Structural and Functional Characterization of CreFH1, the Frataxin Homolog from Chlamydomonas reinhardtii
Abstract
:1. Introduction
2. Results
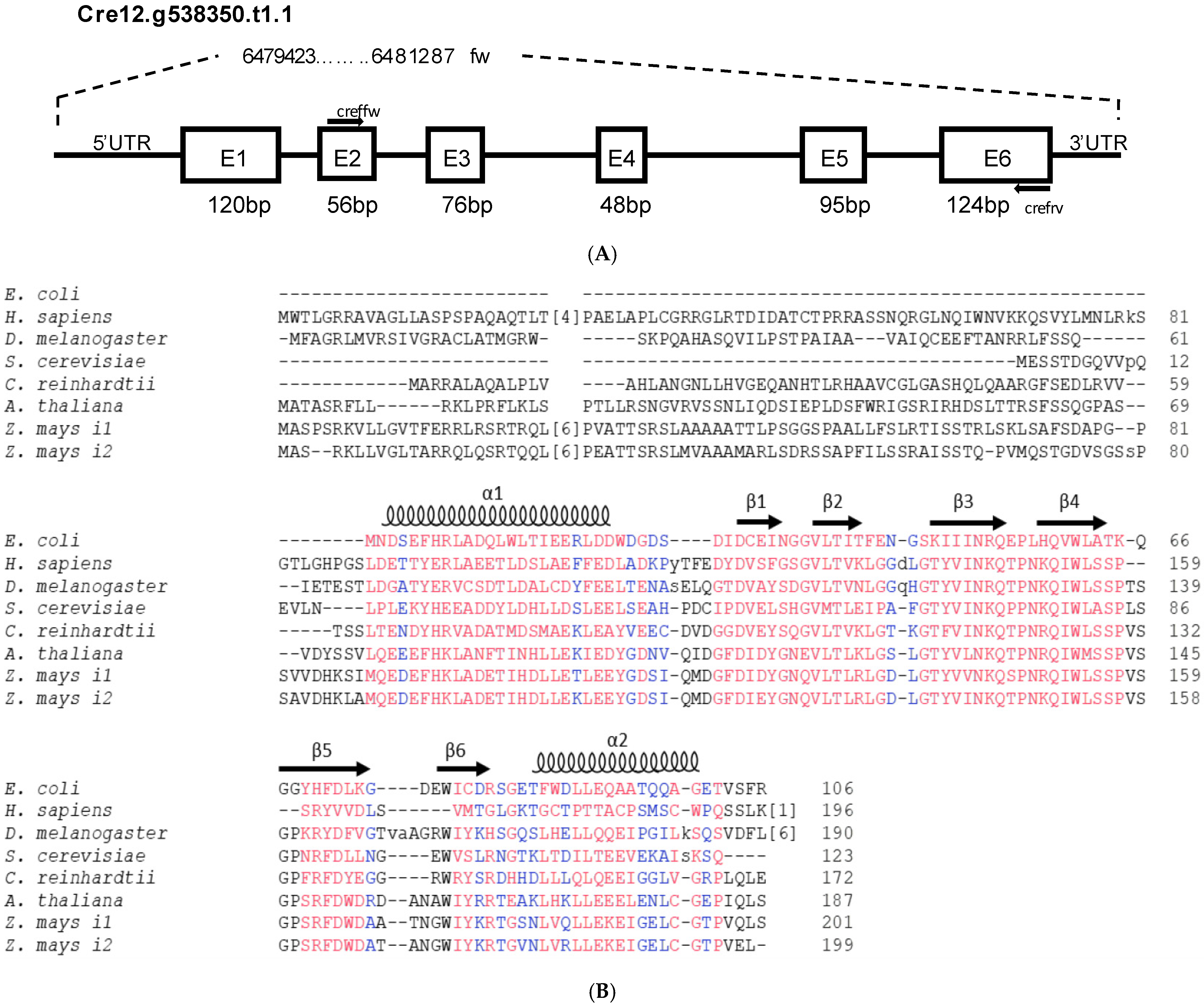
2.1. Gene Structure, Protein Sequence Analysis and Homology Modelling
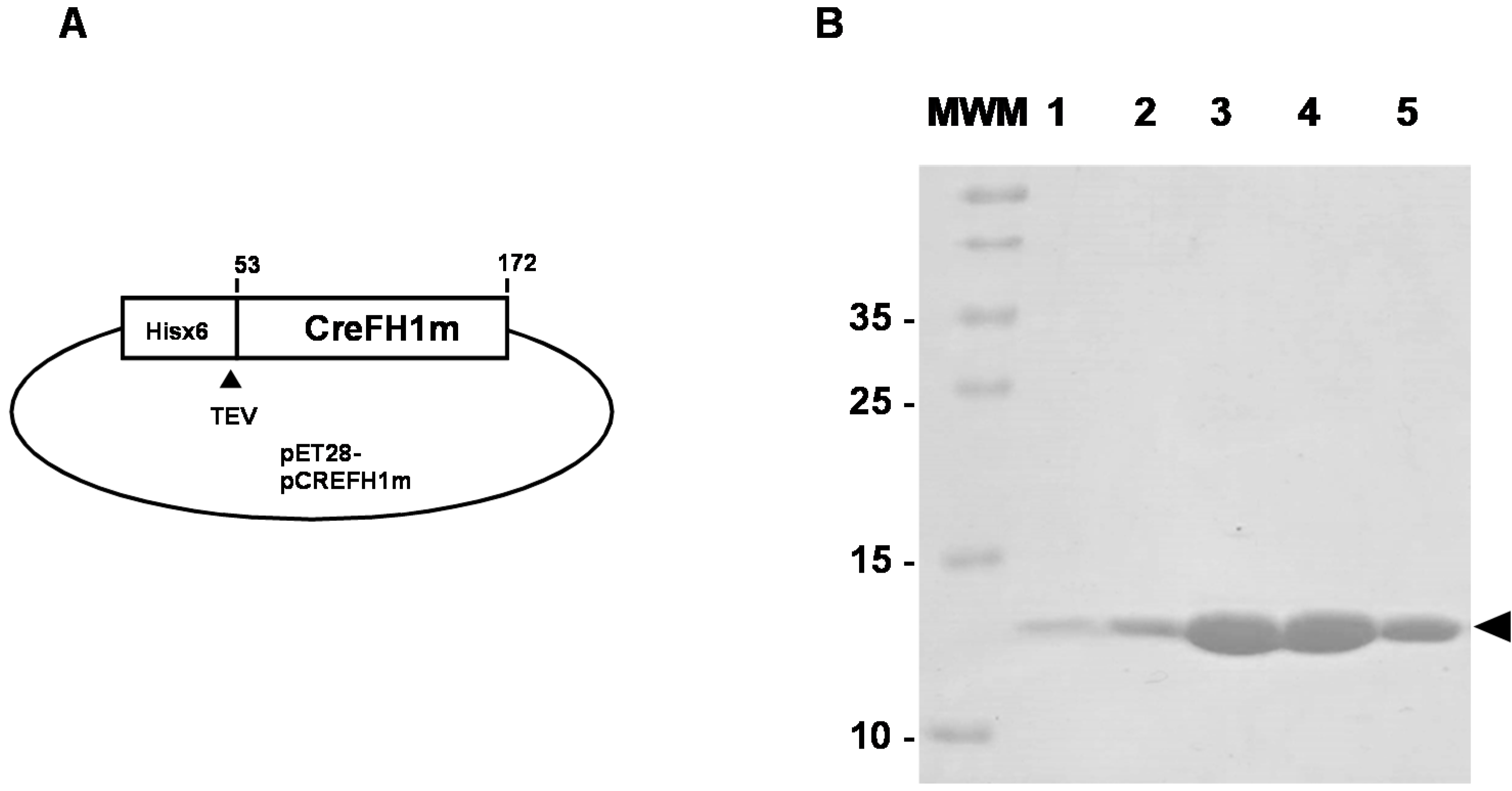
2.2. Cloning, Expression and Purification of CreFH1
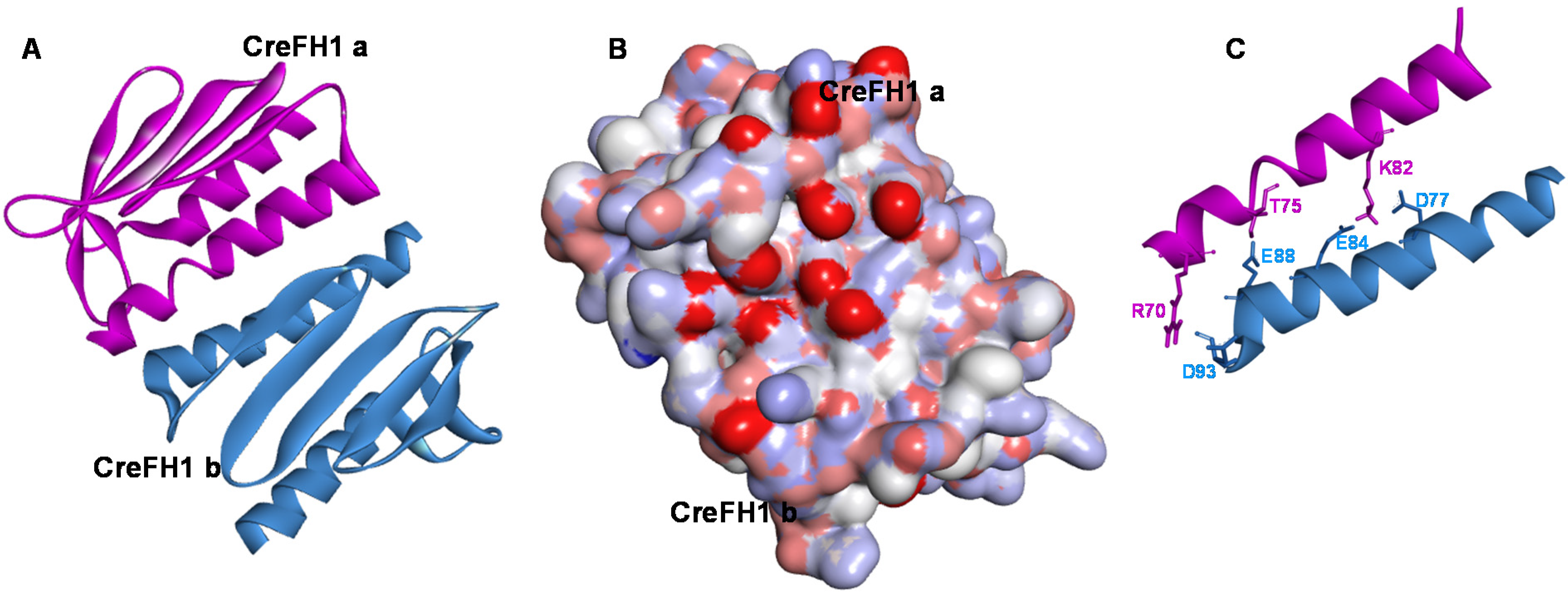
2.3. Oligomerization with Metals
2.4. Attenuation of Fenton Reaction by CreFH1
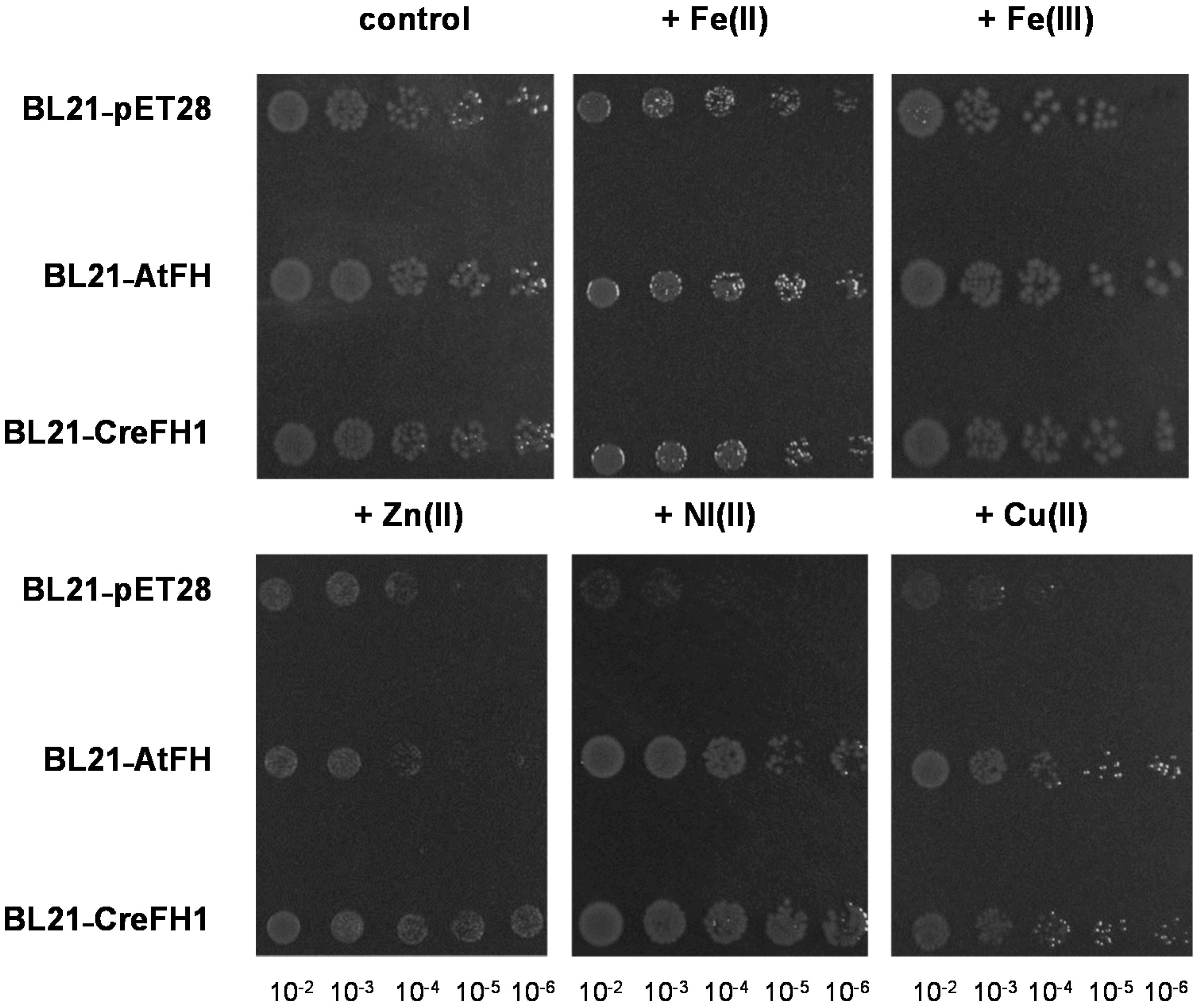
2.5. Effect of Metals and H2O2 in E. coli Overexpressing AtFH and CreFH1
3. Discussion
4. Materials and Methods
4.1. Algal Strains and Culture Conditions
4.2. Cloning, Expression and Purification of the Chlamydomonas Reinhardtii Frataxin Homolog CreFH1
4.3. Native and SDS-PAGE Analysis
4.4. ESI-TOF MS Molecular Mass Determinations
4.5. Oxidative Degradation Assays
4.6. Effect of Metals and H2O2 on E. coli Cells Overexpressing AtFH or CreFH1
4.7. Additional Methods
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campuzano, V.; Montermni, L.; Molto, M.D.; Pianese, L.; Cossee, M.; Cavalcanti, F.; Monros, E.; Rodius, F.; Duclos, F.; Monticelli, A.; et al. Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 1996, 271, 1423–1427. [Google Scholar] [CrossRef] [PubMed]
- Babcock, M.; de Silva, D.; Oaks, R.; Davis-Kaplan, S.; Jiralerspong, S.; Montermini, L.; Pandolfo, M.; Kaplan, J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 1997, 276, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Isaya, G.; O’Neill, H.A.; Gakh, O.; Park, S.; Mantcheva, R.; Mooney, S.M. Functional studies of frataxin. Acta Paediatr. 2004, 93, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Pastore, A.; Puccio, H. Frataxin: A protein in search for a function. J. Neurochem. 2013, 126 (Suppl. S1), 43–52. [Google Scholar] [CrossRef]
