOX1a and/or AOX1b (renamed to AOX1d) in [9] were induced by different stress conditions such as chilling, drought and high salt, while AOX1c was stably detected, and AOX1e was barely expressed in germinating seeds [9,12,13,14,15,16,17,18]. In stress conditions, AOX relaxes the highly coupled and tensed electron transport process by driving electrons from quinol to oxygen, thereby alleviating tensed conditions and reducing ROS production [19]. These characteristics may allow plants to flexibly deal with the challenge of changing scenarios and induce plasticity, facilitating plant persistence.
In addition to AOX, the plant mitochondrial inner membrane possesses UCPs. The UCPs belong to the superfamily of mitochondrial carrier proteins dissipating the proton electrochemical gradient generated by the respiratory chain complexes [20]. In plants, these proteins are involved in mitochondrial energy flow regulation. They have been suggested to play a critical role in mitigating ROS production by the mitochondrial electron transport chain [21]. Moreover, another terminal oxidase is PTOX, which is located in chloroplasts. PTOX is a key factor for maintaining the plastoquinone (PQ) pool redox balance and functions as a “safety valve” to protect photosynthesis [22]. It is a stress-responsive protein and could protect plants from various harmful stresses [23].
With changing environments, the AOX, PTOX and UCP genes show differential expression patterns and are induced by multiple signaling pathways [24,25,26]. Together, AOX, UCP and PTOX are considered primary defense lines mitigating ROS production, an excess of which causes progressive oxidative damage and ultimately cell death. Thus, these protein systems allow for flexibly dealing with the challenge of several stressors, restoring respiratory activities and correcting metabolism.
Cellular damage manifests when the delicate balance between ROS production and elimination is disturbed upon exposure to severe stress. To minimize the damaging effect of ROS, plants have developed an efficient antioxidant system with two components: enzymatic and non-enzymatic antioxidants. In plants, the non-enzymatic antioxidant ROS-scavenging pathway involves ascorbate and glutathione metabolites mediated by the ascorbate–glutathione cycle (AsA-GSH) in chloroplasts, cytosol, mitochondria and peroxisomes [27,28]. Enzymatic components of the antioxidant defense system comprise several antioxidant enzymes such as superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX), which catalyze ROS degradation; and enzymes of the ascorbate–glutathione (AsA-GSH) cycle, such as ascorbate peroxidase (APX), monodehydroascorbate reductase (MDAR), dehydroascorbate reductase (DHAR) and glutathione reductase (GR), that regenerate soluble antioxidants [29,30].
To mitigate and recover from the damaging effects of adverse environmental conditions, understanding and developing mechanisms such as effective reprogramming of a damaged cell is among the primordial needs. Induced cell reprogramming permanently facilitates plants’ immediate persistence regarding environmental factors’ variability throughout the lifetime. Consequently, it promotes individual cell growth and organism survival. Thus, early reprogramming in response to multiple individual and combined stressors could be a unique positive response across plant species and even across diverse taxonomic classes [31,32,33]. AOX has demonstrated a significant role in plant homeostasis, reprogramming and plant growth adaptation in response to diverse abiotic and biotic stresses [34,35,36,37,38]. AOX has positive physiological roles in certain developmental processes and adaptation to environmental stresses. In doing so, AOX improved the ability of cells to rapidly recover their energy status [19]. Short- and long-term fine-tuning of AOX at the transcriptional level was essential for positive performance effects [7,39]. Finally, adaptive plant robustness in the field was shown to connect to the capacity for efficient cell reprogramming, which could be measured already at the level of seeds [34,38]. Prediction of plant holobiont robustness could be linked in a technically simple way to AOX by inhibiting its activity. These tests promoted its use in seed screening for diverse species and also for low-cost on-farm seed selection, and they are awaiting broader validation [34,38,40,41,42].
Recently, our group demonstrated that transcript accumulation of genes linked to early cell reprogramming under stress related primarily to ROS/RNS balancing and energy status connected to cell restructuration and cell cycle regulation [33,43,44,45,46]. Our approach is in conformity with the view that “optimization of adaptive potential requires reconfiguration of developmental attributes to allow growth adjustment and stress avoidance” [47]. Thus, in the current study, we explored a selected gene set in public transcriptomic data of two rice cultivars with contrasting responses to salt stress (Pokkali tolerant and IR29 susceptible) during 24 h following salt stress treatment to advance knowledge on the relevance of early cell reprogramming to general plant plasticity and robustness. Pokkali is most famous for salt tolerance but is appropriate for our approach because this traditional cultivar has a broad spectrum of resilience [48,49]. Here, we focused on gene expression involved in ROS formation (AOX, UCP and PTOX), impacting ATP production (PFK, ADH and COX) and associated with the antioxidant systems (APX, MDHAR, DHAR, GR, CAT, SOD and GPX) in different cell compartments in order to gain insight into early cell reprogramming of salinity tolerance in rice. The results are discussed in relation to a connected view of redox homeostasis and energy supply as critical traits for salinity tolerance.
2. Material and Methods
2.1. Gene Expression Analyses of RNA-Seq Data
This study used publicly available RNA-seq data of two rice genotypes with contrasting responses to salt stress (Pokkali tolerant and IR29 susceptible) [50]. Both genotypes were grown in growth chambers to the three-leaf stage. The salt stress treatment was applied by watering 2-week-old seedlings with 300 mM NaCl solution or by normal watering in control plants. Shoots (stem and leaves) were harvested at 1, 2, 5, 10 and 24 h post-treatment to obtain transcriptomic data [50]. The transcriptomic data are available in SRA database from Genbank (NCBI) under the following Bioproject numbers: PRJEB4671 (Pokkali) and PRJEB4672 (IR29).
The expression analysis of target genes in transcriptomic data, with three replicates for each sample, was performed in three steps: (1) mapping of reads by the Magic-Blast software [51]; (2) quantification of mapped reads using the HTseq program [52]; and (3) normalization of read amount in all samples. Thus, in the mapping of the reads, the target cDNAs were aligned against RNA-seq data. After quantification of the mapped reads, the normalization of reads among different samples was carried out using the RPKM (reads per kilobase of transcript per million mapped reads) method [53] according to the following equation: RPKM = (number of mapped reads × 109)/(number of sequences in each database X number of nucleotides of each gene).
The target genes were associated with glycolysis, fermentation, aerobic respiration, anti-ROS formation and antioxidants. Thus, these genes included seven gene members encoding cytosolic PFK (phosphofructokinase) [54] to represent total PFK in glycolysis. Four ADH (alcohol dehydrogenase) genes [55] denoted total ADH in alcohol fermentation, while mitochondrial aerobic respiration was represented by total COX (cytochrome c oxidase), with eleven gene members [56]. In addition, total AOX (alternative oxidase) with four genes [9,11] and UCP (UCP 1 and 2) [57] were the systems involved in mitochondrial ROS formation control, while a single PTOX gene, as per Tamiru et al. [58], represented the system regulating ROS formation in the chloroplast (Table S1). Regarding antioxidant enzymes, multiple gene families of APX, MDHAR, DHAR, GR, SOD, GPX and CAT that encode proteins with different subcellular destinations, such as cytosol, peroxisomes, mitochondria and chloroplasts (Supplementary Tables S1 and S2), were evaluated. In general, all antioxidant genes presented at least one member associated with each compartment, except GPX (without members related to cytosol) and CAT (without members related to chloroplasts and mitochondria). To infer the glycolysis pathway, we analyzed genes encoding phosphofructokinase (PFK) enzyme, as it is well known that its regulation is highly influenced by the energy status of the cell.
