Dissecting the Chloroplast Proteome of the Potato (Solanum Tuberosum L.) and Its Comparison with the Tuber Amyloplast Proteome
Abstract
:1. Introduction
2. Results
2.1. Identification of Potato Chloroplastic Proteins
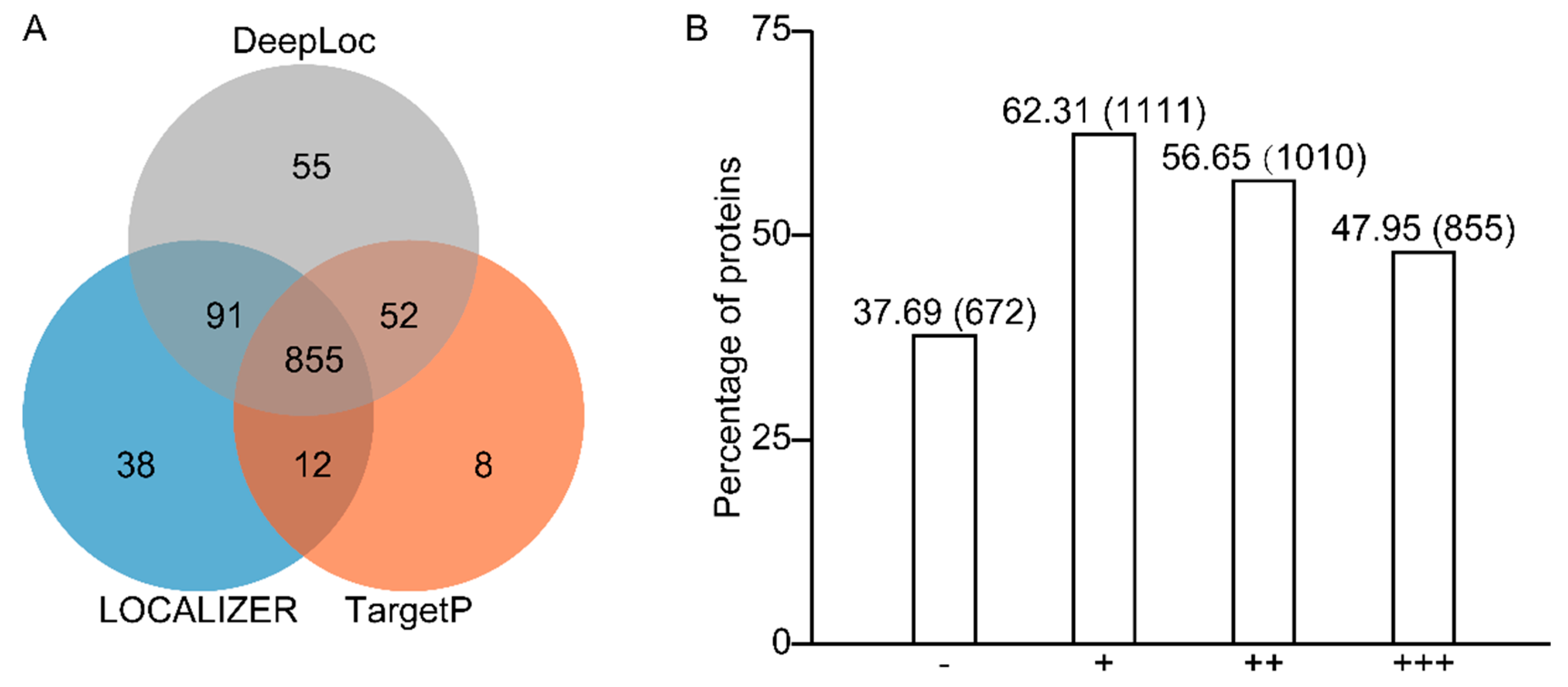
2.2. Prediction Programs for Chloroplast Localization Recognize 62% of All Identified Proteins
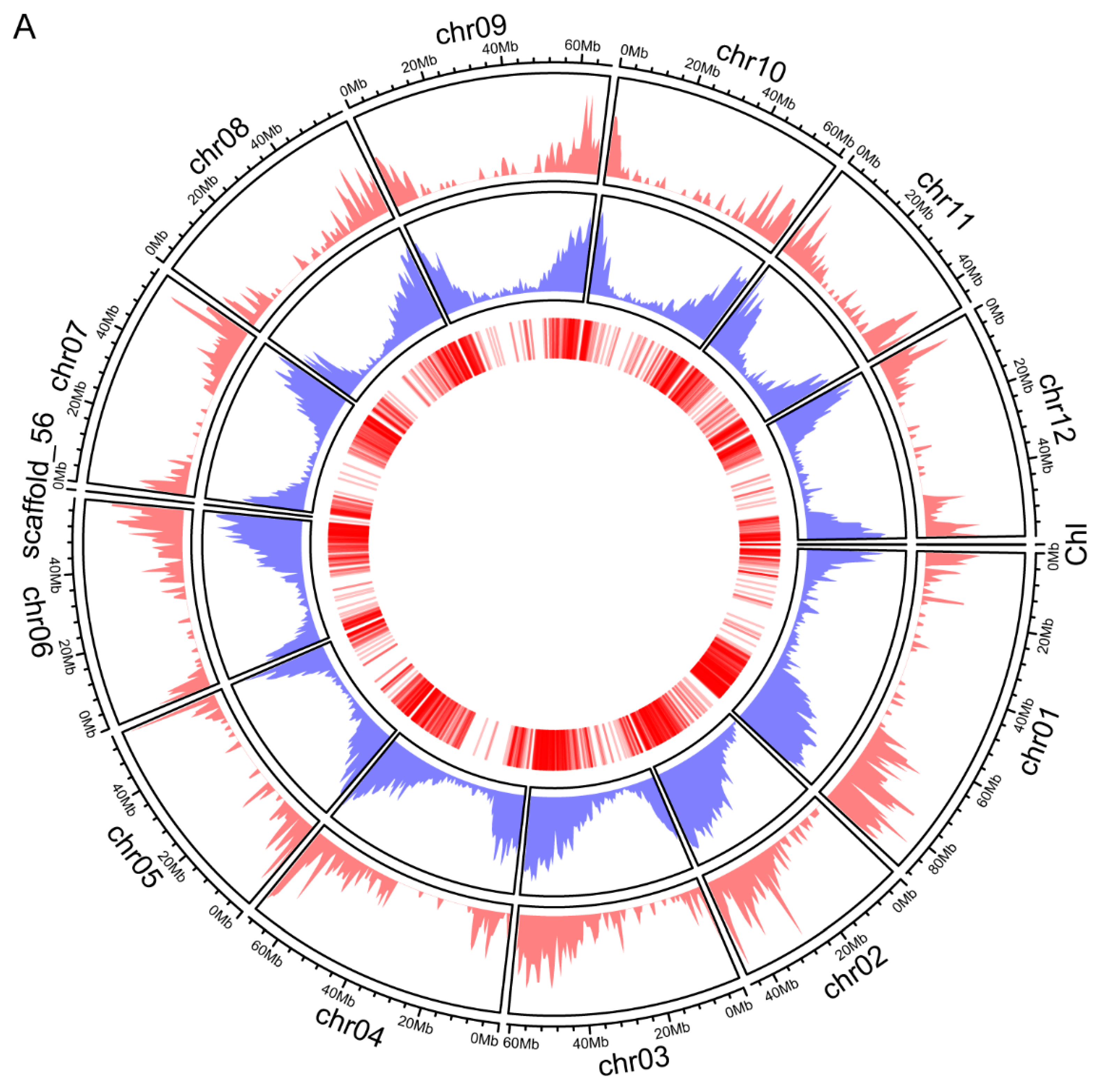
2.3. The Distribution of Genes Encoded the Identified Proteins across the Chromosomes
2.4. GO Analysis of the Identified Proteins
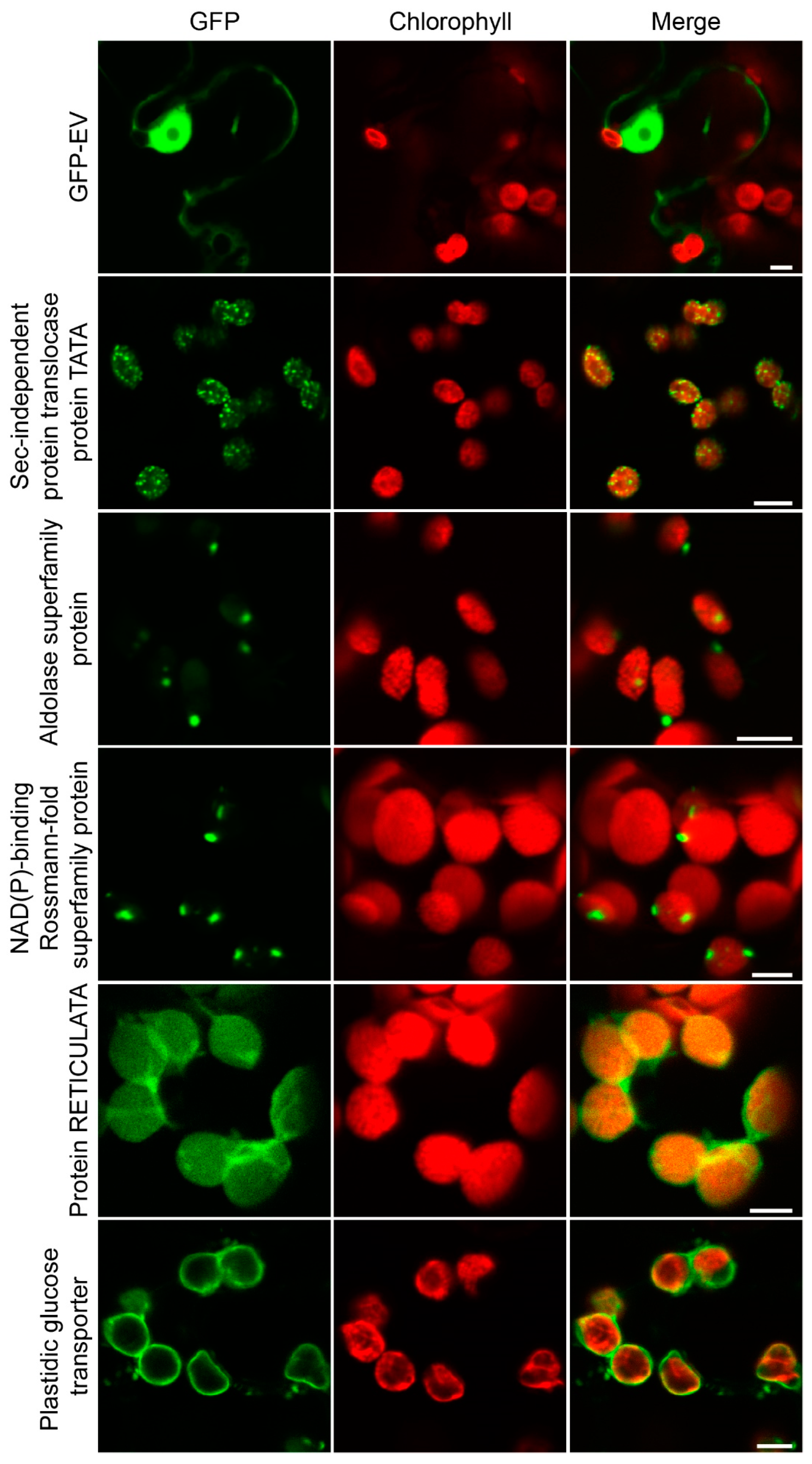
2.5. Evaluation of the Chloroplast Proteome by Transient Fluorescence Assay in Tobacco Epidermal Cells
2.6. Posttranslational Modification Analysis of the Potato Chloroplast Proteins
2.7. Comparative Proteomics between Chloroplast and Amyloplast
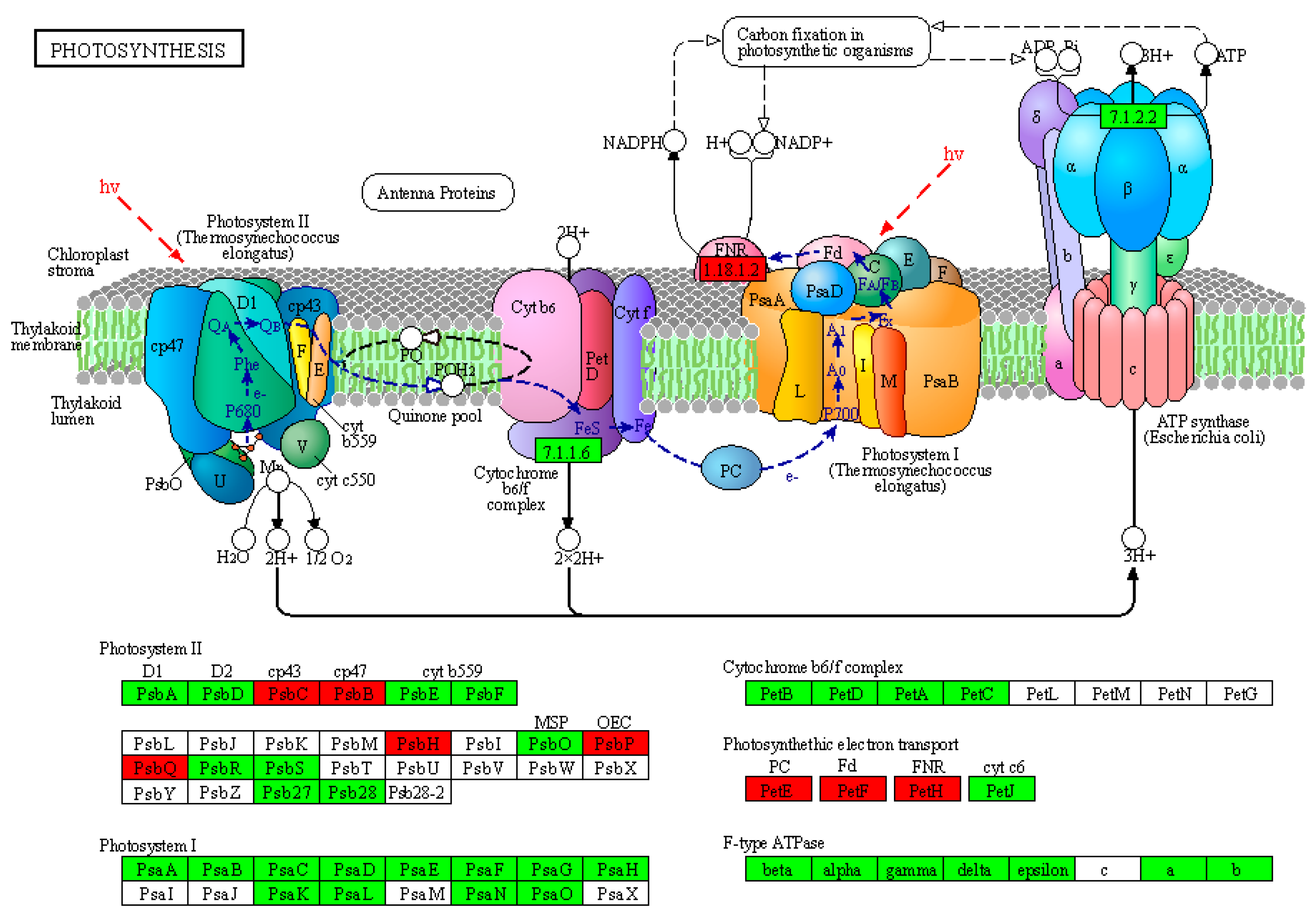
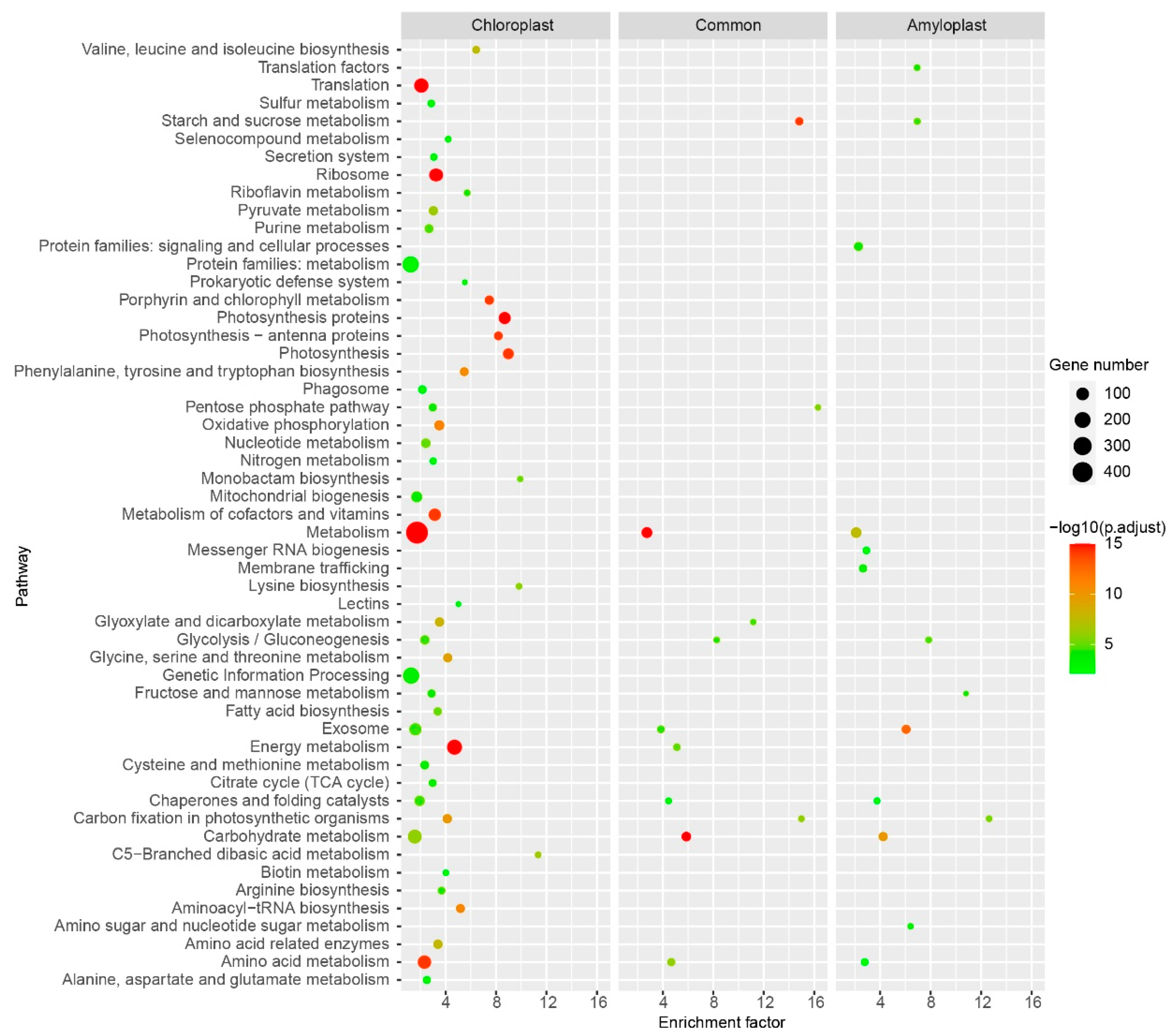
2.8. KEGG Pathway Illustrating Similarities and Differences between Chloroplast and Amyloplast Proteome
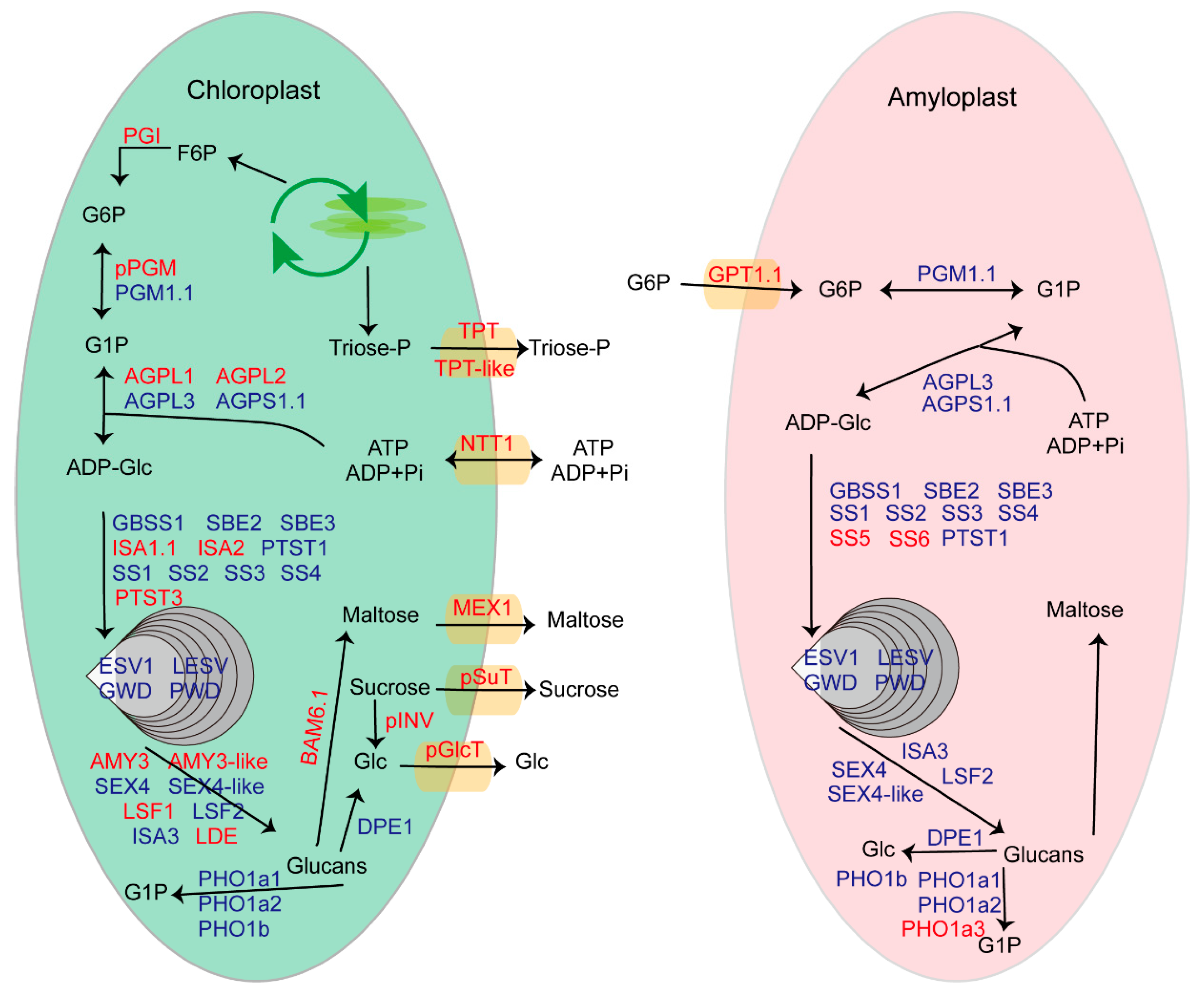
2.9. Reconstruction of the Snapshots along Starch Metabolism Pathway in Chloroplasts and Amyloplasts
3. Discussion
4. Materials and Methods
4.1. Plant Material Preparation
4.2. Chloroplast Isolation
4.3. Protein Extraction and Preparation
4.4. MS Analysis
4.5. Database Searching and Protein Identification
4.6. Bioinformatic Analysis
4.7. Subcellular Localization Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.; Zhou, B.; Li, J.; Tang, H.; Tang, J.; Yang, Z. Formation and Change of Chloroplast-Located Plant Metabolites in Response to Light Conditions. Int. J. Mol. Sci. 2018, 19, 654. [Google Scholar] [CrossRef] [Green Version]
