Pangenomics and Crop Genome Adaptation in a Changing Climate
Abstract
:1. Introduction
2. Main
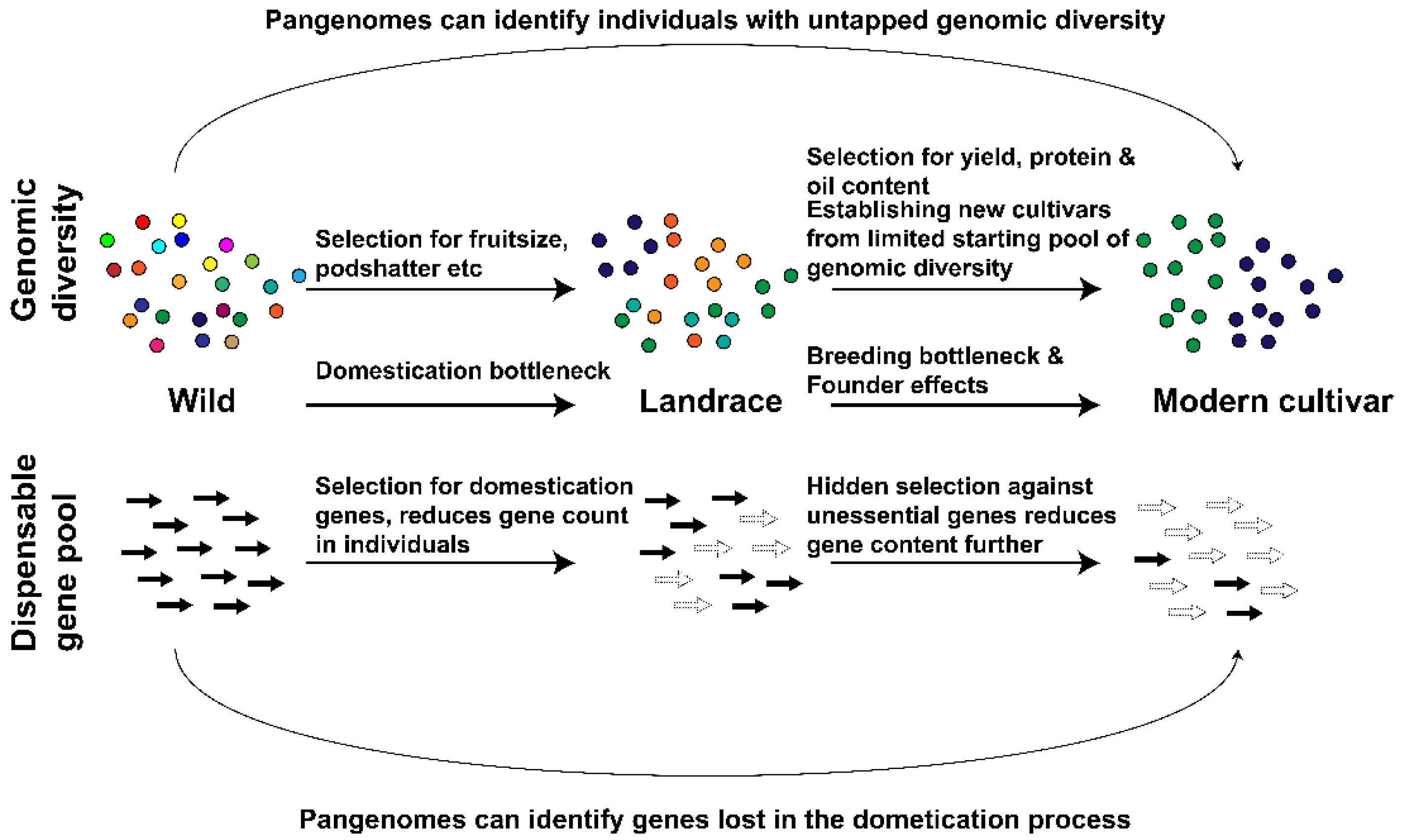
2.1. Pangenomes Highlight Individuals with Unused Genomic Diversity
2.2. Pangenomes Enable the Recovery of Genes Lost during Domestication and Breeding Improvements
2.3. Accelerated Breeding and Targeted Genome Editing Can Alleviate the Impact of Reduced Genomic Diversity and Loss of Genes in Modern Cultivars
3. Conclusions and Outlook
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xu, Y.; Ramanathan, V.; Victor, D.G. Global Warming Will Happen Faster than we Think. 2018. Available online: https://www.nature.com/articles/d41586-018-07586-5 (accessed on 30 May 2022).