2.2. Prediction of Subcellular Localization of Antioxidant Proteins
For the prediction of the subcellular localization from corresponding deduced antioxidant proteins (listed in Table S2), the following tools were used: TargetP-2.0. Available online: http://www.cbs.dtu.dk/services/TargetP (accessed on 20 March 2022). MitoProtII. Available online: https://ihg.gsf.de/ihg/mitoprot.html (accessed on 22 March 2022). DeepLoc-1.0. Available online: http://www.cbs.dtu.dk/services/DeepLoc (accessed on 24 March 2022) and Plant-mSubP. Available online: http://bioinfo.usu.edu/Plant-mSubP (accessed on 25 March 2022) [59,60,61]. In addition, experimental confirmation of subcellular localization was available for proteins of some gene members of APX [62,63] MDHAR [64,65,66] CAT [67], SOD [68].
2.3. Statistical Analysis
The statistical analyses of gene expression data were performed using the GraphPad Prism 9.0 software. The results were expressed as means (RPKM values) ± standard deviation (SD) from three biological replicates. The data obtained were subjected to analysis of variance (ANOVA) using the GraphPad Prism 9.0 software, and Bonferroni’s test compared averages at 5% probability.
3. Results
3.1. Tolerant Genotype Shows Elevated Transcript Levels of PFK (Glycolysis) and ADH (Fermentation)
In Figure 1, total transcript levels of PFK, ADH and COX are shown to indicate the status of glycolysis, fermentation and aerobic respiration in two rice genotypes differing in salt stress tolerance [Pokkali (tolerant) and IR29 (susceptible)].
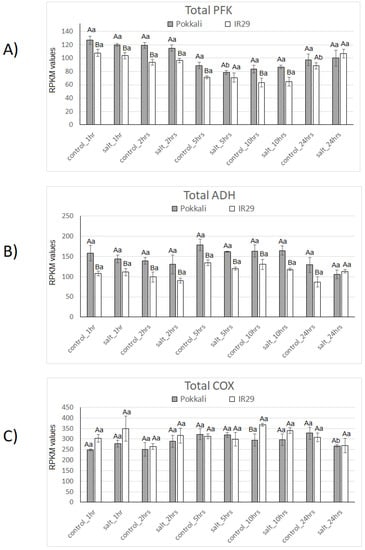
Figure 1.
Expression profiles of Total PFK (A), ADH (B) and COX (C) genes in Pokkali and IR29 genotypes of Oryza sativa under salt stress. Data represent RPKM means with standard deviations from 3 biological replicates. For each gene, different capital letters indicate significant differences (at p < 0.05) between genotypes (Pokkali and IR29), while lowercase letters designate significant differences between treatments (control and stress) at the same time point and genotype, according to Bonferroni’s test.
In general, higher mRNA levels of total PFK and ADH were observed in the tolerant genotype across the time points, with a significant difference in most cases. The only exception was the point salt stress at 24 h, in which similar levels of both transcripts were detected for Pokkali and IR29 genotypes (Figure 1A,B). Regarding the salt stress effect, a significant difference in the controls was found in PFK expression, with a decrease at 5 h in Pokkali and an increase at 24 h in IR29 (Figure 1A,B).
Interestingly, the differential gene expression pattern observed for total PFK and ADH among both rice genotypes differed from the pattern of total COX transcripts (Figure 1C). Overall, similar COX transcript levels were observed in both genotypes across all time points, except in the time point control 10 h. Significantly higher COX mRNA levels were observed in the IR29 genotype. A single significant COX mRNA decrease occurred at 24 h in the Pokkali genotype under salt stress.
3.2. Alternative Oxidase Expression Is Preponderant in Tolerant Genotype among Different ROS Formation Control Systems
The total transcript levels of AOX, UCP and PTOX, are shown in Figure 2 to provide insight into energy-dissipating systems in mitochondria (AOX and UCP) and plastids (PTOX) in rice genotypes under salt stress.
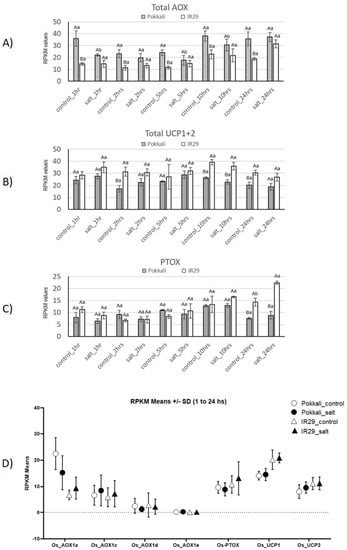
Figure 2.
Expression profiles of Total AOX (A), UCP (B) and PTOX (C) genes in Pokkali and IR29 genotypes of Oryza sativa under salt stress. Data represent RPKM (D) means with standard deviations from 3 biolog-ical replicates. For each gene, different capital letters indicate significant differences (at p < 0.05) between genotypes (Pokkali and IR29), while lowercase letters designate significant differences between treatments (control and stress) at the same time point and genotype, according to Bonferroni’s test.
Curiously, higher total AOX transcripts (Figure 2A) were observed in the tolerant (Pokkali) genotype compared to the susceptible (IR29) one, with significant data in the majority of cases. This difference can be associated with a higher expression of AOX1a (Figure 2D). Concerning the salt stress effect, a total AOX mRNA decrease (significant) was observed at times 1, 5 and 10 h in Pokkali, while some AOX mRNA increase was observed at 24 h in IR29 (not significant) (Figure 2A).
For UCP, higher mRNA levels were detected in the susceptible IR29 genotype than in the tolerant one in both control and salt conditions, although the values were not significant at most time points. No significant salt stress effect was observed (Figure 2B). This difference between genotypes can be connected with higher UCP1 expression in IR29 (Figure 2D).
Concerning PTOX, differences between the two genotypes and two treatments were found mainly at 24 h. At this time point, significantly higher PTOX mRNA levels were observed in IR29 in response to salt stress (Figure 2C).
3.3. Antioxidant Gene Expression Indicates Redox Status Compartmentalization in Rice Genotypes under Salinity
The expression analyses of different gene members of APX, MDHAR, DHAR, GR, SOD, GPX and CAT indicated the redox status of different subcellular compartments such as cytosol, peroxisomes, mitochondria and chloroplasts. In general, the main increase responses to salt stress differed between genotypes, with IR29 responding from 2 h while Pokkali responded from 10 h (Figure 3).
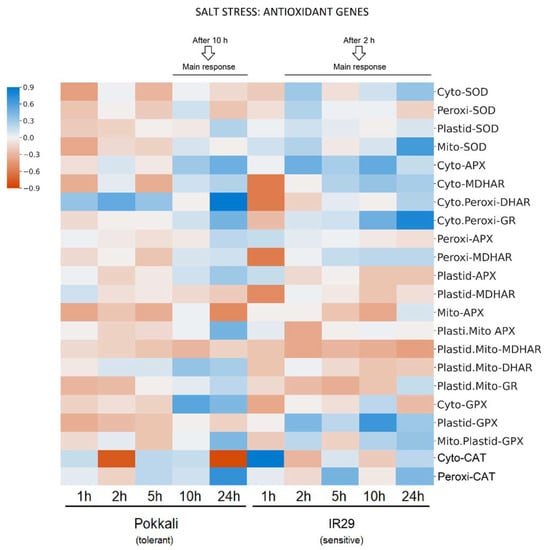
Figure 3.