- Martin, W.; Rujan, T.; Richly, E.; Hansen, A.; Cornelsen, S.; Lins, T.; Leister, D.; Stoebe, B.; Hasegawa, M.; Penny, D. Evolutionary Analysis of Arabidopsis, Cyanobacterial, and Chloroplast Genomes Reveals Plastid Phylogeny and Thousands of Cyanobacterial Genes in the Nucleus. Proc. Natl. Acad. Sci. USA 2002, 99, 12246–12251. [Google Scholar] [CrossRef] [Green Version]
- Villarejo, A.; Burén, S.; Larsson, S.; Déjardin, A.; Monné, M.; Rudhe, C.; Karlsson, J.; Jansson, S.; Lerouge, P.; Rolland, N.; et al. Evidence for a Protein Transported through the Secretory Pathway En Route to the Higher Plant Chloroplast. Nat. Cell Biol. 2005, 7, 1224–1231. [Google Scholar] [CrossRef]
- Kang, Z.-H.; Wang, G.-X. Redox Regulation in the Thylakoid Lumen. J. Plant Physiol. 2016, 192, 28–37. [Google Scholar] [CrossRef]
- Li, H.-M.; Chiu, C.-C. Protein Transport into Chloroplasts. Annu. Rev. Plant Biol. 2010, 61, 157–180. [Google Scholar] [CrossRef]
- Shi, L.-X.; Theg, S.M. The Chloroplast Protein Import System: From Algae to Trees. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Res. 2013, 1833, 314–331. [Google Scholar] [CrossRef] [Green Version]
- Dunkley, T.P.; Hester, S.; Shadforth, I.P.; Runions, J.; Weimar, T.; Hanton, S.L.; Griffin, J.L.; Bessant, C.; Brandizzi, F.; Hawes, C.; et al. Mapping the Arabidopsis Organelle Proteome. Proc. Natl. Acad. Sci. USA 2006, 103, 6518–6523. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, S.; Kamal, A.H.; Hossain, Z. Wheat Proteomics: Proteome Modulation and Abiotic Stress Acclimation. Front. Plant Sci. 2014, 5, 684. [Google Scholar] [CrossRef] [Green Version]
