Phytochemical Characterization of Cannabis sativa L. Chemotype V Reveals Three New Dihydrophenanthrenoids That Favorably Reprogram Lipid Mediator Biosynthesis in Macrophages
Abstract
:1. Introduction
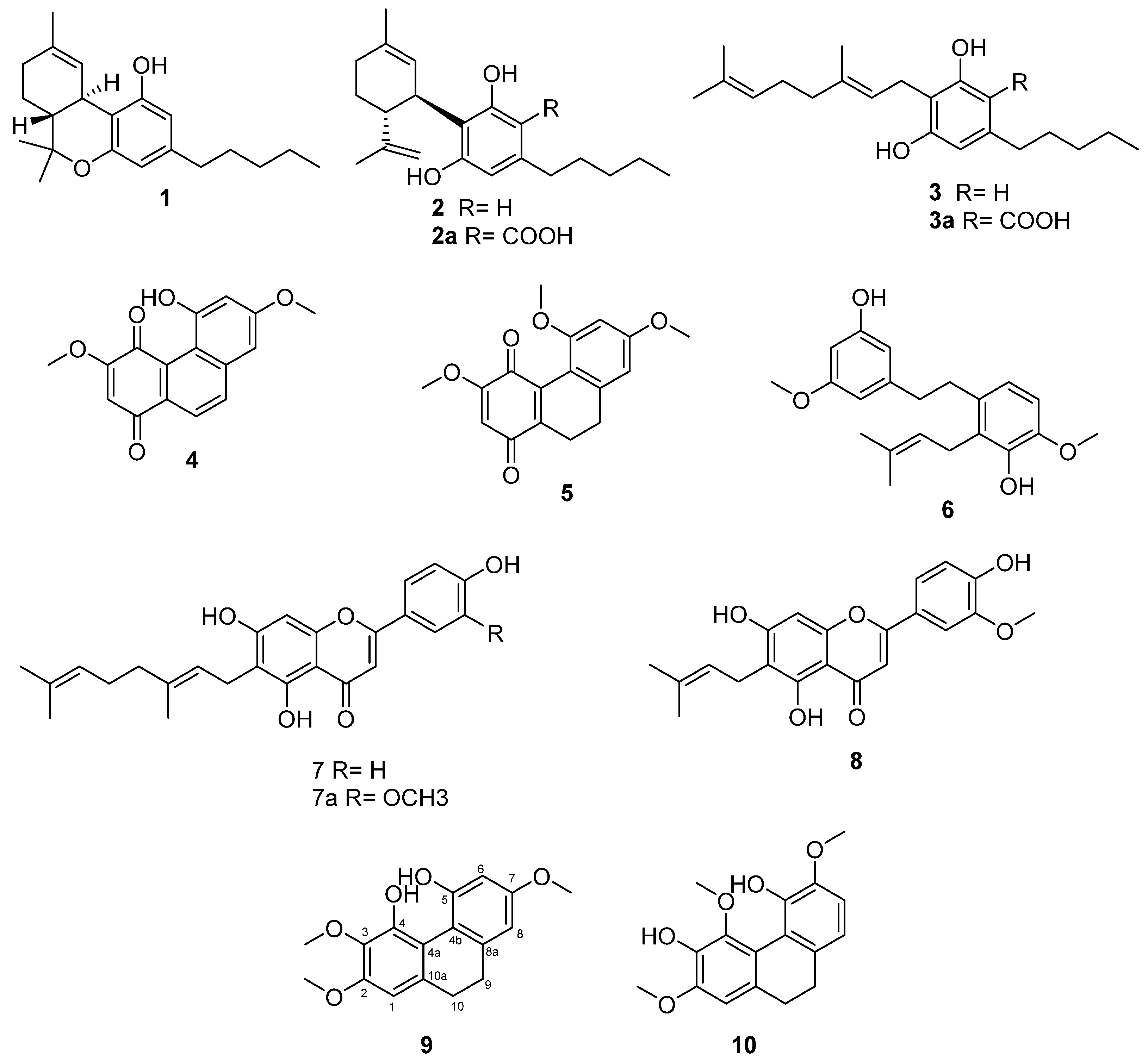
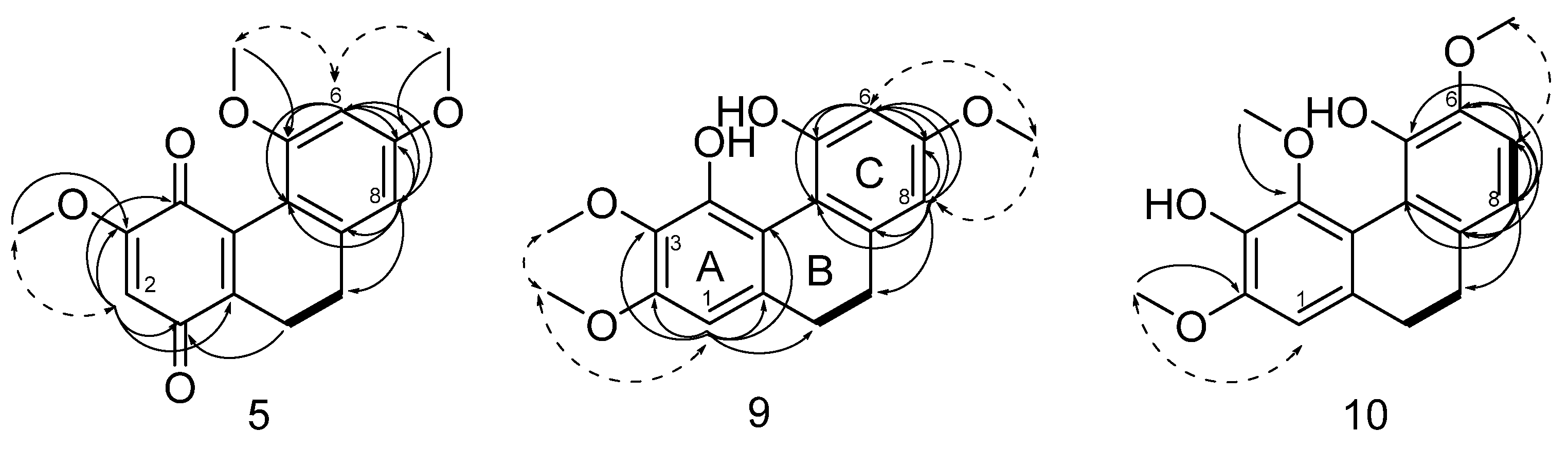
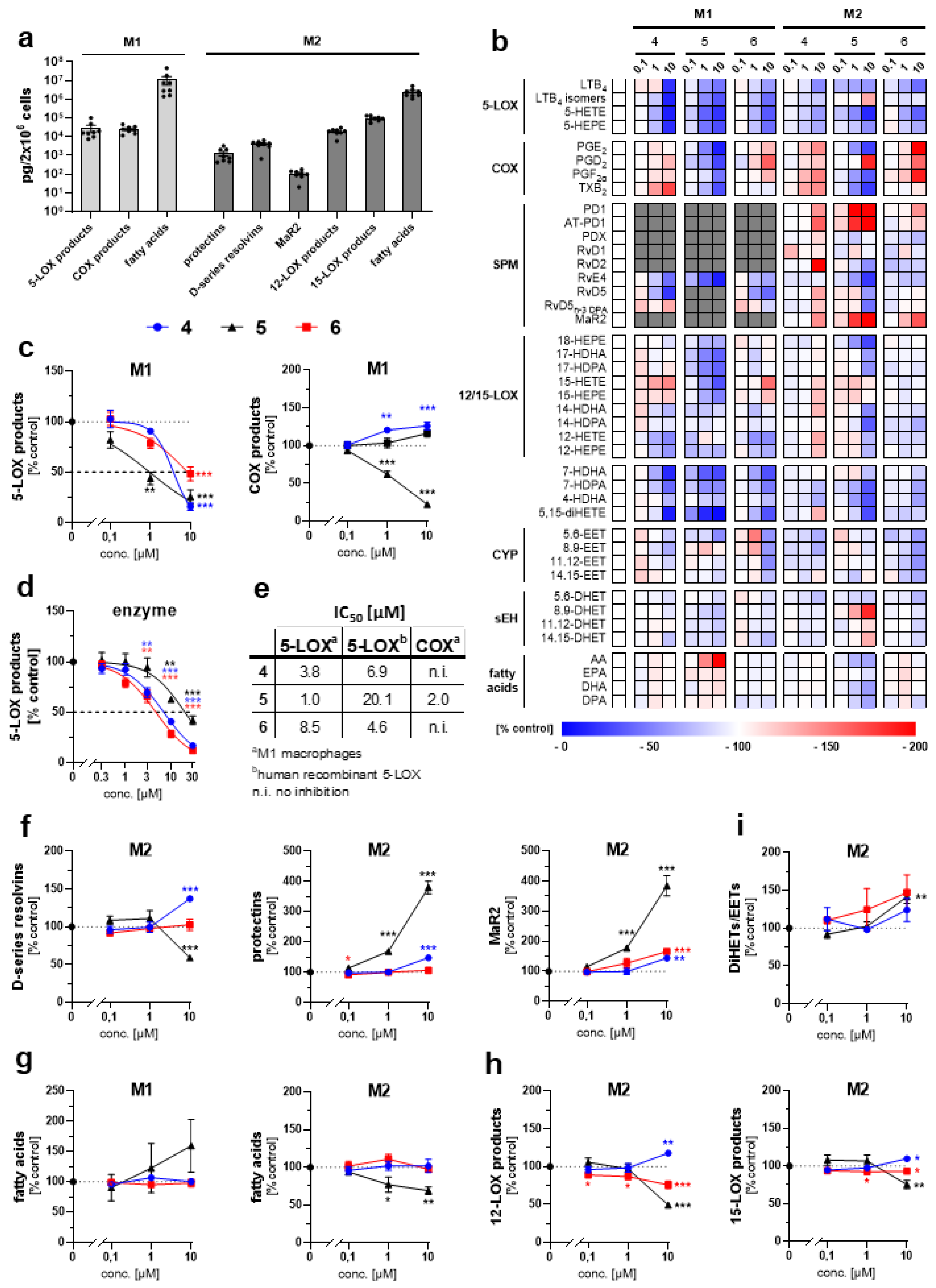
2. Results and Discussion
3. Material and Method
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Spectroscopic Data
3.5. Isolation of Peripheral Blood Mononuclear Cells (PBMC) from Human Blood
3.6. Monocyte Differentiation and Macrophage Polarization
3.7. Sample Preparation and Metabololipidomic Profiling of Lipid Mediators
3.8. Determination of 5-Lipoxygenase Activity
3.9. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pollastro, F.; Minassi, A.; Fresu, L.G. Cannabis phenolics and their bioactivities. Curr. Med. Chem. 2018, 25, 1160–1185. [Google Scholar] [CrossRef]
- Attard, T.M.; Bainier, C.; Reinaud, M.; Lanot, A.; McQueen-Mason, S.J.; Hunt, A.J. Utilisation of supercritical fluids for the effective extraction of waxes and Cannabidiol (CBD) from hemp wastes. Ind. Crops Prod. 2018, 12, 38–46. [Google Scholar] [CrossRef]
- Chandra, S.; Radwan, M.M.; Majumdar, C.G.; Church, J.C.; Freeman, T.P.; ElSohly, M.A. New trends in cannabis potency in USA and Europe during the last decade (2008–2017). Eur. Arch Psychiatry Clin. Neurosci. 2019, 269, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Aizpurua-Olaizola, O.; Soydaner, U.; Öztürk, E.; Schibano, D.; Simsir, Y.; Navarro, P.; Etxebarria, N.; Usobiaga, A. Evolution of the cannabinoid and terpene content during the growth of Cannabis sativa plants from different chemotypes. J. Nat. Prod. 2016, 79, 324–331. [Google Scholar] [CrossRef]
- Russo, E.B. The Case for the Entourage Effect and Conventional Breeding of Clinical Cannabis: No “Strain”, No Gain Front. Plant Sci. 2019, 9, 1969. [Google Scholar] [CrossRef]