- Martin, M.; Colman, M.J.; Gomez-Casati, D.F.; Lamattina, L.; Zabaleta, E.J. Nitric oxide accumulation is required to protect against iron-mediated oxidative stress in frataxin-deficient Arabidopsis plants. FEBS Lett. 2009, 583, 542–548. [Google Scholar] [CrossRef] [Green Version]
- Ristow, M.; Pfister, M.F.; Yee, A.J.; Schubert, M.; Michael, L.; Zhang, C.Y.; Ueki, K.; Michael, M.D., 2nd; Lowell, B.B.; Kahn, C.R. Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. Proc. Natl. Acad. Sci. USA 2000, 97, 12239–12243. [Google Scholar] [CrossRef] [Green Version]
- Bulteau, A.L.; O’Neill, H.A.; Kennedy, M.C.; Ikeda-Saito, M.; Isaya, G.; Szweda, L.I. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 2004, 305, 242–245. [Google Scholar] [CrossRef] [Green Version]
- Bencze, K.Z.; Kondapalli, K.C.; Cook, J.D.; McMahon, S.; Millan-Pacheco, C.; Pastor, N.; Stemmler, T.L. The structure and function of frataxin. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 269–291. [Google Scholar] [CrossRef] [Green Version]
- Busi, M.V.; Gomez-Casati, D.F. Exploring frataxin function. IUBMB Life 2012, 64, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Casati, D.F.; Busi, M.V.; Pagani, M.A. Plant Frataxin in Metal Metabolism. Front. Plant Sci. 2018, 9, 1706. [Google Scholar] [CrossRef]
- Schoenfeld, R.A.; Napoli, E.; Wong, A.; Zhan, S.; Reutenauer, L.; Morin, D.; Buckpitt, A.R.; Taroni, F.; Lonnerdal, B.; Ristow, M.; et al. Frataxin deficiency alters heme pathway transcripts and decreases mitochondrial heme metabolites in mammalian cells. Hum. Mol. Genet. 