- Pechanova, O.; Takáč, T.; Šamaj, J.; Pechan, T. Maize Proteomics: An Insight into the Biology of an Important Cereal Crop. Proteomics 2013, 13, 637–662. [Google Scholar] [CrossRef]
- Wang, X.; Komatsu, S. Plant Subcellular Proteomics: Application for Exploring Optimal Cell Function in Soybean. J. Proteom. 2016, 143, 45–56. [Google Scholar] [CrossRef]
- Tomizioli, M.; Lazar, C.; Brugière, S.; Burger, T.; Salvi, D.; Gatto, L.; Moyet, L.; Breckels, L.M.; Hesse, A.M.; Lilley, K.S.; et al. Deciphering Thylakoid Sub-Compartments Using a Mass Spectrometry-Based Approach. Mol. Cell. Proteom. 2014, 13, 2147–2167. [Google Scholar] [CrossRef] [Green Version]
- Ferro, M.; Salvi, D.; Rivière-Rolland, H.; Vermat, T.; Seigneurin-Berny, D.; Grunwald, D.; Garin, J.; Joyard, J.; Rolland, N. Integral Membrane Proteins of the Chloroplast Envelope: Identification and Subcellular Localization of New Transporters. Proc. Natl. Acad. Sci. USA 2002, 99, 11487–11492. [Google Scholar] [CrossRef] [Green Version]
- Tamburino, R.; Vitale, M.; Ruggiero, A.; Sassi, M.; Sannino, L.; Arena, S.; Costa, A.; Batelli, G.; Zambrano, N.; Scaloni, A.; et al. Chloroplast Proteome Response to Drought Stress and Recovery in Tomato (Solanum Lycopersicum L.). BMC Plant Biol. 2017, 17, 40. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liang, W.; Xing, J.; Tan, F.; Chen, Y.; Huang, L.; Cheng, C.L.; Chen, W. Dynamics of Chloroplast Proteome in Salt-Stressed Mangrove Kandelia candel (L.) Druce. J. Proteome Res. 2013, 12, 5124–5136. [Google Scholar] [CrossRef]