- De Meijer, E.P.; Hammond, K.M.M.; Sutton, A.G. The inheritance of chemical phenotype in Cannabis sativa L. (IV). Euphytica 2009, 168, 95–112. [Google Scholar] [CrossRef]
- Upton, R.; ElSohly, M. (Eds.) Cannabis Inflorescence (Cannabis spp.): Standards of Identity, Analysis, and Quality Control; American Herbal Pharmacopoeia: Soquel, CA, USA, 2013. [Google Scholar]
- Pagani, A.; Scala, F.; Chianese, G.; Grassi, G.; Appendino, G.; Taglialatela-Scafati, O. Cannabioxepane, a novel tetracyclic cannabinoid from hemp, Cannabis sativa L. Tetrahedron 2011, 67, 3369–3373. [Google Scholar] [CrossRef]
- Magwere, T. Escaping immune surveillance in cancer: Is denbinobin the panacea? Br. J. Pharmacol. 2009, 157, 1172–1174. [Google Scholar] [CrossRef]
- Brenneisen, R. Forensic Science and Medicine: Marijuana and the Cannabinoids; ElSohly, M.A., Humana Press Inc., Eds.; Springer: Totowa, NJ, USA, 2007; pp. 17–49. [Google Scholar]
- Allegrone, G.; Pollastro, F.; Magagnini, G.; Taglialatela-Scafati, O.; Julia Seegers, J.; Koeberle, A.; Werz, O.; Appendino, G. The Bibenzyl Canniprene Inhibits the Production of Pro-Inflammatory Eicosanoids and Selectively Accumulates in Some Cannabis sativa Strains. J. Nat. Prod. 2017, 80, 731–734. [Google Scholar] [CrossRef]
- Werz, O.; Seegers, J.; Schaible, A.M.; Weinigel, C.; Barz, D.; Koeberle, A.; Allegrone, G.; Pollastro, F.; Zampieri, L.; Grassi, G.; et al. Cannflavins from hemp sprouts, a novel cannabinoid-free hemp food product, target microsomal prostaglandin E 2 synthase-1 and 5-lipoxygenase. PharmaNutrition 2014, 2, 53–60. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Luo, J.G.; Wan, C.X.; Zhou, Z.B.; Kong, L.Y. Four New Flavonoids with α-Glucosidase Inhibitory Activities from Morus alba var. tatarica. Chem. Biodiv. 2015, 12, 1768–1776. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.T.; Zhang, J.C.; Zhang, H.; Liu, Q.C.; Zhao, Y.; Hou, Y.F.; Bai, L.; Zhang, L.; Liu, X.Q.; Liu, X.Y.; et al. Bioactive spirans and other constituents from the leaves of Cannabis sativa f. sativa. J. Asian Nat. Prod. Res. 2017, 19, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Sànchez-Duffhues, G.; Calzado, M.A.; de Vinuesa, A.G.; Caballero, F.J.; Ech-Chahad, A.; Appendino, G.; Krohn, K.; Fiebich, B.L.; Muñoz, E. Denbinobin, a naturally occurring 1,4-phenanthrenequinone, inhibits HIV-1 replication through an NF-kB-dependent pathway. Biochem. Pharmacol. 2008, 76, 1240–1250. [Google Scholar] [CrossRef]
- Kuo, C.T.; Hsu, M.J.; Chen, B.C.; Chen, C.C.; Teng, C.M.; Pan, S.L.; Lin, C.H. Denbinobin induces apoptosis in human lung adenocarcinoma cells via Akt inactivation, Bad activation, and mitochondrial dysfunction. Toxicol. Lett. 2008, 177, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Sànchez-Duffhues, G.; Calzado, M.A.; de Vinuesa, A.G.; Appendino, G.; Fiebich, B.L.; Loock, U.; Lefarth-Risse, A.; Krohn, K.; Muñoz, E. Denbinobin inhibits nuclear factor-kB and induces apoptosis via reactive oxygen species generation in human leukemic cells. Biochem. Pharmacol. 2009, 77, 1401–1409. [Google Scholar] [CrossRef]
- Tóth, B.; Hohmann, J.; Vasas, A. Phenanthrenes: A promising group of plant secondary metabolites. J. Nat. Prod. 2018, 81, 661–678. [Google Scholar] [CrossRef]
- Kovács, A.; Vasas, A.; Hohmann, J. Natural phenanthrenes and their biological activity. Phytochemistry 2007, 69, 1084–1110. [Google Scholar] [CrossRef]
- Mahboubi-Rabbani, M.; Zarghi, A. Lipoxygenase inhibitors as cancer chemopreventives: Discovery, recent developments and future perspectives. Curr. Med. Chem. 2021, 28, 1143–1175. [Google Scholar] [CrossRef]
- Land, W.G. Prologue: The “Long Arm” of DAMPs in Shaping Adaptive Immune Responses and Tissue Repairing Processes. In Damage-Associated Molecular Patterns in Human Diseases; Springer: Cham, Switzerland, 2018; pp. 717–722. [Google Scholar] [CrossRef]