2005, 14, 3787–3799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Napoli, E.; Morin, D.; Bernhardt, R.; Buckpitt, A.; Cortopassi, G. Hemin rescues adrenodoxin, heme a and cytochrome oxidase activity in frataxin-deficient oligodendroglioma cells. Biochim. Biophys. Acta 2007, 1772, 773–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maliandi, M.V.; Busi, M.V.; Turowski, V.R.; Leaden, L.; Araya, A.; Gomez-Casati, D.F. The mitochondrial protein frataxin is essential for heme biosynthesis in plants. FEBS J. 2011, 278, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Muhlenhoff, U.; Richhardt, N.; Gerber, J.; Lill, R. Characterization of iron-sulfur protein assembly in isolated mitochondria: A requirement for ATP, NADH, and reduced iron. J. Biol. Chem. 2002, 277, 29810–29816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Gakh, O.; Mooney, S.M.; Isaya, G. The ferroxidase activity of yeast frataxin. J. Biol. Chem. 2002, 277, 38589–38595. [Google Scholar] [CrossRef] [Green Version]
- Yoon, T.; Cowan, J.A. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 2003, 125, 6078–6084. [Google Scholar] [CrossRef]
- Armas, A.M.; Balparda, M.; Terenzi, A.; Busi, M.V.; Pagani, M.A.; Gomez-Casati, D.F. Ferrochelatase activity of plant frataxin. Biochimie 2019, 156, 118–122. [Google Scholar] [CrossRef]
- Du, J.; Zhou, Y.; Li, Y.; Xia, J.; Chen, Y.; Chen, S.; Wang, X.; Sun, W.; Wang, T.; Ren, X.; et al. Identification of Frataxin as a regulator of ferroptosis. Redox Biol. 2020, 32, 101483. [Google Scholar] [CrossRef]
- Chen, O.S.; Hemenway, S.; Kaplan, J. Inhibition of Fe-S cluster biosynthesis decreases mitochondrial iron export: Evidence that Yfh1p affects Fe-S cluster synthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 12321–12326. [Google Scholar] [CrossRef] [Green Version]