- Stensballe, A.; Hald, S.; Bauw, G.; Blennow, A.; Welinder, K.G. The Amyloplast Proteome of Potato Tuber. FEBS J. 2008, 275, 1723–1741. [Google Scholar] [CrossRef]
- Helle, S.; Bray, F.; Verbeke, J.; Devassine, S.; Courseaux, A.; Facon, M.; Tokarski, C.; Rolando, C.; Szydlowski, N. Proteome Analysis of Potato Starch Reveals the Presence of New Starch Metabolic Proteins as Well as Multiple Protease Inhibitors. Front. Plant Sci. 2018, 9, 746. [Google Scholar] [CrossRef]
- Van Harsselaar, J.K.; Lorenz, J.; Senning, M.; Sonnewald, U.; Sonnewald, S. Genome-Wide Analysis of Starch Metabolism Genes in Potato (Solanum Tuberosum L.). BMC Genom. 2017, 18, 37. [Google Scholar] [CrossRef] [Green Version]
- Li, H.-M.; Chen, L.-J. A Novel Chloroplastic Outer Membrane-Targeting Signal That Functions at Both Termini of Passenger Polypeptides. J. Biol. Chem. 1997, 272, 10968–10974. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.J.; Jung, J.D.; Park, H.W.; Kim, J.H.; Cha, H.W.; Min, S.R.; Jeong, W.J.; Liu, J.R. The Complete Chloroplast Genome Sequences of Solanum Tuberosum and Comparative Analysis with Solanaceae Species Identified the Presence of a 241-Bp Deletion in Cultivated Potato Chloroplast DNA Sequence. Plant Cell Rep. 2006, 25, 1369–1379. [Google Scholar] [CrossRef]
- Longoni, F.P.; Goldschmidt-Clermont, M. Thylakoid Protein Phosphorylation in Chloroplasts. Plant Cell Physiol. 2021, 62, 1094–1107. [Google Scholar] [CrossRef]