- Liu, K.; Tian, L.X.; Tang, X.; Wang, J.; Tang, W.Q.; Ma, Z.F.; Chen, T.; Liang, H.P. Neutrophilic granule protein (NGP) attenuates lipopolysaccharide-induced inflammatory responses and enhances phagocytosis of bacteria by macrophages. Cytokine 2020, 128, 155001. [Google Scholar] [CrossRef]
- Serhan, C.N.; Levy, B.D. Resolvins in inflammation: Emergence of the pro-resolving superfamily of mediators. J. Clin. Investig. 2018, 128, 2657–2669. [Google Scholar] [CrossRef]
- Basil, M.; Levy, B. Specialized pro-resolving mediators: Endogenous regulators of infection and inflammation. Nat. Rev. Immunol. 2016, 16, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Buckley, C.D.; Gilroy, D.W.; Serhan, C.N. Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity 2014, 40, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Hoover, R.L.; Williams, J.D.; Sperling, R.I.; Ravalese, J., III; Spur, B.W.; Dwight, R.; Robinson, D.R.; Corey, E.J.; Lewis, R.A.; et al. Effect of dietary enrichment with eicosapentaenoic and docosahexaenoic acids on in vitro neutrophil and monocyte leukotriene generation and neutrophil function. NEJM 1985, 312, 1217–1224. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Gupta, S.K.; Perretti, M.; Godson, C.; Brennan, E.; Li, Y.; Soehnlein, O.; Shimizui, T.; Werz, O.; Chiurchiù, V.; et al. The atlas of inflammation resolution (AIR). Mol. Asp. Med. 2020, 74, 100894. [Google Scholar] [CrossRef]
- Deng, B.; Wang, C.W.; Arnardottir, H.H.; Li, Y.; Cheng, C.Y.C.; Dalli, J.; Serhan, C.N. Maresin biosynthesis and identification of maresin 2, a new anti-inflammatory and pro-resolving mediator from human macrophages. PLoS ONE 2014, 9, e102362. [Google Scholar] [CrossRef]
- Wagner, K.M.; McReynolds, C.B.; Schmidt, W.K.; Hammock, B.D. Soluble epoxide hydrolase as a therapeutic target for pain, inflammatory and neurodegenerative diseases. Pharmacol. Ther. 2017, 180, 62–76. [Google Scholar] [CrossRef]