- Gerber, J.; Muhlenhoff, U.; Lill, R. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003, 4, 906–911. [Google Scholar] [CrossRef] [Green Version]
- Tsai, C.L.; Barondeau, D.P. Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry 2010, 49, 9132–9139. [Google Scholar] [CrossRef] [PubMed]
- Turowski, V.R.; Busi, M.V.; Gomez-Casati, D.F. Structural and functional studies of the mitochondrial cysteine desulfurase from Arabidopsis thaliana. Mol. Plant 2012, 5, 1001–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, N.G.; Yu, X.; Feng, X.; Bailey, H.J.; Martelli, A.; Nabhan, J.F.; Strain-Damerell, C.; Bulawa, C.; Yue, W.W.; Han, S. Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism. Nat. Commun. 2019, 10, 2210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, K.; Frederick, R.O.; Tonelli, M.; Markley, J.L. Interactions of iron-bound frataxin with ISCU and ferredoxin on the cysteine desulfurase complex leading to Fe-S cluster assembly. J. Inorg. Biochem. 2018, 183, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.P.; Srivastava, S.; Kumar, S.K.P.; Sinha, D.; D’Silva, P. Mapping Key Residues of ISD11 Critical for NFS1-ISD11 Subcomplex Stability: Implications in the development of mitochondrial disorder, COXPD19. J. Biol. Chem. 2015, 290, 25876–25890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, Y.; Napoli, E.; Cortopassi, G. Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum. Mol. Genet. 2007, 16, 929–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, T.; Cowan, J.A. Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J. Biol. Chem. 2004, 279, 25943–25946. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Gakh, O.; O’Neill, H.A.; Mangravita, A.; Nichol, H.; Ferreira, G.C.; Isaya, G. Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation. J. Biol. Chem. 2003, 278, 31340–31351. [Google Scholar] [CrossRef] [Green Version]