- Patzke, K.; Prananingrum, P.; Klemens, P.A.; Trentmann, O.; Rodrigues, C.M.; Keller, I.; Fernie, A.R.; Geigenberger, P.; Bölter, B.; Lehmann, M.; et al. The Plastidic Sugar Transporter Psut Influences Flowering and Affects Cold Responses. Plant Physiol. 2019, 179, 569–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feike, D.; Seung, D.; Graf, A.; Bischof, S.; Ellick, T.; Coiro, M.; Soyk, S.; Eicke, S.; Mettler-Altmann, T.; Lu, K.J.; et al. The Starch Granule-Associated Protein Early Starvation1 Is Required for the Control of Starch Degradation in Arabidopsis Thaliana Leaves. Plant Cell 2016, 28, 1472–1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seung, D.; Boudet, J.; Monroe, J.; Schreier, T.B.; David, L.C.; Abt, M.; Lu, K.J.; Zanella, M.; Zeeman, S.C. Homologs of Protein Targeting to Starch Control Starch Granule Initiation in Arabidopsis Leaves. Plant Cell 2017, 29, 1657–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niittylä, T.; Messerli, G.; Trevisan, M.; Chen, J.; Smith, A.M.; Zeeman, S.C. A Previously Unknown Maltose Transporter Essential for Starch Degradation in Leaves. Science 2004, 303, 87–89. [Google Scholar] [CrossRef]
- Cho, M.H.; Lim, H.; Shin, D.H.; Jeon, J.S.; Bhoo, S.H.; Park, Y.I.; Hahn, T.R. Role of the Plastidic Glucose Translocator in the Export of Starch Degradation Products from the Chloroplasts in Arabidopsis Thaliana. New Phytol. 2011, 190, 101–112. [Google Scholar] [CrossRef]