- Werz, O.; Gerstmeier, J.; Libreros, S.; De la Rosa, X.; Werner, M.; Norris, P.C.; Chiang, N.; Serhan, C.N. Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat. Commun. 2018, 9, 59. [Google Scholar] [CrossRef]
- Neukirch, K.; Alsabil, K.; Dinh, C.P.; Bilancia, R.; Raasch, M.; Ville, A.; Cerqua, I.; Viault, G.; Bréard, D.; Pace, S.; et al. Exploration of Long-Chain Vitamin E Metabolites for the Discovery of a Highly Potent, Orally Effective, and Metabolically STable 5-LOX Inhibitor that Limits Inflammation. J. Med. Chem. 2021, 64, 11496–11526. [Google Scholar] [CrossRef]
- Van Anh, T.T.; Mostafa, A.; Rao, Z.; Pace, S.; Schwaiger, S.; Kretzer, C.; Temmla, V.; Giesel, C.; Jordan, P.M.; Bilancia, R.; et al. From Vietnamese plants to a biflavonoid that relieves inflammation by triggering the lipid mediator class switch to resolution. Acta Pharm. Sin. B 2021, 11, 1629–1647. [Google Scholar] [CrossRef]
- Rao, Z.; Caprioglio, D.; Gollowitzer, A.; Kretzer, C.; Imperio, D.; Collado, J.A.; Waltl, L.; Lackner, S.; Appendino, G.; Muñoz, E.; et al. Rotational constriction of curcuminoids impacts 5-lipoxygenase and mPGES-1 inhibition and evokes a lipid mediator class switch in macrophages. Biochem. Pharmacol. 2022, 203, 115202. [Google Scholar] [CrossRef]
| Chemotype | Major Cannabinoids | Classification |
|---|---|---|
| I | Drug-type plants (narcotic) with high content of the psychotropic Δ9-THC | Drug-type plant (narcotic) |
| II | Medicinal cannabis with Δ9-THC/CBD 1:1 | Fiber-type |
| III | Industrial fiber hemp with CBD as predominant and a minimum content of Δ9-THC (from 0.2% w/w to 0.6% w/w) | Fiber-type |
| IV | Industrial fiber hemp with CBG as predominant cannabinoid | Fiber-type |
| V | Industrial fiber hemp with almost no cannabinoids | Fiber-type |
| 5 | 9 | 10 | |
|---|---|---|---|