- Armas, A.M.; Balparda, M.; Terenzi, A.; Busi, M.V.; Pagani, M.A.; Gomez-Casati, D.F. Iron-Sulfur Cluster Complex Assembly in the Mitochondria of Arabidopsis thaliana. Plants 2020, 9, 1171. [Google Scholar] [CrossRef]
- Gakh, O.; Park, S.; Liu, G.; Macomber, L.; Imlay, J.A.; Ferreira, G.C.; Isaya, G. Mitochondrial iron detoxification is a primary function of frataxin that limits oxidative damage and preserves cell longevity. Hum. Mol. Genet. 2006, 15, 467–479. [Google Scholar] [CrossRef] [Green Version]
- Karlberg, T.; Schagerlof, U.; Gakh, O.; Park, S.; Ryde, U.; Lindahl, M.; Leath, K.; Garman, E.; Isaya, G.; Al-Karadaghi, S. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure 2006, 14, 1535–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busi, M.V.; Zabaleta, E.J.; Araya, A.; Gomez-Casati, D.F. Functional and molecular characterization of the frataxin homolog from Arabidopsis thaliana. FEBS Lett. 2004, 576, 141–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busi, M.V.; Maliandi, M.V.; Valdez, H.; Clemente, M.; Zabaleta, E.J.; Araya, A.; Gomez-Casati, D.F. Deficiency of Arabidopsis thaliana frataxin alters activity of mitochondrial Fe-S proteins and induces oxidative stress. Plant J. Cell Mol. Biol. 2006, 48, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Turowski, V.R.; Aknin, C.; Maliandi, M.V.; Buchensky, C.; Leaden, L.; Peralta, D.A.; Busi, M.V.; Araya, A.; Gomez-Casati, D.F. Frataxin Is Localized to Both the Chloroplast and Mitochondrion and Is Involved in Chloroplast Fe-S Protein Function in Arabidopsis. PLoS ONE 2015, 