- Abt, M.R.; Pfister, B.; Sharma, M.; Eicke, S.; Bürgy, L.; Neale, I.; Seung, D.; Zeeman, S.C. Starch Synthase5, a Noncanonical Starch Synthase-Like Protein, Promotes Starch Granule Initiation in Arabidopsis. Plant Cell 2020, 32, 2543–2565. [Google Scholar] [CrossRef]
- Kleffmann, T.; Russenberger, D.; von Zychlinski, A.; Christopher, W.; Sjölander, K.; Gruissem, W.; Baginsky, S. The Arabidopsis Thaliana Chloroplast Proteome Reveals Pathway Abundance and Novel Protein Functions. Curr. Biol. 2004, 14, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Joyard, J.; Ferro, M.; Masselon, C.; Seigneurin-Berny, D.; Salvi, D.; Garin, J.; Rolland, N. Chloroplast Proteomics and the Compartmentation of Plastidial Isoprenoid Biosynthetic Pathways. Mol. Plant 2009, 2, 1154–1180. [Google Scholar] [CrossRef]
- Miras, S.; Salvi, D.; Ferro, M.; Grunwald, D.; Garin, J.; Joyard, J.; Rolland, N. Non-Canonical Transit Peptide for Import into the Chloroplast. J. Biol. Chem. 2002, 277, 47770–47778. [Google Scholar] [CrossRef] [Green Version]
- Zybailov, B.; Rutschow, H.; Friso, G.; Rudella, A.; Emanuelsson, O.; Sun, Q.; van Wijk, K.J. Sorting Signals, N-Terminal Modifications and Abundance of the Chloroplast Proteome. PLoS ONE 2008, 3, e1994. [Google Scholar] [CrossRef] [Green Version]
- Salvi, D.; Bournais, S.; Moyet, L.; Bouchnak, I.; Kuntz, M.; Bruley, C.; Rolland, N. At_Chloro: The First Step When Looking for Information About Subplastidial Localization of Proteins. In Plastids; Springer: Berlin/Heidelberg, Germany, 2018; pp. 395–406. [Google Scholar]