Heat map showing the gene expression of antioxidant enzymes in different cellular compartments of Oryza sativa genotypes under salt stress. The analyzed genes were APX, MDHAR, DHAR, GR, SOD, GPX and CAT in Pokkali and IR29 genotypes. The data represent log2 fold changes of salt treatment values at 1, 2, 5, 10 and 24 h in relation to the respective control conditions. In heat maps, the colors blue and orange represent up- and down-regulated genes, respectively. Statistical analyses of the RPKM means with standard deviations from 3 biological replicates are show in Supplementary Tables S3 and S4.
For genes encoding proteins to the cytosol, three transcripts (APX, DHAR and SOD) presented significant changes in response to salt stress compared to the control in both genotypes. APX transcripts significantly increased in Pokkali at 10 and 24 h and in IR29 at 2 and 10 h. On the other hand, DHAR increased in Pokkali (1 to 24 h) and decreased in IR29 (1 h), while SOD decreased in Pokkali (1 and 5 h) and increased in IR29 (2 to 24 h). In addition, cytosolic CAT mRNA decreased in Pokkali and increased in IR29 (1 h). Also, cytosolic GPX mRNA increased in Pokkali (non-significant) and remained stable in IR29.
In peroxisomes, both MDHAR and DHAR transcripts significantly decreased in IR29 at 1 h, while DHAR transcripts increased in Pokkali at times 1 to 24 h. In response to salt stress, CAT transcripts increased significantly in Pokkali (24 h) and in IR29 (5 and 24 h). Also, peroxisomal SOD decreases in Pokkali and increases in IR29 were observed (not significant).
In chloroplasts, GPX revealed significant changes in response to salinity in both genotypes. GPX transcripts increased in IR29 at times 2 to 24 h and decreased in Pokkali at 1 to 24 h. In addition, plastid.mito APX transcripts increased significantly in Pokkali (24 h) and decreased in IR29 (2 h). Likewise, SOD increases were observed in both genotypes (not significant). GPX mRNA increases were observed in both genotypes (not significant). In addition, SOD increased in IR29 and decreased in Pokkali (not significant). Plastid.mito DHAR increased in Pokkali (10 and 24 h), while a slight decrease was observed in IR29.
4. Discussion
In this research, we support the hypothesis that salt tolerance in rice involves AOX linked to rapidly induced alternative energy production via glycolysis-driven aerobic fermentation. Following the current insight that transcript-level changes during early cell reprogramming can be critically relevant [33,43,44,45] for predicting later performance, we explored salt-induced transcript accumulation changes for the selected gene sets for up to 24 h. Thus, we selected transcriptomic data from two rice genotypes with known contrasting salinity stress tolerances in the field (Pokkali and IR29) treated at the seedling stage with 300 mM NaCl. Recently, our group confirmed that AOX might play a relevant role under mild stress (e.g., at watering for seed germination) and also under severe stress conditions (e.g., induction of somatic embryogenesis) and demonstrated that this was connected to temporarily enhanced fermentation [34,38]. Here, we advanced our knowledge of redox homeostasis (AOX/antioxidant enzymes) and energy supply (glycolysis/fermentation) under severe salt stress in rice. Singh et al. [69] pointed out the importance of mild salt stress as critical for the reproductive stage. However, these authors pointed also to the danger of higher salt concentrations under higher temperatures and low relative humidity when transpiration increases [70].
In fact, the higher transcript levels of AOX, PFK and ADH in the tolerant (Pokkali) compared to susceptible (IR29) genotype (Figure 1 and Figure 2) support a critical involvement of AOX and glycolysis/fermentation in salt stress tolerance. Corroborating these findings, AOX expression variation was also observed in Vigna unguiculata cultivars, contrasting in salt/drought stress tolerance [8]. In rice, generally, AOX1a and AOX1d are the stress-responsive genes (9, 12, 13, 14, 15, 16, 17, 18). However, regarding the present experiment (seedling stage under 300 mM NaCl for 24 h) the higher AOX expression in the tolerant genotype was due to AOX1a (Figure 2D). In this regard, very recently, Challabathula et al. [71] also observed this peculiarity of higher AOX1a expression in rice cultivars tolerant to drought and salinity. Among other metabolic pathways, glycolysis transcripts increased under salinity in stress-tolerant rice cultivars [72]. More recently, Bharadwaj et al. [38] showed that adaptive reprogramming during early seed germination requires enhanced fermentation, and it involves a critical role of AOX to maintain metabolic homeostasis. Also, Costa et al. [43,44,45] showed that variable ROS/RNS rebalancing and temporarily increased aerobic fermentation appear to generally combine stress-defense mechanisms in humans. These traits are connected to cell restructuration and can discriminate stress factors and distinguish genotypes of cell origins.
Furthermore, Zheng et al. [73] identified AOX pathway involvement with waterlogging tolerance in watermelon, which was associated with increased fermentation instead of aerobic respiration. They compared two contrasting genotypes, YL (tolerant) and Zaojia8424 (sensitive) and observed a strong increase of AOX and ADH transcripts in the tolerant genotype over 24 h. Also, higher AOX and ADH mRNA levels were always detected in the tolerant genotype at all analysis times (until 72 h). Considering the relevance of aerobic respiration during early hours after stress perception, these authors observed a strong decrease in COX transcripts in both genotypes over up to 24 h of stress. In rice, we detected similar COX mRNA levels in both genotypes at the majority of the evaluated time points up to 24 h (Figure 1). Overall, these data support our findings in rice genotypes, indicating a critical role of AOX in stress tolerance followed by efficient respiration and mitigating oxidative impairment in the tolerant genotype (Pokkali).
Because AOX function avoids ROS formation, it is also important to investigate the antioxidant systems among genotypes with contrasting tolerances. According to Lakra et al. [74], the Pokkali genotype has a more efficient antioxidant system than other, salt-susceptible genotypes. In our data, the susceptible IR29 genotype, overall, revealed early antioxidant response from 1 or 2 h compared to Pokkali, which responded from 10 h (Figure 3). This early response could be due to the higher oxidative stress in IR29 compared to Pokkali. In fact, SOD transcripts in IR29 increased in response to salt stress in all cell compartments, while in Pokkali SOD expression increased only in the chloroplast (Figure 3 and Figure 4), supporting disseminated O2− overproduction in IR29. In this context, it is of interest that Costa et al. [46] observed an earlier and higher transcript increase of ASC-GSH cycle genes in susceptible soybean genotypes compared to the tolerant ones in response to different biotic and abiotic stresses.
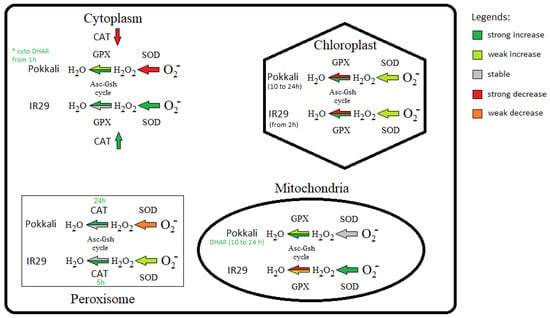
Figure 4.