10, e0141443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchensky, C.; Sanchez, M.; Carrillo, M.; Palacios, O.; Capdevila, M.; Dominguez-Vera, J.M.; Busi, M.V.; Atrian, S.; Pagani, M.A.; Gomez-Casati, D.F. Identification of two frataxin isoforms in Zea mays: Structural and functional studies. Biochimie 2017, 140, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Maliandi, M.V.; Busi, M.V.; Clemente, M.; Zabaleta, E.J.; Araya, A.; Gomez-Casati, D.F. Expression and one-step purification of recombinant Arabidopsis thaliana frataxin homolog (AtFH). Protein Expr. Purif. 2007, 51, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Vazzola, V.; Losa, A.; Soave, C.; Murgia, I. Knockout of frataxin gene causes embryo lethality in Arabidopsis. FEBS Lett. 2007, 581, 667–672. [Google Scholar] [CrossRef] [Green Version]
- Hippler, M.; Redding, K.; Rochaix, J.D. Chlamydomonas genetics, a tool for the study of bioenergetic pathways. Biochim. Biophys. Acta 1998, 1367, 1–62. [Google Scholar] [CrossRef]
- Glaesener, A.G.; Merchant, S.S.; Blaby-Haas, C.E. Iron economy in Chlamydomonas reinhardtii. Front. Plant Sci. 2013, 4, 337. [Google Scholar] [CrossRef] [Green Version]
- Godman, J.; Balk, J. Genome analysis of Chlamydomonas reinhardtii reveals the existence of multiple, compartmentalized iron-sulfur protein assembly machineries of different evolutionary origins. Genetics 2008, 179, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Casati, D.F.; Busi, M.V.; Barchiesi, J.; Pagani, M.A.; Marchetti-Acosta, N.S.; Terenzi, A. Fe-S Protein Synthesis in Green Algae Mitochondria. Plants 2021, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Phytozome v12. Available online: https://phytozome.jgi.doe.gov/pz/portal.html (accessed on 6 May 2022).