- Abberton, M.; Batley, J.; Bentley, A.; Bryant, J.; Cai, H.; Cockram, J.; Costa de Oliveira, A.; Cseke, L.J.; Dempewolf, H.; De Pace, C. Global agricultural intensification during climate change: A role for genomics. Plant Biotechnol. J. 2016, 14, 1095–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batley, J.; Edwards, D. The application of genomics and bioinformatics to accelerate crop improvement in a changing climate. Curr. Opin. Plant Biol. 2016, 30, 78–81. [Google Scholar] [CrossRef] [Green Version]
- Rosenzweig, C.; Iglesius, A.; Yang, X.-B.; Epstein, P.R.; Chivian, E. Climate change and extreme weather events-Implications for food production, plant diseases, and pests. Glob. Change Hum. Health 2001, 2, 2. [Google Scholar]
- Pugh, T.; Müller, C.; Elliott, J.; Deryng, D.; Folberth, C.; Olin, S.; Schmid, E.; Arneth, A. Climate analogues suggest limited potential for intensification of production on current croplands under climate change. Nat. Commun. 2016, 7, 12608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parry, M.; Rosenzweig, C.; Livermore, M. Climate change, global food supply and risk of hunger. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 2125–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.; Somanathan, E.; Dey, S. Global warming and local air pollution have reduced wheat yields in India. Clim. Change 2017, 140, 593–604. [Google Scholar] [CrossRef]
- Moore Frances, C.; Lobell David, B. The fingerprint of climate trends on European crop yields. Proc. Natl. Acad. Sci. USA 2015, 112, 2670–2675. [Google Scholar] [CrossRef] [Green Version]
- Potopová, V.; Zahradníček, P.; Štěpánek, P.; Türkott, L.; Farda, A.; Soukup, J. The impacts of key adverse weather events on the field-grown vegetable yield variability in the Czech Republic from 1961 to 2014. Int. J. Climatol. 2017, 37, 1648–1664. [Google Scholar] [CrossRef]
- van Dijk, M.; Morley, T.; Rau, M.L.; Saghai, Y. A meta-analysis of projected global food demand and population at risk of hunger for the period 2010–2050. Nature Food 2021, 2, 494–501. [Google Scholar] [CrossRef]
- Elzinga, J.A.; Atlan, A.; Biere, A.; Gigord, L.; Weis, A.E.; Bernasconi, G. Time after time: Flowering phenology and biotic interactions. Trends Ecol. Evol. 2007, 22, 432–439. [Google Scholar] [CrossRef] [Green Version]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef] [Green Version]
- Gepts, P. Crop domestication as a long-term selection experiment. In Plant Breeding Reviews; Wiley: Hoboken, NJ, USA, 2004; Volume 24, pp. 1–44. [Google Scholar]
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005, 8, 135–141. [Google Scholar] [CrossRef]
- Kejnovsky, E.; Hawkins, J.; Feschotte, C.; Wendel, J.; Greilhuber, J.; Dolezel, J.; Leitch, I. Plant Genome Diversity; Springer: Vienna, Austria, 2012; Volume 10, pp. 973–978. [Google Scholar]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Cai, K.; Zhang, G.; Zeng, F. Metabolite profiling of barley grains subjected to water stress: To explain the genotypic difference in drought-induced impacts on malting quality. Front. Plant Sci. 2017, 8, 1547. [Google Scholar] [CrossRef] [PubMed]
- Zeder, M.A.; Emshwiller, E.; Smith, B.D.; Bradley, D.G. Documenting domestication: The intersection of genetics and archaeology. Trends Genet. 2006, 22, 139–155. [Google Scholar] [CrossRef]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef]