Schematic representation of antioxidant enzymes in different cellular compartments of Oryza sativa genotypes under salt stress. Significant values denoted substantial transcript increases or decreases, while non-significant values indicated weak increases or decreases according to Supplementary Tables S3 and S4.
In addition, given that APX, GPX and CAT are the main H2O2 scavenging enzymes in plants [75], our data indicated that rice genotypes under salinity used different enzymatic pathways to control H2O2 concentration in cell compartments (Figure 4). Apparently, Pokkali preferably activates ASC-GSH cycle genes to scavenge H2O2 via APX from cytosol and organelles (mitochondria and chloroplasts), while IR29 seems to rather activate the ASC-GSH cycle in cytosol and GPX in organelles (Figure 3 and Figure 4). Also, the ascorbate recycling via ASC-GSH cycle appeared more active in Pokkali because cytosolic/organelles DHAR mRNA levels increased only in this genotype (Figure 3). In this regard, stress-tolerant genotypes in Glycine max also differed from susceptible genotypes increasing transcripts involved in ascorbate regeneration [46]. Some works showed that DHAR overexpression successfully conducted transgenic plants to abiotic stress tolerance, such as in response to aluminum and cold [76]. Among these enzymatic systems, APX appears to be the pivotal antioxidant enzyme to maintain the H2O2 balance because it has higher H2O2 affinity, acting the same in very slow protein concentration [77,78], whereas CAT is more associated with H2O2 detoxification [79]. Thus, while higher CAT could act in peroxisomes, scavenging toxic H2O2 in both genotypes (Figure 3 and Figure 4), the cytosolic CAT transcript decrease observed only in Pokkali suggests that cytosolic H2O2 concentration is regulated mainly by the ASC-GSH cycle via APX. However, the higher antioxidant efficiency observed in Pokkali (tolerant) can start much earlier through alternative pathway activity to avoid ROS formation, denoted by the higher AOX mRNA levels observed in this genotype compared to IR29 (susceptible). Supporting our findings, very recently, Challabathula et al. [71] observed that increased AOX1a mRNA levels with an efficient antioxidant system were essential for tolerant rice cultivars to maintain lower ROS, higher photosynthesis rates and stress tolerance.
Interestingly, higher AOX, PFK and ADH transcript levels already occur in Pokkali control plants, suggesting that this genotype could also be more resistant to other stress conditions. In this context, some studies show Pokkali as more resistant to lead (Pb) accumulation [48] or as having a similar expression profile of abiotic inducible genes in response to multiple stresses such as NaCl, ABA, polyethylene glycol (PEG) or cold (4 °C) [80]. Thus, Pokkali could have a resourceful genetic background [81] or an intrinsic environmental feature involved in stress tolerance. In this sense, recently, Sampangi-Ramaiah et al. [82] identified a salt-tolerant endophyte (Fusarium sp.,) in Pokkali, which could confer salt tolerance when colonizing the salt-sensitive rice variety IR64. Nevertheless, it was possible to detect this endophyte in the Pokkali genotype in our transcriptomic data, but in a small amount (data not shown). However, we identified Fusarium also in IR29 at a similarly low amount (data not shown). Fungal endophyte diversity in plants depends nonexclusively on genotypes and their effects on the surrounding environment. Natural environmental contexts importantly influence plant endophyte diversity, mainly in the rhizosphere, and thus, depending on agricultural management, also in experimental conditions [83]. Bharadwaj et al. [38] suggested that microbiota can provide a sink for stress-induced higher levels of sucrose and, in this way, might help to alleviate oxidative stress through overloaded mitochondria as a consequence of enhanced glycolysis. In this sense, genotype-compatible endophytes can complement fermentation and alternative respiration through their effect on maintaining host metabolic and energetic homeostasis. Furthermore, it was suggested that endophyte-born AOX genes could complement plant AOX capacities as an added value that evolved under plants’ holobiont natures [38,84,85,86]. However, preliminary observations of Bharadwaj et al. [38] indicated also that more robust plant genotypes can act more independently on microbiota assistance.
Our studies suggest that developing functional marker-assisted rice breeding or genetically engineered respectively edited rice plants by targeting AOX and glycolysis/fermentation-related genes could be among the promising strategies to confer salt stress and sustain rice productivity. However, such strategies require considering carefully the following general and specific state-of-the-art insights: (1) AOX genes have shown to be highly polymorphic in exon regions and even more pronounced in non-exon regions [86,87,88,89,90,91,92,93] In general, causative polymorphic sites within a gene were found to have low degree of conservation and phenotypic variation in a target trait can be linked to a diverse sequence polymorphisms [94]. (2) The relevance of AOX is due to its link to coordinating early plasticity provoked by continuously acting, ever-changing diverse environmental conditions, where salinity is only one among many stressors. This flexibility can be expected to rely on allelic polymorphisms [93] and flexible switching between polymorphic sites [95] depending on environmental and metabolic conditions; thus, diversity in AOX genes might be a desired trait per se; (3) as also shown in the present research, AOX is embedded in complex networking contexts [96] that evolved in unique, complex systems/organisms. This also includes the level of cells and their unique context in the plant body-shaped tissue and organ landscapes and concerns cell-free spaces (apoplasts). Thus, the species-specific role of target cells for defined agronomic traits needs to be considered [97], and (4) gene technology and gene editing have technical obstacles (reviewed, e.g, [98]) because they require in vitro culture as a first step. However, this means applying strong stress [86,91,99] and typically requires tissue and cell disruption from established networks. In contrast to functional marker-assisted selection at the seed and plant level, this bears at least the risk of undetected somaclonal variations through epigenetic and genetic side effects, which might change intrinsic, deeper phenotype characteristics of the original plant that can escape breeders’ awareness due to their focus on restricted agronomic or quality selection criteria.
In conclusion, our data support the relevant involvement of alternative pathways and glycolysis/fermentation in the more efficient stress response observed in a salt stress-tolerant rice genotype. This response is primarily associated with adaptive ROS balancing by AOX (via AOX1a expression), effective tuning of the antioxidant system and, secondarily, rapid energy production (via fermentation). Both contribute to sustaining and optimizing respiration. We cannot exclude the possibility that this intrinsic feature observed for stress-tolerance performance could have been modified by host–endophyte interactions, as we confirmed the holobiont nature of both genotypes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11162145/s1, Table S1: List of Oryza sativa genes used to advance in cell reprograming under salt stress; Table S2: Predict /experimental subcellular localization of antioxidant proteins from Oryza sativa; Table S3: Means of RPKM values ± SD (standard deviation) of antioxidant transcripts during salt stress in rice genotype (Pokkali). Statistical analysis (t test) was applied in relation to the controls (water) of each time point 1, 2, 5, 10 or 24 h. Up and down regulated genes are in green and red, respectively. Significant differences from the controls are indicated by * at p < 0.05; Table S4. Means of RPKM values ± SD (standard deviation) of antioxidant transcripts during salt stress in rice genotype (ir29). Statistical analysis (t test) was applied in relation to the controls (water) of each time point 1, 2, 5, 10 or 24 h. Up and down regulated genes are in green and red, respectively. Significant differences from the controls are indicated by * at p < 0.05.