- CD Search. Available online: https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 7 May 2022).
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- TargetP-2.0 Server. Available online: http://www.cbs.dtu.dk/services/TargetP/ (accessed on 29 April 2022).
- Cho, S.J.; Lee, M.G.; Yang, J.K.; Lee, J.Y.; Song, H.K.; Suh, S.W. Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family. Proc. Natl. Acad. Sci. USA 2000, 97, 8932–8937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhe-Paganon, S.; Shigeta, R.; Chi, Y.I.; Ristow, M.; Shoelson, S.E. Crystal structure of human frataxin. J. Biol. Chem. 2000, 275, 30753–30756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Alam, S.L.; Proteasa, S.V.; Zhang, Y.; Lesuisse, E.; Dancis, A.; Stemmler, T.L. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry 2004, 43, 16254–16262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musco, G.; Stier, G.; Kolmerer, B.; Adinolfi, S.; Martin, S.; Frenkiel, T.; Gibson, T.; Pastore, A. Towards a structural understanding of Friedreich’s ataxia: The solution structure of frataxin. Structure 2000, 8, 695–707. [Google Scholar] [CrossRef] [Green Version]
- Nair, M.; Adinolfi, S.; Pastore, C.; Kelly, G.; Temussi, P.; Pastore, A. Solution structure of the bacterial frataxin ortholog, CyaY: Mapping the iron binding sites. Structure 2004, 12, 2037–2048. [Google Scholar] [CrossRef] [Green Version]
- Jpred 4. Available online: http://www.compbio.dundee.ac.uk/jpred/ (accessed on 30 April 2022).
- ProSA-web server. Available online: https://prosa.services.came.sbg.ac.at/prosa.php (accessed on 30 April 2022).
- Verify 3D. Available online: https://servicesn.mbi.ucla.edu/Verify3D/ (accessed on 30 April 2022).
- Bowie, J.U.; Luthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [Green Version]
- Luthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins 1993, 17, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Kundrotas, P.J.; Alexov, E. Predicting 3D structures of transient protein-protein complexes by homology. Biochim. Biophys. Acta 2006, 1764, 1498–1511. [Google Scholar] [CrossRef]
- Hu, Z.; Ma, B.; Wolfson, H.; Nussinov, R. Conservation of polar residues as hot spots at protein interfaces. Proteins 2000, 39, 331–342. [Google Scholar] [CrossRef]
- @Tome v.3. Available online: http://atome.cbs.cnrs.fr/ATOME_V3/index.html (accessed on 30 April 2022).
- SuperPose server v. 1.0. Available online: http://superpose.wishartlab.com/ (accessed on 30 April 2022).
- Cavadini, P.; O’Neill, H.A.; Benada, O.; Isaya, G. Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum. Mol. Genet. 2002, 11, 217–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bou-Abdallah, F.; Adinolfi, S.; Pastore, A.; Laue, T.M.; Dennis Chasteen, N. Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. J. Mol. Biol. 2004, 341, 605–615. [Google Scholar] [CrossRef]
- Gakh, O.; Adamec, J.; Gacy, A.M.; Twesten, R.D.; Owen, W.G.; Isaya, G. Physical evidence that yeast frataxin is an iron storage protein. Biochemistry 2002, 41, 6798–6804. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.; Palacios, O.; Buchensky, C.; Sabio, L.; Gomez-Casati, D.F.; Pagani, M.A.; Capdevila, M.; Atrian, S.; Dominguez-Vera, J.M. Copper redox chemistry of plant frataxins. J. Inorg. Biochem. 2018, 180, 135–140. [Google Scholar] [CrossRef]
- van Zundert, G.C.P.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [Green Version]
- Seguin, A.; Bayot, A.; Dancis, A.; Rogowska-Wrzesinska, A.; Auchere, F.; Camadro, J.M.; Bulteau, A.L.; Lesuisse, E. Overexpression of the yeast frataxin homolog (Yfh1): Contrasting effects on iron-sulfur cluster assembly, heme synthesis and resistance to oxidative stress. Mitochondrion 2009, 9, 130–138. [Google Scholar] [CrossRef]