- Bayer, P.E.; Valliyodan, B.; Hu, H.; Marsh, J.I.; Yuan, Y.; Vuong, T.D.; Patil, G.; Song, Q.; Batley, J.; Varshney, R.K. Sequencing the USDA core soybean collection reveals gene loss during domestication and breeding. Plant Genome 2021, 15, e20109. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.-A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-Genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176.e113. [Google Scholar] [CrossRef]
- Gao, L.; Gonda, I.; Sun, H.; Ma, Q.; Bao, K.; Tieman, D.M.; Burzynski-Chang, E.A.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L.; et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef]
- Li, J.; Yuan, D.; Wang, P.; Wang, Q.; Sun, M.; Liu, Z.; Si, H.; Xu, Z.; Ma, Y.; Zhang, B.; et al. Cotton pan-genome retrieves the lost sequences and genes during domestication and selection. Genome Biol. 2021, 22, 119. [Google Scholar] [CrossRef]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Zhan, Q.; Lu, Y.; Zhang, L.; Huang, T.; et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshney, R.K.; Roorkiwal, M.; Sun, S.; Bajaj, P.; Chitikineni, A.; Thudi, M.; Singh, N.P.; Du, X.; Upadhyaya, H.D.; Khan, A.W.; et al. A chickpea genetic variation map based on the sequencing of 3366 genomes. Nature 2021, 599, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Ruperao, P.; Thirunavukkarasu, N.; Gandham, P.; Selvanayagam, S.; Govindaraj, M.; Nebie, B.; Manyasa, E.; Gupta, R.; Das, R.R.; Odeny, D.A.; et al. Sorghum Pan-Genome Explores the Functional Utility for Genomic-Assisted Breeding to Accelerate the Genetic Gain. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Hübner, S.; Bercovich, N.; Todesco, M.; Mandel, J.R.; Odenheimer, J.; Ziegler, E.; Lee, J.S.; Baute, G.J.; Owens, G.L.; Grassa, C.J.; et al. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat. Plants 2019, 5, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Hufford, M.B.; Seetharam, A.S.; Woodhouse, M.R.; Chougule, K.M.; Ou, S.; Liu, J.; Ricci, W.A.; Guo, T.; Olson, A.; Qiu, Y.; et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. bioRxiv 2021. [Google Scholar] [CrossRef]
- Golicz, A.A.; Bayer, P.E.; Barker, G.C.; Edger, P.P.; Kim, H.; Martinez, P.A.; Chan, C.K.K.; Severn-Ellis, A.; McCombie, W.R.; Parkin, I.A.P.; et al. The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Commun. 2016, 7, 13390. [Google Scholar] [CrossRef] [Green Version]
- Hurgobin, B.; Golicz, A.A.; Bayer, P.E.; Chan, C.-K.K.; Tirnaz, S.; Dolatabadian, A.; Schiessl, S.V.; Samans, B.; Montenegro, J.D.; Parkin, I.A.P.; et al. Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol. J. 2018, 16, 1265–1274. [Google Scholar] [CrossRef] [Green Version]
- Bayer, P.E.; Scheben, A.; Golicz, A.A.; Yuan, Y.; Faure, S.; Lee, H.; Chawla, H.S.; Anderson, R.; Bancroft, I.; Raman, H.; et al. Modelling of gene loss propensity in the pangenomes of three Brassica species suggests different mechanisms between polyploids and diploids. Plant Biotechnol. J. 2021, 19, 2488–2500. [Google Scholar] [CrossRef]
- Khush, G.S. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef]
- Eyre-Walker, A.; Gaut, R.L.; Hilton, H.; Feldman, D.L.; Gaut, B.S. Investigation of the bottleneck leading to the domestication of maize. Proc. Natl. Acad. Sci. USA 1998, 95, 4441–4446. [Google Scholar] [CrossRef] [Green Version]