Author Contributions
J.H.C. and S.A. conceived the basic idea and planned the study. S.A. performed data analyses, helped in writing the basic draft and prepared the final manuscript for submission. T.A.G. contributed to statistical analyses. R.d.S.M., K.L.L.T. and M.C.B. helped S.A. with basic data preparation. J.H.C. revised the data analysis and results interpretation and wrote and advanced the final manuscript. B.A.-S. coordinated final manuscript discussion and revision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The raw data used to obtain the results presented in this paper are available in SRA database from Genbank (NCBI) under bioproject numbers: PRJEB4671 (Pokkali) and PRJEB4672 (IR29).
Acknowledgments
S.A., T.A.G., K.L.L.T. and M.C.B. are grateful to CAPES for the doctoral fellowship. J.H.C. is grateful to CNPq for the researcher fellowship (CNPq grant 313899/2021-5).
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Otlewska, A.; Migliore, M.; Dybka-Stępień, K.; Manfredini, A.; Struszczyk-Świta, K.; Napoli, R.; Białkowska, A.; Canfora, L.; Pinzari, F. When salt meddles between plant, soil, and microorganisms. Front. Plant Sci. 2020, 1429. [Google Scholar] [CrossRef] [PubMed]
- Xavier, A.I.S.; Arbage, A.P.; da Silva, M.R.; Ribas, G.G.; Meus, L.D.; Santos, G.A.D.A.D.; Streck, N.A.; Zanon, A.J. Economic and productive analysis of irrigated rice crops using a multicase study. Pesqui. Agropecuária Bras. 2021, 56. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review. Plants 2019, 8, 34. [Google Scholar] [CrossRef]
- Yichie, Y.; Brien, C.; Berger, B.; Roberts, T.H.; Atwell, B.J. Salinity tolerance in Australian wild Oryza species varies widely and matches that observed in O. sativa. Rice 2018, 11, 66. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of Genes Involved in the Response of Arabidopsis to Simultaneous Biotic and Abiotic Stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef]
- Perez, I.E.; Brown, P.J. The role of ROS signaling in cross-tolerance: From model to crop. Front. Plant Sci. 2014, 5, 754. [Google Scholar] [CrossRef] [PubMed]
- Clifton, R.; Lister, R.; Parker, K.L.; Sappl, P.G.; Elhafez, D.; Millar, A.H.; Day, D.; Whelan, J. Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 2005, 58, 193–212. [Google Scholar] [CrossRef]
- Costa, J.H.; Jolivet, Y.; Hasenfratz-Sauder, M.P.; Orellano, E.G.; Lima MD, G.S.; Dizengremel, P.; de Melo, D.F. Alternative oxidase regulation in roots of Vigna unguiculata cultivars differing in drought/salt tolerance. J. Plant Physiol. 2007, 164, 718–727. [Google Scholar] [CrossRef]
- Costa, J.H.; McDonald, A.E.; Arnholdt-Schmitt, B.; de Melo, D.F. A classification scheme for alternative oxidases reveals the taxonomic distribution and evolutionary history of the enzyme in angiosperms. Mitochondrion 2014, 19, 172–183. [Google Scholar] [CrossRef]
- Costa, J.H.; dos Santos, C.P.; Lima, B.D.S.E.; Netto, A.N.M.; Saraiva, K.D.D.C.; Arnholdt-Schmitt, B. In silico identification of alternative oxidase 2 (AOX2) in monocots: A new evolutionary scenario. J. Plant Physiol. 2017, 210, 58–63. [Google Scholar] [CrossRef]
- Considine, M.; Holtzapffel, R.C.; Day, D.A.; Whelan, J.; Millar, A.H. Molecular Distinction between Alternative Oxidase from Monocots and Dicots. Plant Physiol. 2002, 129, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Saisho, D.; Nakazono, M.; Tsutsumi, N.; Hirai, A. Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature. Gene 1997, 203, 121–129. [Google Scholar] [CrossRef]
- Ohtsu, K.; Ito, Y.; Saika, H.; Nakazono, M.; Tsutsumi, N.; Hirai, A. ABA-Independent Expression of Rice Alternative Oxidase Genes under Environmental Stresses. Plant Biotechnol. 2002, 19, 187–190. [Google Scholar] [CrossRef][Green Version]
- Saika, H.; Ohtsu, K.; Hamanaka, S.; Nakazono, M.; Tsutsumi, N.; Hirai, A. AOX1c, a novel rice gene for alternative oxidase; Comparison with rice AOX1a and AOX1b. Genes Genet. Syst. 2002, 77, 31–38. [Google Scholar] [CrossRef]
- Feng, H.; Houa, X.; Li, X.; Sun, K.; Wang, R.; Zhang, T.; Ding, Y. Cell death of rice roots under salt stress may be mediated by cyanide-resistant respiration. Z. Für Nat. C 2013, 68, 39–46. [Google Scholar] [CrossRef]
- Feng, H.Q.; Li, H.Y.; Sun, K. Enhanced expression of alternative oxidase genes is involved in the tolerance of rice (Oryza sativa L.) seedlings to drought stress. Z. Für. Nat. C 2009, 64, 704–710. [Google Scholar] [CrossRef]
- Li, C.R.; Liang, D.D.; Li, J.; Duan, Y.B.; Li, H.A.; Yang, Y.C.; Qin, R.Y.; Li, L.I.; Wei, P.C.; Yang, J.B. Unravelling mitochondrial retrograde regulation in the abiotic stress induction of rice Alternative Oxidase 1 genes. Plant Cell Environ. 2013, 36, 775–788. [Google Scholar] [CrossRef]
- Wanniarachchi, V.R.; Dametto, L.; Sweetman, C.; Shavrukov, Y.; Day, D.A.; Jenkins, C.L.D.; Soole, K.L. Alternative Respiratory Pathway Component Genes (AOX and ND) in Rice and Barley and Their Response to Stress. Int. J. Mol. Sci. 2018, 19, 915. [Google Scholar] [CrossRef]
- Jayawardhane, J.; Cochrane, D.W.; Vyas, P.; Bykova, N.V.; Vanlerberghe, G.C.; Igamberdiev, A.U. Roles for plant mitochondrial alternative oxidase under normoxia, hypoxia, and reoxygenation conditions. Front. Plant Sci. 2020, 11, 566. [Google Scholar] [CrossRef]
- Czobor, A.; Hajdinák, P.; Németh, B.; Piros, B.; Németh, A.; Szarka, A. Comparison of the response of alternative oxidase and uncoupling proteins to bacterial elicitor induced oxidative burst. PLoS ONE 2019, 14, e0210592. [Google Scholar] [CrossRef]
- Haferkamp, I.; Schmitz-Esser, S. The Plant Mitochondrial Carrier Family: Functional and Evolutionary Aspects. Front. Plant Sci. 2012, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fu, A. The Plastid Terminal Oxidase is a Key Factor Balancing the Redox State of Thylakoid Membrane. Enzymes 2016, 40, 143–171. [Google Scholar] [PubMed]
- Stepien, P.; Johnson, G.N. Plastid terminal oxidase requires translocation to the grana stacks to act as a sink for electron transport. Proc. Natl. Acad. Sci. USA 2018, 115, 9634–9639. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Noguchi, K. Differential Gene Expression Profiles of the Mitochondrial Respiratory Components in Illuminated Arabidopsis Leaves. Plant Cell Physiol. 2009, 50, 1449–1462. [Google Scholar] [CrossRef] [PubMed]