- Noguera, M.E.; Roman, E.A.; Rigal, J.B.; Cousido-Siah, A.; Mitschler, A.; Podjarny, A.; Santos, J. Structural characterization of metal binding to a cold-adapted frataxin. J. Biol. Inorg. Chem. JBIC A Publ. Soc. Biol. Inorg. Chem. 2015, 20, 653–664. [Google Scholar] [CrossRef]
- Armas, A.M.; Balparda, M.; Turowski, V.R.; Busi, M.V.; Pagani, M.A.; Gomez-Casati, D.F. Altered levels of mitochondrial NFS1 affect cellular Fe and S contents in plants. Plant Cell Rep. 2019, 38, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Adinolfi, S.; Trifuoggi, M.; Politou, A.S.; Martin, S.; Pastore, A. A structural approach to understanding the iron-binding properties of phylogenetically different frataxins. Hum. Mol. Genet. 2002, 11, 1865–1877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamec, J.; Rusnak, F.; Owen, W.G.; Naylor, S.; Benson, L.M.; Gacy, A.M.; Isaya, G. Iron-dependent self-assembly of recombinant yeast frataxin: Implications for Friedreich ataxia. Am. J. Hum. Genet. 2000, 67, 549–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahlgren, E.C.; Fekry, M.; Wiemann, M.; Soderberg, C.A.; Bernfur, K.; Gakh, O.; Rasmussen, M.; Hojrup, P.; Emanuelsson, C.; Isaya, G.; et al. Iron-induced oligomerization of human FXN81-210 and bacterial CyaY frataxin and the effect of iron chelators. PLoS ONE 2017, 12, e0188937. [Google Scholar] [CrossRef] [Green Version]
- Ravindra Kumar, S.; Imlay, J.A. How Escherichia coli tolerates profuse hydrogen peroxide formation by a catabolic pathway. J. Bacteriol. 2013, 195, 4569–4579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, S.; Imlay, J.A. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 2007, 282, 929–937. [Google Scholar] [CrossRef] [Green Version]
- Peralta, D.A.; Araya, A.; Busi, M.V.; Gomez-Casati, D.F. The E3 ubiquitin-ligase SEVEN IN ABSENTIA like 7 mono-ubiquitinates glyceraldehyde-3-phosphate dehydrogenase 1 isoform in vitro and is required for its nuclear localization in Arabidopsis thaliana. Int. J. Biochem. Cell Biol. 2016, 70, 48–56. [Google Scholar] [CrossRef]
- Tropea, J.E.; Cherry, S.; Waugh, D.S. Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol. Biol. 2009, 498, 297–307. [Google Scholar] [CrossRef]
- Gomez Casati, D.F.; Sesma, J.I.; Iglesias, A.A. Structural and kinetic characterization of NADP-dependent, non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from celery leaves. Plant Sci. 2000, 154, 107–115. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pons, J.L.; Labesse, G. @TOME-2: A new pipeline for comparative modeling of protein-ligand complexes. Nucleic Acids Res. 2009, 37, W485–W491. [Google Scholar] [CrossRef] [PubMed]
- Maiti, R.; Van Domselaar, G.H.; Zhang, H.; Wishart, D.S. SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Res. 2004, 32, W590–W594. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terenzi, A.; Pagani, M.A.; Gomez-Casati, D.F.; Busi, M.V. Structural and Functional Characterization of CreFH1, the Frataxin Homolog from Chlamydomonas reinhardtii. Plants 2022, 11, 1931. https://doi.org/10.3390/plants11151931
Terenzi A, Pagani MA, Gomez-Casati DF, Busi MV. Structural and Functional Characterization of CreFH1, the Frataxin Homolog from Chlamydomonas reinhardtii. Plants. 2022; 11(15):1931. https://doi.org/10.3390/plants11151931
Chicago/Turabian StyleTerenzi, Agustina, Maria A. Pagani, Diego F. Gomez-Casati, and Maria V. Busi. 2022. "Structural and Functional Characterization of CreFH1, the Frataxin Homolog from Chlamydomonas reinhardtii" Plants 11, no. 15: 1931. https://doi.org/10.3390/plants11151931
APA StyleTerenzi, A., Pagani, M. A., Gomez-Casati, D. F., & Busi, M. V. (2022). Structural and Functional Characterization of CreFH1, the Frataxin Homolog from Chlamydomonas reinhardtii. Plants, 11(15), 1931. https://doi.org/10.3390/plants11151931