- Hyten, D.L.; Song, Q.; Zhu, Y.; Choi, I.-Y.; Nelson, R.L.; Costa, J.M.; Specht, J.E.; Shoemaker, R.C.; Cregan, P.B. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 2006, 103, 16666–16671. [Google Scholar] [CrossRef] [Green Version]
- Carter, T.E., Jr.; Nelson, R.L.; Sneller, C.H.; Cui, Z. Genetic diversity in soybean. Soybeans Improv. Prod. Uses 2004, 16, 303–416. [Google Scholar]
- Valliyodan, B.; Cannon, S.B.; Bayer, P.E.; Shu, S.; Brown, A.V.; Ren, L.; Jenkins, J.; Chung, C.Y.-L.; Chan, T.-F.; Daum, C.G.; et al. Construction and comparison of three reference-quality genome assemblies for soybean. Plant J. 2019, 100, 1066–1082. [Google Scholar] [CrossRef]
- Wysmierski, P.T.; Vello, N.A. The genetic base of Brazilian soybean cultivars: Evolution over time and breeding implications. Genet. Mol. Biol. 2013, 36, 547–555. [Google Scholar] [CrossRef]
- Sood, S.; Kuraparthy, V.; Bai, G.; Gill, B.S. The major threshability genes soft glume (sog) and tenacious glume (Tg), of diploid and polyploid wheat, trace their origin to independent mutations at non-orthologous loci. Theor. Appl. Genet. 2009, 119, 341–351. [Google Scholar] [CrossRef]
- Simons, K.J.; Fellers, J.P.; Trick, H.N.; Zhang, Z.; Tai, Y.-S.; Gill, B.S.; Faris, J.D. Molecular characterization of the major wheat domestication gene Q. Genetics 2006, 172, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Charmet, G. Wheat domestication: Lessons for the future. Comptes Rendus Biol. 2011, 334, 212–220. [Google Scholar] [CrossRef]
- Berger, J.D.; Buirchell, B.J.; Luckett, D.J.; Nelson, M.N. Domestication bottlenecks limit genetic diversity and constrain adaptation in narrow-leafed lupin (Lupinus angustifolius L.). Theor. Appl. Genet. 2012, 124, 637–652. [Google Scholar] [CrossRef]
- Grau Nersting, L.; Bode Andersen, S.; von Bothmer, R.; Gullord, M.; Bagger Jørgensen, R. Morphological and molecular diversity of Nordic oat through one hundred years of breeding. Euphytica 2006, 150, 327–337. [Google Scholar] [CrossRef]
- Parzies, H.; Spoor, W.; Ennos, R. Genetic diversity of barley landrace accessions (Hordeum vulgare ssp. vulgare) conserved for different lengths of time in ex situ gene banks. Heredity 2000, 84, 476–486. [Google Scholar] [CrossRef]
- Peroni, N.; Hanazaki, N. Current and lost diversity of cultivated varieties, especially cassava, under swidden cultivation systems in the Brazilian Atlantic Forest. Agric. Ecosyst. Environ. 2002, 92, 171–183. [Google Scholar] [CrossRef]
- Prashanth, S.; Parani, M.; Mohanty, B.; Talame, V.; Tuberosa, R.; Parida, A. Genetic diversity in cultivars and landraces of Oryza sativa subsp. indica as revealed by AFLP markers. Genome 2002, 45, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Pressoir, G.; Berthaud, J. Patterns of population structure in maize landraces from the Central Valleys of Oaxaca in Mexico. Heredity 2004, 92, 88–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberdi, M.; Bravo, L.A.; Gutiérrez, A.; Gidekel, M.; Corcuera, L.J. Ecophysiology of Antarctic vascular plants. Physiol. Plant. 2002, 115, 479–486. [Google Scholar] [CrossRef]
- Amtmann, A.; Bohnert, H.J.; Bressan, R.A. Abiotic stress and plant genome evolution. Search for new models. Plant Physiol. 2005, 138, 127–130. [Google Scholar] [CrossRef] [Green Version]