- Maia, R.A.; da Cruz Saraiva, K.D.; Roque, A.L.; Thiers, K.L.; Dos Santos, C.P.; da Silva, J.H.; Feijó, D.F.; Arnholdt-Schmitt, B.; Costa, J.H. Differential expression of recently duplicated PTOX genes in Glycine max during plant development and stress conditions. J. Bioenerg. Biomembr. 2019, 51, 355–370. [Google Scholar] [CrossRef]
- Oliver, P.; Picó, C.; Palou, A. Differential expression of genes for uncoupling proteins 1, 2 and 3 in brown and white adipose tissue depots during rat development. Experientia 2001, 58, 470–476. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Intracellular Redox Compartmentation and ROS-Related Communication in Regulation and Signaling. Plant Physiol. 2016, 171, 1581–1592. [Google Scholar] [CrossRef]
- Noctor, G.; Reichheld, J.-P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. In Seminars in Cell & Developmental Biology; Academic Press: London, UK, 2018; Volume 80, pp. 3–12. [Google Scholar]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Liu, L.; Li, J. Communications Between the Endoplasmic Reticulum and Other Organelles During Abiotic Stress Response in Plants. Front. Plant Sci. 2019, 10, 749. [Google Scholar] [CrossRef]
- Arnholdt-Schmitt, B.; Costa, J.H.; de Melo, D.F. AOX—A functional marker for efficient cell reprogramming under stress? Trends Plant Sci. 2006, 11, 281–287. [Google Scholar] [CrossRef]
- Pandey, P.; Ramegowda, V.; Senthil-Kumar, M. Shared and unique responses of plants to multiple individual stresses and stress combinations: Physiological and molecular mechanisms. Front. Plant Sci. 2015, 6, 723. [Google Scholar] [CrossRef] [PubMed]
- Arnholdt-Schmitt, B.; Mohanapriya, G.; Bharadwaj, R.; Noceda, C.; Macedo, E.S.; Sathishkumar, R.; Gupta, K.J.; Sircar, D.; Kumar, S.R.; Srivastava, S.; et al. From Plant Survival Under Severe Stress to Anti-Viral Human Defense—A Perspective That Calls for Common Efforts. Front. Immunol. 2021, 12, 673723. [Google Scholar] [CrossRef] [PubMed]
- Mohanapriya, G.; Bharadwaj, R.; Noceda, C.; Costa, J.H.; Kumar, S.R.; Sathishkumar, R.; Thiers, K.L.; Santos Macedo, E.; Silva, S.; Annicchiarico, P.; et al. Alternative oxidase (AOX) senses stress levels to coordinate auxin-induced reprogramming from seed germination to somatic embryogenesis—A role relevant for seed vigor prediction and plant robustness. Front. Plant Sci. 2019, 10, 1134. [Google Scholar] [CrossRef] [PubMed]
- Rasmusson, A.G.; Fernie, A.R.; van Dongen, J. Alternative oxidase: A defence against metabolic fluctuations? Physiol. Plant. 2009, 137, 371–382. [Google Scholar] [CrossRef]
- Vanlerberghe, G.C. Alternative Oxidase: A Mitochondrial Respiratory Pathway to Maintain Metabolic and Signaling Homeostasis during Abiotic and Biotic Stress in Plants. Int. J. Mol. Sci. 2013, 14, 6805–6847. [Google Scholar] [CrossRef]
- Rogov, A.G.; Sukhanova, E.I.; Uralskaya, L.A.; Aliverdieva, D.A.; Zvyagilskaya, R.A. Alternative oxidase: Distribution, induction, properties, structure, regulation, and functions. Biochemistry 2017, 79, 1615–1634. [Google Scholar] [CrossRef]
- Bharadwaj, R.; Noceda, C.; Mohanapriya, G.; Kumar, S.R.; Thiers, K.L.; Costa, J.H.; Macedo, E.S.; Kumari, A.; Gupta, K.J.; Srivastava, S.; et al. Adaptive Reprogramming During Early Seed Germination Requires Temporarily Enhanced Fermentation-A Critical Role for Alternative Oxidase Regulation That Concerns Also Microbiota Effectiveness. Front. Plant Sci. 2021, 1888. [Google Scholar] [CrossRef]
- Murphy, A.M.; Zhou, T.; Carr, J.P. An update on salicylic acid biosynthesis, its induction and potential exploitation by plant viruses. Curr. Opin. Virol. 2020, 42, 8–17. [Google Scholar] [CrossRef]
- Arnholdt-Schmitt, B.; Valadas, V.; Döring, M. Functional marker development is challenged by the ubiquity of endophytes--a practical perspective. Brief. Funct. Genom. 2014, 15, 16–21. [Google Scholar] [CrossRef]
- Arnholdt-Schmitt, B.; Hansen, L.D.; Nogales, A. Calorespirometry, oxygen isotope analysis and functional-marker-assisted selection (‘CalOxy-FMAS’) for genotype screening: A novel concept and tool kit for predicting stable plant growth performance and functional marker identification. Brief. Funct. Genom. 2015, 15, 10–15. [Google Scholar] [CrossRef]
- Arnholdt-Schmitt, B.; Mohanapriya, G.; Sathishkumar, R.; Macedo, E.S.; Costa, J.H. Predicting Biomass Production from Plant Robustness and Germination Efficiency by Calorespirometry. In Biofuels: Greenhouse Gas Mitigation and Global Warming, Kumar, A., Ogita, S., Yau, Y., Eds.; Next Generation Biofuels and Role of Biotechnology; Springer: New Delhi, India, 2018; pp. 81–94. [Google Scholar]
- Costa, J.H.; Mohanapriya, G.; Bharadwaj, R.; Noceda, C.; Thiers, K.L.; Aziz, S.; Srivastava, S.; Oliveira, M.; Gupta, K.J.; Kumari, A.; et al. ROS/RNS balancing, aerobic fermentation regulation and cell cycle control–a complex early trait (‘CoV-MAC-TED’) for combating SARS-CoV-2-induced cell reprogramming. Front. Immunol. 2021, 12, 673692. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.H.; Aziz, S.; Noceda, C.; Arnholdt-Schmitt, B. Major Complex Trait for Early De Novo Programming ‘CoV-MAC-TED’Detected in Human Nasal Epithelial Cells Infected by Two SARS-CoV-2 Variants Is Promising to Help in Designing Therapeutic Strategies. Vaccines 2021, 9, 1399. [Google Scholar] [CrossRef]
- Costa, J.H.; Aziz, S.; Arnholdt-Schmitt, B. Transcriptome data from human nasal epithelial cells infected by H3N2 influenza virus indicate early unbalanced ROS/RNA levels, temporarily increased aerobic fermentation linked to enhanced α-tubulin and rapid energy-dependent IRF9-marked immunization. bioRxiv 2021. [Google Scholar] [CrossRef]
- Costa, J.H.; Roque, A.L.M.; Aziz, S.; dos Santos, C.P.; Germano, T.A.; Batista, M.C.; Thiers, K.L.L.; Saraiva, K.D.D.C.; Arnholdt-Schmitt, B. Genome-wide identification of ascorbate-glutathione cycle gene families in soybean (Glycine max) reveals gene duplication events and specificity of gene members linked to development and stress conditions. Int. J. Biol. Macromol. 2021, 187, 528–543. [Google Scholar] [CrossRef] [PubMed]