| Position | δH, mult (J in Hz) | δH, mult (J in Hz) | δH, mult (J in Hz) |
| 1 | 6.62, s | 6.63, s | |
| 2 | 5.95, s | ||
| 3 | |||
| 4 | |||
| 5 | |||
| 6 | 6.53, d, 2.40 | 6.44, d 2.70 | |
| 7 | 6.87, d, 8.2 | ||
| 8 | 6.53, d, 2.40 | 6.50, d 2.70 | 6.82, d, 8.2 |
| 9 | 2.67, t, 7.3 | 2.64, bs | 2.62, bs |
| 10 | 2.51, t 7.3 | 2.64, bs | 2.62, bs |
| OMe-2 | 3.84, s | 3.87, s | |
| OMe-3 | 3.87, s | 3.63, s | |
| OMe-4 | 3.79, s | ||
| OMe-5 | 3.76, s | ||
| OMe-6 | 3.86, s | ||
| OMe-7 | 3.85, s | 3.81, s | |
| OH-5 | 9.09, s |
| 5 | 9 | 10 | |
|---|---|---|---|
| Position | δC, Type | δC, Type | δC, Type |
| 1 | 185.2, C | 99.1, CH | 104.4, CH |
| 2 | 105.6, CH | 151.3, C | 152.1, C |
| 3 | 160.1, C | 137.0, C | 147.4, C |
| 4 | 179.2, C | 150.3, C | 136.6, C |
| 4a | 138.0, C | 117.3, C | 114.7, C |
| 4b | 112.0, C | 113.3, C | 120.7, C |
| 5 | 158.0, C | 155.7, C | 141.5, C |
| 6 | 97.0, CH | 102.0, CH | 147.6, C |
| 7 | 162.0, C | 160.4, C | 109.6, CH |
| 8 | 105.0, CH | 106.4, CH | 118.9, CH |
| 8a | 138.8, C | 127.5, C | 132.2, C |
| 9 | 28.3, CH2 | 31.2, CH2 | 30.3, CH2 |
| 10 | 20.1, CH2 | 21.8, CH2 | 31.3, CH2 |
| 10a | 140.1, C | 142.7, C | 136.7, C |
| OMe-2 | 55.3, CH3 | 55.2, CH3 | |
| OMe-3 | 55.7, CH3 | 61.2, CH3 | |
| OMe-4 | 59.7, CH3 | ||
| OMe-5 | 55.2, CH3 | ||
| OMe-6 | 55.5, CH3 | ||
| OMe-7 | 54.9, CH3 | 54.5, CH3 |
| Q1 | Q3 | ID | DP (V) | EP (V) | CE (eV) | CXP (V) |
|---|---|---|---|---|---|---|
| 327.3 | 116.1 | d8-5S-HETE | −80.0 | −10.0 | −17.0 | −10.0 |
| 339.3 | 197.2 | d4-LTB4 | −80.0 | −10.0 | −22.0 | −13.0 |
| 355.3 | 193.2 | d4-PGE2 | −80.0 | −10.0 | −25.0 | −16.0 |
| 356.3 | 115.2 | d5-LXA4 | −80.0 | −10.0 | −19.0 | −14.0 |
| 380.3 | 141.2 | d5-RvD2 | −80.0 | −10.0 | −23.0 | −14.0 |
| 311.3 | 267.1 | d8-AA | −100.0 | −10.0 | −16.0 | −18.0 |
| 335.2 | 195.1 | LTB4 isomers | −80.0 | −10.0 | −22.0 | −13.0 |