- Von Willert, D.; Eller, B.; Werger, M.; Brinckmann, E. Desert succulents and their life strategies. Vegetatio 1990, 90, 133–143. [Google Scholar] [CrossRef]
- Hartman, G.; Gardner, M.; Hymowitz, T.; Naidoo, G. Evaluation of perennial Glycine species for resistance to soybean fungal pathogens that cause Sclerotinia stem rot and sudden death syndrome. Crop Sci. 2000, 40, 545–549. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Yuan, C.; Herman, T.; Hartman, G. Accessions of perennial Glycine species with resistance to multiple types of soybean cyst nematode (Heterodera glycines). Plant Dis. 2017, 101, 1201–1206. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [Green Version]
- Ayenan, M.A.T.; Danquah, A.; Hanson, P.; Asante, I.K.; Danquah, E.Y. Identification of new sources of heat tolerance in cultivated and wild tomatoes. Euphytica 2021, 217, 33. [Google Scholar] [CrossRef]
- Tenorio, F.; Ye, C.; Redoña, E.; Sierra, S.; Laza, M.; Argayoso, M. Screening rice genetic resources for heat tolerance. Sabrao J. Breed. Genet. 2013, 45, 371–381. [Google Scholar]
- Li, G.; Wang, L.; Yang, J.; He, H.; Jin, H.; Li, X.; Ren, T.; Ren, Z.; Li, F.; Han, X.; et al. A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 2021, 53, 574–584. [Google Scholar] [CrossRef]
- Montenegro, J.D.; Golicz, A.A.; Bayer, P.E.; Hurgobin, B.; Lee, H.; Chan, C.K.K.; Visendi, P.; Lai, K.; Doležel, J.; Batley, J. The pangenome of hexaploid bread wheat. Plant J. 2017, 90, 1007–1013. [Google Scholar] [CrossRef] [Green Version]
- Yao, W.; Li, G.; Zhao, H.; Wang, G.; Lian, X.; Xie, W. Exploring the rice dispensable genome using a metagenome-like assembly strategy. Genome Biol. 2015, 16, 187. [Google Scholar] [CrossRef] [Green Version]
- Bayer, P.E.; Hu, H.; Petereit, J.; Derbyshire, M.C.; Varshney, R.K.; Valliyodan, B.; Nguyen, H.T.; Batley, J.; Edwards, D. Yield is negatively correlated with nucleotide-binding leucine-rich repeat gene content in soybean. bioRxiv 2021. [Google Scholar] [CrossRef]
- Xu, K.; Mackill, D.J. A major locus for submergence tolerance mapped on rice chromosome 9. Mol. Breed. 1996, 2, 219–224. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Fukao, T.; Ronald, P.; Ismail, A.; Heuer, S.; Mackill, D. Submergence tolerant rice: SUB1’s journey from landrace to modern cultivar. Rice 2010, 3, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [Green Version]
- Pucciariello, C.; Perata, P. Quiescence in rice submergence tolerance: An evolutionary hypothesis. Trends Plant Sci. 2013, 18, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Shimelis, H.; Laing, M. Timelines in conventional crop improvement: Pre-breeding and breeding procedures. Aust. J. Crop Sci. 2012, 6, 1542–1549. [Google Scholar]
- Ahmar, S.; Gill, R.A.; Jung, K.-H.; Faheem, A.; Qasim, M.U.; Mubeen, M.; Zhou, W. Conventional and molecular techniques from simple breeding to speed breeding in crop plants: Recent advances and future outlook. Int. J. Mol. Sci. 2020, 21, 2590. [Google Scholar] [CrossRef] [Green Version]
- Gerald, N.; Frei, U.K.; Lübberstedt, T. Accelerating plant breeding. Trends Plant Sci. 2013, 18, 667–672. [Google Scholar]
- Osakabe, Y.; Osakabe, K. Genome editing with engineered nucleases in plants. Plant Cell Physiol. 2015, 56, 389–400. [Google Scholar] [CrossRef] [Green Version]
