100 Years of Chromosome Research in Rye, Secale L.
Abstract
1. Introduction
2. Karyology
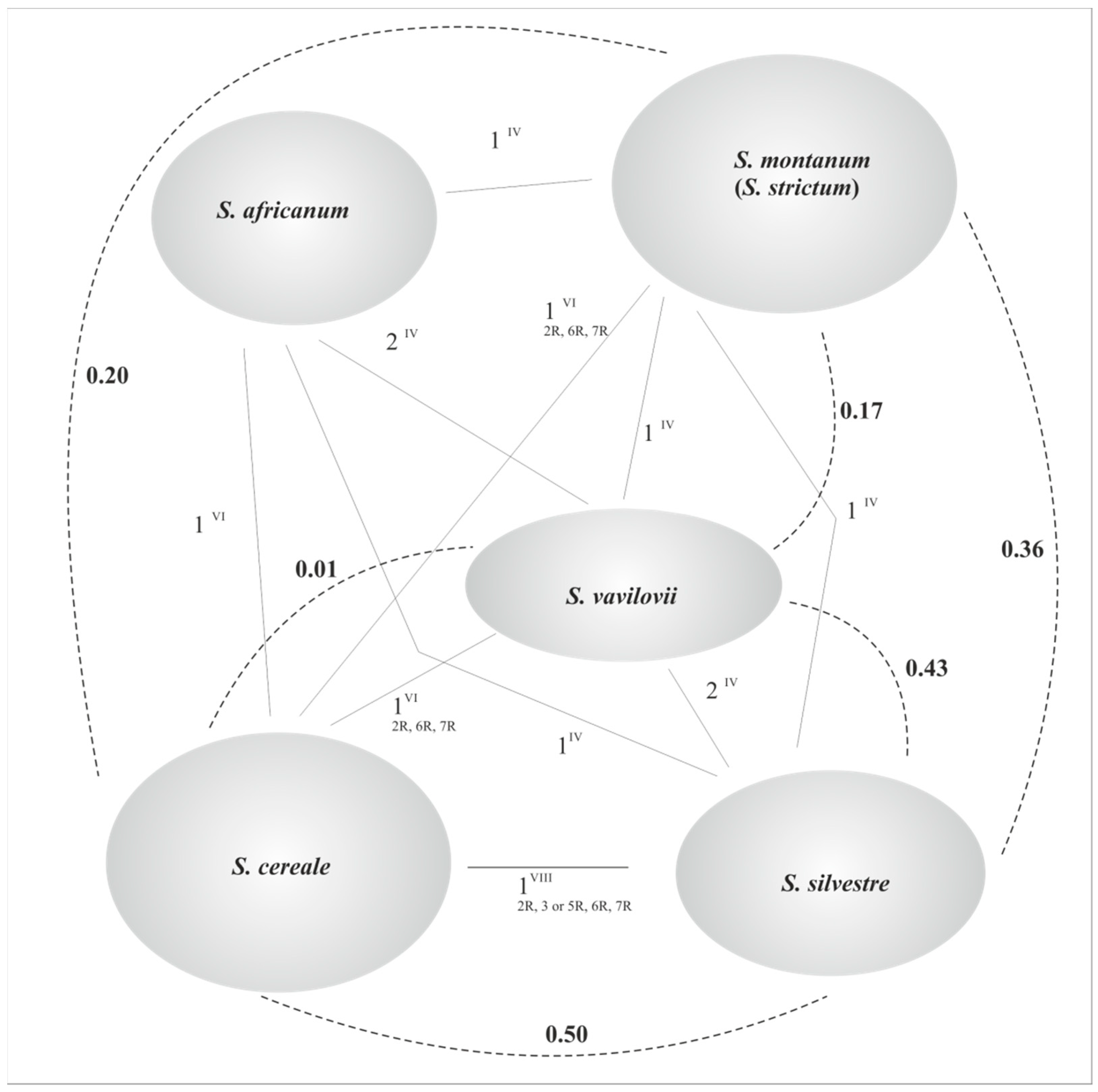
Wild Species
3. Polyploidy