| 335.2 | 195.1 | LTB4 | −80.0 | −10.0 | −22.0 | −13.0 |
| 319.2 | 115.1 | 5-HETE | −80.0 | −10.0 | −21.0 | −12.0 |
| 317.2 | 115.1 | 5-HEPE | −80.0 | −10.0 | −18.0 | −12.0 |
| 351.2 | 271.0 | PGE2 | −120.0 | −10.0 | −20.0 | −13.0 |
| 351.3 | 233.1 | PGD2 | −80.0 | −10.0 | −16.0 | −15.0 |
| 353.3 | 193.1 | PGF2α | −80.0 | −10.0 | −34.0 | −11.0 |
| 369.3 | 169.1 | TXB2 | −80.0 | −10.0 | −22.0 | −15.0 |
| 375.2 | 215.1 | RvD1 | −80.0 | −10.0 | −26.0 | −13.0 |
| 375.2 | 175.1 | RvD2 | −80.0 | −10.0 | −30.0 | −13.0 |
| 333.3 | 115.1 | RvE4 | −80.0 | −10.0 | −22.0 | −13.0 |
| 359.2 | 199.1 | RvD5 | −80.0 | −10.0 | −21.0 | −13.0 |
| 361.2 | 143.0 | RvD5n-3DPA | −110.0 | −10.0 | −23.0 | −25.0 |
| 359.2 | 250.1 | Maresin 1 | −80.0 | −10.0 | −20.0 | −16.0 |
| 359.2 | 221.0 | Maresin 2 | −80.0 | −10.0 | −20.0 | −12.0 |
| 359.2 | 153.1 | PD1/PDX/AT-PD1 | −80.0 | −10.0 | −21.0 | −9.0 |
| 343.2 | 245.1 | 17-HDHA | −80.0 | −10.0 | −17.0 | −14.0 |
| 345.2 | 247.1 | 17-HDPA | −80.0 | −10.0 | −17.0 | −14.0 |
| 319.2 | 219.1 | 15-HETE | −80.0 | −10.0 | −19.0 | −12.0 |
| 317.2 | 219.1 | 15-HEPE | −80.0 | −10.0 | −18.0 | −12.0 |
| 343.2 | 205.1 | 14-HDHA | −80.0 | −10.0 | −17.0 | −14.0 |
| 345.2 | 207.1 | 14-HDPA | −80.0 | −10.0 | −17.0 | −14.0 |
| 319.2 | 179.1 | 12-HETE | −80.0 | −10.0 | −21.0 | −12.0 |
| 317.2 | 179.1 | 12-HEPE | −80.0 | −10.0 | −19.0 | −12.0 |
| 317.2 | 259.1 | 18-HEPE | −80.0 | −10.0 | −16.0 | −23.0 |
| 343.2 | 141.1 | 7-HDHA | −80.0 | −10.0 | −18.0 | −15.0 |
| 345.2 | 143.1 | 7-HDPA | −80.0 | −10.0 | −18.0 | −15.0 |
| 343.2 | 101.1 | 4-HDHA | −80.0 | −10.0 | −17.0 | −15.0 |
| 335.2 | 201.0 | 5,15-diHETE | −50.0 | −10.0 | −30.0 | −13.0 |
| 319.2 | 191.1 | 5.6-EET | −50.0 | −5.0 | −20.0 | −15.0 |
| 319.2 | 167.0 | 8.9-EET | −40.0 | −5.0 | −20.0 | −10.0 |
| 319.2 | 167.2 | 11.12-EET | −40.0 | −10.0 | −20.0 | −10.0 |
| 319.2 | 219.2 | 14.15-EET | −60.0 | −10.0 | −20.0 | −10.0 |