- Pabuayon, I.C.M.; Kitazumi, A.; Gregorio, G.B.; Singh, R.K.; Reyes, B.G.D.L. Contributions of Adaptive Plant Architecture to Transgressive Salinity Tolerance in Recombinant Inbred Lines of Rice: Molecular Mechanisms Based on Transcriptional Networks. Front. Genet. 2020, 11, 594569. [Google Scholar] [CrossRef]
- Arce, L.; Yllano, O. Sensitivity and tolerance of Pokkali and IRRI-112 rice (Oryza sativa) varieties to lead (pb) stress. Univ. Res. J. 2008, 11, 1–15. [Google Scholar]
- Nguyen, H.T.T.; Das Bhowmik, S.; Long, H.; Cheng, Y.; Mundree, S.; Hoang, L.T.M. Rapid Accumulation of Proline Enhances Salinity Tolerance in Australian Wild Rice Oryza australiensis Domin. Plants 2021, 10, 2044. [Google Scholar] [CrossRef]
- Geniza, M.J. Transcriptomic Analyses of Crop Plants Using Next-Generation Sequencing Technology; Oregon State University: Corvallis, OR, USA, 2017. [Google Scholar]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinform. 2019, 20, 405. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Mustroph, A.; Stock, J.; Hess, N.; Aldous, S.; Dreilich, A.; Grimm, B. Characterization of the Phosphofructokinase Gene Family in Rice and Its Expression Under Oxygen Deficiency Stress. Front. Plant Sci. 2013, 4, 125. [Google Scholar] [CrossRef]
- Terada, R.; Johzuka-Hisatomi, Y.; Saitoh, M.; Asao, H.; Iida, S. Gene Targeting by Homologous Recombination as a Biotechnological Tool for Rice Functional Genomics. Plant Physiol. 2007, 144, 846–856. [Google Scholar] [CrossRef]
- Mansilla, N.; Racca, S.; Gras, D.E.; Gonzalez, D.H.; Welchen, E. The Complexity of Mitochondrial Complex IV: An Update of Cytochrome c Oxidase Biogenesis in Plants. Int. J. Mol. Sci. 2018, 19, 662. [Google Scholar] [CrossRef]
- Watanabe, A.; Hirai, A. Two uncoupling protein genes of rice (Oryza sativa L.): Molecular study reveals the defects in the pre-mRNA processing for the heat-generating proteins of the subtropical cereal. Planta 2002, 215, 90–100. [Google Scholar] [CrossRef]
- Tamiru, M.; Abe, A.; Utsushi, H.; Yoshida, K.; Takagi, H.; Fujisaki, K.; Undan, J.R.; Rakshit, S.; Takaichi, S.; Jikumaru, Y.; et al. The tillering phenotype of the rice plastid terminal oxidase (PTOX) loss-of-function mutant is associated with strigolactone deficiency. New Phytol. 2013, 202, 116–131. [Google Scholar] [CrossRef]
- Claros, M.G.; Vincens, P. Computational Method to Predict Mitochondrially Imported Proteins and their Targeting Sequences. JBIC J. Biol. Inorg. Chem. 1996, 241, 779–786. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Sahu, S.S.; Loaiza, C.D.; Kaundal, R. Plant-mSubP: A computational framework for the prediction of single- and multi-target protein subcellular localization using integrated machine-learning approaches. AoB Plants 2019, 12, plz068. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Menezes-Benavente, L.; Galvão, V.C.; Margis, R.; Margis-Pinheiro, M. Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta 2006, 224, 300–314. [Google Scholar] [CrossRef]
- Guan, Q.; Takano, T.; Liu, S. Genetic Transformation and Analysis of Rice OsAPx2 Gene in Medicago sativa. PLoS ONE 2012, 7, e41233. [Google Scholar] [CrossRef]
- Morgante, C.V.; Rodrigues, R.A.O.; Marbach, P.A.S.; Borgonovi, C.M.; Moura, D.S.; Silva-Filho, M.C. Conservation of dual-targeted proteins in Arabidopsis and rice points to a similar pattern of gene-family evolution. Mol. Genet. Genom. 2009, 281, 525–538. [Google Scholar] [CrossRef]
- Liu, J.; Sun, X.; Xu, F.; Zhang, Y.; Zhang, Q.; Miao, R.; Zhang, J.; Liang, J.; Xu, W. Suppression of OsMDHAR4 enhances heat tolerance by mediating H2O2-induced stomatal closure in rice plants. Rice 2018, 11, 38. [Google Scholar] [CrossRef]
- Wu, T.-M.; Lin, W.-R.; Kao, Y.-T.; Hsu, Y.-T.; Yeh, C.-H.; Hong, C.-Y.; Kao, C.H. Identification and characterization of a novel chloroplast/mitochondria co-localized glutathione reductase 3 involved in salt stress response in rice. Plant Mol. Biol. 2013, 83, 379–390. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, Y.; Xie, Z.; Li, X.; He, Z.-H.; Peng, X.-X. Association–Dissociation of Glycolate Oxidase with Catalase in Rice: A Potential Switch to Modulate Intracellular H 2 O 2 Levels. Mol. Plant 2016, 9, 737–748. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, C.; Ai, P.; Cui, X.; Zhang, Z. ALM1, encoding a Fe-superoxide dismutase, is critical for rice chloroplast biogenesis and drought stress response. Crop J. 2020, 9, 1018–1029. [Google Scholar] [CrossRef]
- Singh, R.K.; Kota, S.; Flowers, T.J. Salt tolerance in rice: Seedling and reproductive stage QTL mapping come of age. Theor. Appl. Genet. 2021, 134, 3495–3533. [Google Scholar] [CrossRef]
- Singh, R.K.; Adorada, D.L.; Magsino, C.; Roque, Z.; Tamayo, N.; Gregorio, G.B. Effect of relative humidity and temperature on salinity tolerance of rice. In Plant Breeding, Genetics and Biotechnology (PBGB) Division Biennial Report 2004–2005; IRRI: Manila, Philippines, 2004; pp. 19–21. [Google Scholar]
- Challabathula, D.; Analin, B.; Mohanan, A.; Bakka, K. Differential modulation of photosynthesis, ROS and antioxidant enzyme activities in stress-sensitive and -tolerant rice cultivars during salinity and drought upon restriction of COX and AOX pathways of mitochondrial oxidative electron transport. J. Plant Physiol. 2021, 268, 153583. [Google Scholar] [CrossRef]
- Walia, H.; Wilson, C.; Condamine, P.; Liu, X.; Ismail, A.M.; Zeng, L.; Wanamaker, S.I.; Mandal, J.; Xu, J.; Cui, X.; et al. Comparative Transcriptional Profiling of Two Contrasting Rice Genotypes under Salinity Stress during the Vegetative Growth Stage. Plant Physiol. 2005, 139, 822–835. [Google Scholar] [CrossRef]
- Zheng, J.; Ying, Q.; Fang, C.; Sun, N.; Si, M.; Yang, J.; Zhu, B.; Ruan, Y.-L.; Zhu, Z.; He, Y. Alternative oxidase pathway is likely involved in waterlogging tolerance of watermelon. Sci. Hortic. 2020, 278, 109831. [Google Scholar] [CrossRef]