- Bruley, C.; Dupierris, V.; Salvi, D.; Rolland, N.; Ferro, M. At_Chloro: A Chloroplast Protein Database Dedicated to Sub-Plastidial Localization. Front. Plant Sci. 2012, 3, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolland, N.; Curien, G.; Finazzi, G.; Kuntz, M.; Maréchal, E.; Matringe, M.; Ravanel, S.; Seigneurin-Berny, D. The Biosynthetic Capacities of the Plastids and Integration between Cytoplasmic and Chloroplast Processes. Annu. Rev. Genet. 2012, 46, 233–264. [Google Scholar] [CrossRef] [PubMed]
- Nelson, N.; Junge, W. Structure and Energy Transfer in Photosystems of Oxygenic Photosynthesis. Annu. Rev. Biochem. 2015, 84, 659–683. [Google Scholar] [CrossRef]
- Grieco, M.; Jain, A.; Ebersberger, I.; Teige, M. An Evolutionary View on Thylakoid Protein Phosphorylation Uncovers Novel Phosphorylation Hotspots with Potential Functional Implications. J. Exp. Bot. 2016, 67, 3883–3896. [Google Scholar]
- Bahaji, A.; Li, J.; Sánchez-López, Á.M.; Baroja-Fernández, E.; Muñoz, F.J.; Ovecka, M.; Almagro, G.; Montero, M.; Ezquer, I.; Etxeberria, E.; et al. Starch Biosynthesis, Its Regulation and Biotechnological Approaches to Improve Crop Yields. Biotechnol. Adv. 2014, 32, 87–106. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on Mrna Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Kubis, S.E.; Lilley, K.S.; Jarvis, P. Isolation and Preparation of Chloroplasts from Arabidopsis Thaliana Plants. Methods Mol. Biol. 2008, 425, 171–186. [Google Scholar]