| 337.2 | 145.0 | 5.6-DHET | −70.0 | −5.0 | −20.0 | −10.0 |
| 337.2 | 127.1 | 8.9-DHET | −60.0 | −5.0 | −30.0 | −15.0 |
| 337.2 | 167.1 | 11.12-DHET | −30.0 | −5.0 | −30.0 | −15.0 |
| 337.2 | 207.1 | 14.15-DHET | −60.0 | −5.0 | −20.0 | −10.0 |
| 303.3 | 259.1 | AA | −100.0 | −10.0 | −16.0 | −18.0 |
| 301.3 | 257.1 | EPA | −100.0 | −10.0 | −16.0 | −18.0 |
| 327.3 | 283.1 | DHA | −100.0 | −10.0 | −16.0 | −18.0 |
| 329.3 | 285.1 | DPA | −100.0 | −10.0 | −16.0 | −18.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salamone, S.; Waltl, L.; Pompignan, A.; Grassi, G.; Chianese, G.; Koeberle, A.; Pollastro, F. Phytochemical Characterization of Cannabis sativa L. Chemotype V Reveals Three New Dihydrophenanthrenoids That Favorably Reprogram Lipid Mediator Biosynthesis in Macrophages. Plants 2022, 11, 2130. https://doi.org/10.3390/plants11162130
Salamone S, Waltl L, Pompignan A, Grassi G, Chianese G, Koeberle A, Pollastro F. Phytochemical Characterization of Cannabis sativa L. Chemotype V Reveals Three New Dihydrophenanthrenoids That Favorably Reprogram Lipid Mediator Biosynthesis in Macrophages. Plants. 2022; 11(16):2130. https://doi.org/10.3390/plants11162130
Chicago/Turabian StyleSalamone, Stefano, Lorenz Waltl, Anna Pompignan, Gianpaolo Grassi, Giuseppina Chianese, Andreas Koeberle, and Federica Pollastro. 2022. "Phytochemical Characterization of Cannabis sativa L. Chemotype V Reveals Three New Dihydrophenanthrenoids That Favorably Reprogram Lipid Mediator Biosynthesis in Macrophages" Plants 11, no. 16: 2130. https://doi.org/10.3390/plants11162130
APA StyleSalamone, S., Waltl, L., Pompignan, A., Grassi, G., Chianese, G., Koeberle, A., & Pollastro, F. (2022). Phytochemical Characterization of Cannabis sativa L. Chemotype V Reveals Three New Dihydrophenanthrenoids That Favorably Reprogram Lipid Mediator Biosynthesis in Macrophages. Plants, 11(16), 2130. https://doi.org/10.3390/plants11162130