- Lakra, N.; Kaur, C.; Anwar, K.; Singla-Pareek, S.L.; Pareek, A. Proteomics of contrasting rice genotypes: Identification of potential targets for raising crops for saline environment. Plant Cell Environ. 2017, 41, 947–969. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wang, S.; Eltayeb, A.E.; Uddin, I.; Yamamoto, Y.; Tsuji, W.; Takeuchi, Y.; Tanaka, K. Overexpression of dehydroascorbate reductase, but not monodehydroascorbate reductase, confers tolerance to aluminum stress in transgenic tobacco. Planta 2009, 231, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Anjum, N.A.; Gill, S.S.; Gill, R.; Hasanuzzaman, M.; Duarte, A.C.; Pereira, E.; Ahmad, I.; Tuteja, R.; Tuteja, N. Metal/metalloid stress tolerance in plants: Role of ascorbate, its redox couple, and associated enzymes. Protoplasma 2014, 251, 1265–1283. [Google Scholar] [CrossRef]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate Peroxidase and Catalase Activities and Their Genetic Regulation in Plants Subjected to Drought and Salinity Stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef]
- Cuypers, A.; Karen, S.; Jos, R.; Kelly, O.; Els, K.; Tony, R.; Nele, H.; Nathalie, V.; Suzy, V.S.; Frank, V.B.; et al. The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. J. Plant Physiol. 2011, 168, 309–316. [Google Scholar] [CrossRef]
- Basu, S.; Roychoudhury, A. Expression Profiling of Abiotic Stress-Inducible Genes in response to Multiple Stresses in Rice (Oryza sativa L.) Varieties with Contrasting Level of Stress Tolerance. BioMed Res. Int. 2014, 2014, 706890. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi-Nejad, G.; Arzani, A.; Reza, A.M.; Singh, R.K.; Gregorio, G.B. Assessment of rice genotypes for salt tolerance using microsatellite markers associated with the saltol QTL. Afr. J. Biotechnol. 2008, 7, 730–736. [Google Scholar]
- Sampangi-Ramaiah, M.H.; Dey, P.; Jambagi, S.; Kumari, M.M.V.; Oelmüller, R.; Nataraja, K.N.; Ravishankar, K.V.; Ravikanth, G.; Shaanker, R.U. An endophyte from salt-adapted Pokkali rice confers salt-tolerance to a salt-sensitive rice variety and targets a unique pattern of genes in its new host. Sci. Rep. 2020, 10, 3237. [Google Scholar] [CrossRef]
- Materatski, P.; Varanda, C.; Carvalho, T.; Dias, A.B.; Campos, M.D.; Rei, F.; Félix, M.D.R. Spatial and temporal variation of fungal endophytic richness and diversity associated to the phyllosphere of olive cultivars. Fungal Biol. 2018, 123, 66–76. [Google Scholar] [CrossRef]
- Campos, C.; Cardoso, H.; Nogales, A.; Svensson, J.; Lopez-Ráez, J.A.; Pozo, M.J.; Nobre, T.; Schneider, C.; Arnholdt-Schmitt, B. Intra and Inter-Spore Variability in Rhizophagus irregularis AOX Gene. PLoS ONE 2015, 10, e0142339. [Google Scholar] [CrossRef]
- Mercy, L.; Lucic-Mercy, E.; Nogales, A.; Poghosyan, A.; Schneider, C.; Arnholdt-Schmitt, B. A Functional Approach towards Understanding the Role of the Mitochondrial Respiratory Chain in an Endomycorrhizal Symbiosis. Front. Plant Sci. 2017, 8, 417. [Google Scholar] [CrossRef]
- Bedini, A.; Mercy, L.; Schneider, C.; Franken, P.; Lucic-Mercy, E. Unraveling the Initial Plant Hormone Signaling, Metabolic Mechanisms and Plant Defense Triggering the Endomycorrhizal Symbiosis Behavior. Front. Plant Sci. 2018, 9, 1800. [Google Scholar] [CrossRef]
- Frederico, A.M.; Campos, M.D.; Cardoso, H.G.; Imani, J.; Arnholdt-Schmitt, B. Alternative oxidase involvement in Daucus carota somatic embryogenesis. Physiol. Plant. 2009, 137, 498–508. [Google Scholar] [CrossRef]
- Costa, J.H.; Cardoso, H.G.; Campos, M.D.; Zavattieri, A.; Frederico, A.M.; de Melo, D.F.; Arnholdt-Schmitt, B. Daucus carota L.–An old model for cell reprogramming gains new importance through a novel expansion pattern of alternative oxidase (AOX) genes. Plant Physiol. Biochem. 2009, 47, 753–759. [Google Scholar] [CrossRef]
- Polidoros, A.N.; Mylona, P.V.; Arnholdt-Schmitt, B. Aox gene structure, transcript variation and expression in plants. Physiol. Plant. 2009, 137, 342–353. [Google Scholar] [CrossRef]
- Ferreira, L.J.; Azevedo, V.; Maroco, J.; Oliveira, M.M.; Santos, A.P. Salt Tolerant and Sensitive Rice Varieties Display Differential Methylome Flexibility under Salt Stress. PLoS ONE 2015, 10, e0124060. [Google Scholar] [CrossRef]
- Macedo, E.S.; Cardoso, H.G.; Hernández, A.; Peixe, A.A.; Polidoros, A.; Ferreira, A.; Cordeiro, A.; Arnholdt-Schmitt, B. Physiologic responses and gene diversity indicate olive alternative oxidase as a potential source for markers involved in efficient adventitious root induction. Physiol. Plant. 2009, 137, 532–552. [Google Scholar] [CrossRef]
- Nobre, T.; Oliveira, M.; Arnholdt-Schmitt, B. Wild Carrot Differentiation in Europe and Selection at DcAOX1 Gene? PLoS ONE 2016, 11, e0164872. [Google Scholar] [CrossRef]
- Nogales, A.; Nobre, T.; Cardoso, H.G.; Muñoz-Sanhueza, L.; Valadas, V.; Campos, M.D.; Arnholdt-Schmitt, B. Allelic variation on DcAOX1 gene in carrot (Daucus carota L.): An interesting simple sequence repeat in a highly variable intron. Plant Gene 2016, 5, 49–55. [Google Scholar] [CrossRef]
- Cardoso, H.G.; Arnholdt-Schmitt, B. Functional marker development across species in selected traits. In Diagnostics in Plant Breeding; Springer: Dordrecht, The Netherlands, 2013; pp. 467–515. [Google Scholar]
- Ito, K.; Ogata, T.; Seito, T.; Umekawa, Y.; Kakizaki, Y.; Osada, H.; Moore, A.L. Degradation of mitochondrial alternative oxidase in the appendices of Arum maculatum. Biochem. J. 2020, 477, 3417–3431. [Google Scholar] [CrossRef]
- Scheibe, R. Maintaining homeostasis by controlled alternatives for energy distribution in plant cells under changing conditions of supply and demand. Photosynth. Res. 2018, 139, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Ragonezi, C.; Arnholdt-Schmitt, B. Laser Capture Microdissection for Amplification of Alternative Oxidase (AOX) Genes in Target Tissues in Daucus carota L. In Plant Respiration and Internal Oxygen; Humana Press: New York, NY, USA, 2017; pp. 245–252. [Google Scholar]
- Varanda, C.; Félix, M.; Campos, M.; Patanita, M.; Materatski, P. Plant Viruses: From Targets to Tools for CRISPR. Viruses 2021, 13, 141. [Google Scholar] [CrossRef]
- Zavattieri, M.A.; Frederico, A.M.; Lima, M.; Sabino, R.; Arnholdt-Schmitt, B. Induction of somatic embryogenesis as an example of stress-related plant reactions. Electron. J. Biotechnol. 2010, 13, 12–13. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).