- Li, X.; Chai, Y.; Yang, H.; Tian, Z.; Li, C.; Xu, R.; Shi, C.; Zhu, F.; Zeng, Y.; Deng, X.; et al. Isolation and Comparative Proteomic Analysis of Mitochondria from the Pulp of Ripening Citrus Fruit. Hortic. Res. 2021, 8, 31. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The Pride Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, Transient Expression of Fluorescent Fusion Proteins in Tobacco Plants and Generation of Stably Transformed Plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Liu, T.; Wang, E.; Cheng, Y.; Liu, T.; Chen, G.; Guo, M.; Song, B. Dissecting the Chloroplast Proteome of the Potato (Solanum Tuberosum L.) and Its Comparison with the Tuber Amyloplast Proteome. Plants 2022, 11, 1915. https://doi.org/10.3390/plants11151915
Liu S, Liu T, Wang E, Cheng Y, Liu T, Chen G, Guo M, Song B. Dissecting the Chloroplast Proteome of the Potato (Solanum Tuberosum L.) and Its Comparison with the Tuber Amyloplast Proteome. Plants. 2022; 11(15):1915. https://doi.org/10.3390/plants11151915
Chicago/Turabian StyleLiu, Shengxuan, Tengfei Liu, Enshuang Wang, Yunxia Cheng, Tiantian Liu, Guogang Chen, Minrui Guo, and Botao Song. 2022. "Dissecting the Chloroplast Proteome of the Potato (Solanum Tuberosum L.) and Its Comparison with the Tuber Amyloplast Proteome" Plants 11, no. 15: 1915. https://doi.org/10.3390/plants11151915
APA StyleLiu, S., Liu, T., Wang, E., Cheng, Y., Liu, T., Chen, G., Guo, M., & Song, B. (2022). Dissecting the Chloroplast Proteome of the Potato (Solanum Tuberosum L.) and Its Comparison with the Tuber Amyloplast Proteome. Plants, 11(15), 1915. https://doi.org/10.3390/plants11151915