- Scheben, A.; Wolter, F.; Batley, J.; Puchta, H.; Edwards, D. Towards CRISPR/Cas crops—Bringing together genomics and genome editing. New Phytol. 2017, 216, 682–698. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [Green Version]
- Marsh, J.I.; Hu, H.; Gill, M.; Batley, J.; Edwards, D. Crop breeding for a changing climate: Integrating phenomics and genomics with bioinformatics. Theor. Appl. Genet. 2021, 134, 1677–1690. [Google Scholar] [CrossRef]
- Warschefsky, E.; Penmetsa, R.V.; Cook, D.R.; von Wettberg, E.J. Back to the wilds: Tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am. J. Bot. 2014, 101, 1791–1800. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Bandyopadhyay, A.; Kancharla, N.; Javalkote, V.S.; Dasgupta, S.; Brutnell, T.P. CRISPR-Cas12a (Cpf1): A Versatile Tool in the Plant Genome Editing Tool Box for Agricultural Advancement. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014, 32, 577–582. [Google Scholar] [CrossRef]
- Terns, M.P. CRISPR-Based Technologies: Impact of RNA-Targeting Systems. Mol. Cell 2018, 72, 404–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazi, T.A.; Biswas, S.R. CRISPR/dCas system as the modulator of gene expression. Prog. Mol. Biol. Transl. Sci. 2021, 178, 99–122. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.-e.-A.; Mahas, A.; Vanderschuren, H.; Mahfouz, M.M. Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 2020, 21, 289. [Google Scholar] [CrossRef] [PubMed]
- Scheben, A.; Edwards, D. Genome editors take on crops. Science 2017, 355, 1122–1123. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.W.; Garg, V.; Roorkiwal, M.; Golicz, A.A.; Edwards, D.; Varshney, R.K. Super-Pangenome by Integrating the Wild Side of a Species for Accelerated Crop Improvement. Trends Plant Sci. 2020, 25, 148–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxted, N.; Kell, S. Establishment of a Global Network for the In Situ Conservation of Crop Wild Relatives: Status and Needs; FAO Commission on Genetic Resources for Food and Agriculture: Rome, Italy, 2009. [Google Scholar]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Liu, X.; Ge, S.; Jensen, J.D.; Hu, F.; Li, X.; Dong, Y.; Gutenkunst, R.N.; Fang, L.; Huang, L. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012, 30, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, K.; Moreira, J.d.R.; Peres, L.E.P.; Zsögön, A. De novo domestication of wild species to create crops with increased resilience and nutritional value. Curr. Opin. Plant Biol. 2021, 60, 102006. [Google Scholar] [CrossRef]
- Yu, H.; Lin, T.; Meng, X.; Du, H.; Zhang, J.; Liu, G.; Chen, M.; Jing, Y.; Kou, L.; Li, X.; et al. A route to de novo domestication of wild allotetraploid rice. Cell 2021, 184, 1156–1170.e1114. [Google Scholar] [CrossRef]
- Tay Fernandez, C.G.; Nestor, B.J.; Danilevicz, M.F.; Marsh, J.I.; Petereit, J.; Bayer, P.E.; Batley, J.; Edwards, D. Expanding Gene-Editing Potential in Crop Improvement with Pangenomes. Int. J. Mol. Sci. 2022, 23, 2276. [Google Scholar] [CrossRef]
- Bosse, M.; Megens, H.J.; Derks, M.F.L.; de Cara, Á.M.R.; Groenen, M.A.M. Deleterious alleles in the context of domestication, inbreeding, and selection. Evol. Appl. 2019, 12, 6–17. [Google Scholar] [CrossRef] [Green Version]
- Dolatabadian, A.; Bayer, P.E.; Tirnaz, S.; Hurgobin, B.; Edwards, D.; Batley, J. Characterization of disease resistance genes in the Brassica napus pangenome reveals significant structural variation. Plant Biotechnol. J. 2020, 18, 969–982. [Google Scholar] [CrossRef] [Green Version]
- Karim, M.M.; Siddika, A.; Tonu, N.N.; Hossain, D.M.; Meah, M.B.; Kawanabe, T.; Fujimoto, R.; Okazaki, K. Production of high yield short duration Brassica napus by interspecific hybridization between B. oleracea and B. rapa. Breed Sci. 2014, 63, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Mason, A.S.; Farooq, M.A.; Islam, F.; Quezada-Martinez, D.; Hu, D.; Yang, S.; Zou, J.; Zhou, W. Challenges and prospects for a potential allohexaploid Brassica crop. Theor. Appl. Genet. 2021, 134, 2711–2726. [Google Scholar] [CrossRef]
- Stace, C.A. Triticale: A Case of Nomenclatural mistreatment. TAXON 1987, 36, 445–452. [Google Scholar] [CrossRef]
- Ruiz, M.; Oustric, J.; Santini, J.; Morillon, R. Synthetic Polyploidy in Grafted Crops. Front. Plant Sci. 2020, 11, 540894. [Google Scholar] [CrossRef]
- Mason, A.S.; Zhang, J.; Tollenaere, R.; Vasquez Teuber, P.; Dalton-Morgan, J.; Hu, L.; Yan, G.; Edwards, D.; Redden, R.; Batley, J. High-throughput genotyping for species identification and diversity assessment in germplasm collections. Mol. Ecol. Resour. 2015, 15, 1091–1101. [Google Scholar] [CrossRef] [Green Version]
- Rawale, K.S.; Khan, M.A.; Gill, K.S. The novel function of the Ph1 gene to differentiate homologs from homoeologs evolved in Triticum turgidum ssp. dicoccoides via a dramatic meiosis-specific increase in the expression of the 5B copy of the C-Ph1 gene. Chromosoma 2019, 128, 561–570. [Google Scholar] [CrossRef]
- Higgins, E.E.; Howell, E.C.; Armstrong, S.J.; Parkin, I.A.P. A major quantitative trait locus on chromosome A9, BnaPh1, controls homoeologous recombination in Brassica napus. New Phytol. 2021, 229, 3281–3293. [Google Scholar] [CrossRef]
| Crop | Breeding State | Dispensable Genes | Reference |
|---|---|---|---|
| Soybean | Wild (G. soja) | 10.17% | [54] |
| Landrace (G. max) | 9.06% | ||
| Modern cultivar (G. max) | 8.69% | ||
| Cotton | Landrace (G. hirsutum) | 24.14% | [55] |
| Modern cultivar (G. hirsutum) | 23.48% | ||
| Tomato | Wild (S. pimpinellifolium) | 20.98% | [22] |
| Landrace (S. lycopersicum var. cerasiforme) | 18.60% | ||
| Modern cultivar (S. lycopersicum var. lycopersicum-Heirloom) | 16.11% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petereit, J.; Bayer, P.E.; Thomas, W.J.W.; Tay Fernandez, C.G.; Amas, J.; Zhang, Y.; Batley, J.; Edwards, D. Pangenomics and Crop Genome Adaptation in a Changing Climate. Plants 2022, 11, 1949. https://doi.org/10.3390/plants11151949
Petereit J, Bayer PE, Thomas WJW, Tay Fernandez CG, Amas J, Zhang Y, Batley J, Edwards D. Pangenomics and Crop Genome Adaptation in a Changing Climate. Plants. 2022; 11(15):1949. https://doi.org/10.3390/plants11151949
Chicago/Turabian StylePetereit, Jakob, Philipp E. Bayer, William J. W. Thomas, Cassandria G. Tay Fernandez, Junrey Amas, Yueqi Zhang, Jacqueline Batley, and David Edwards. 2022. "Pangenomics and Crop Genome Adaptation in a Changing Climate" Plants 11, no. 15: 1949. https://doi.org/10.3390/plants11151949
APA StylePetereit, J., Bayer, P. E., Thomas, W. J. W., Tay Fernandez, C. G., Amas, J., Zhang, Y., Batley, J., & Edwards, D. (2022). Pangenomics and Crop Genome Adaptation in a Changing Climate. Plants, 11(15), 1949. https://doi.org/10.3390/plants11151949