3.1. Genetic Control of Chromosome Behavior
3.2. Manipulation of Chiasma Frequency
3.3. Manipulation of Chromosome Structure
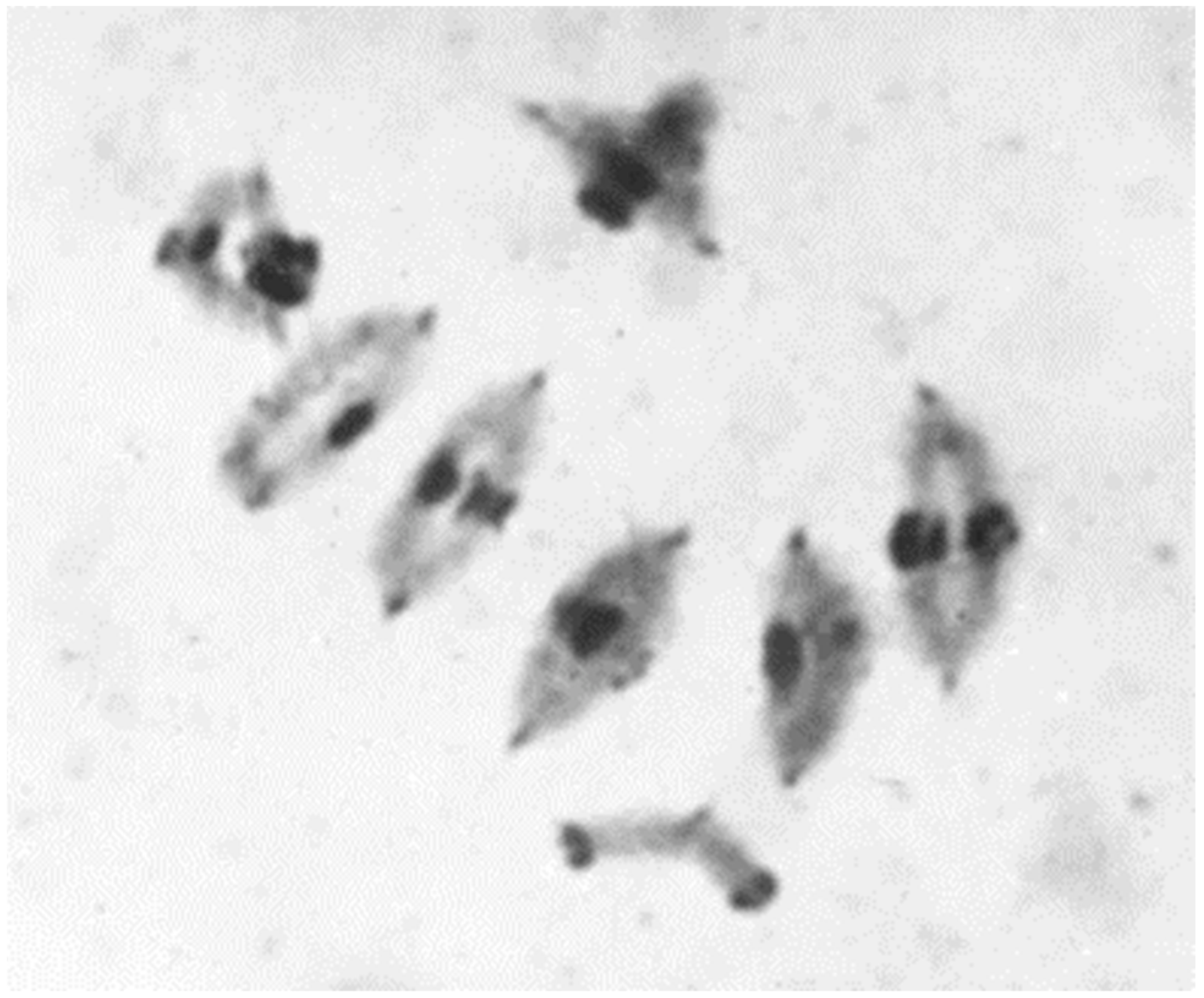
3.4. Trisomics and Telo-Trisomics
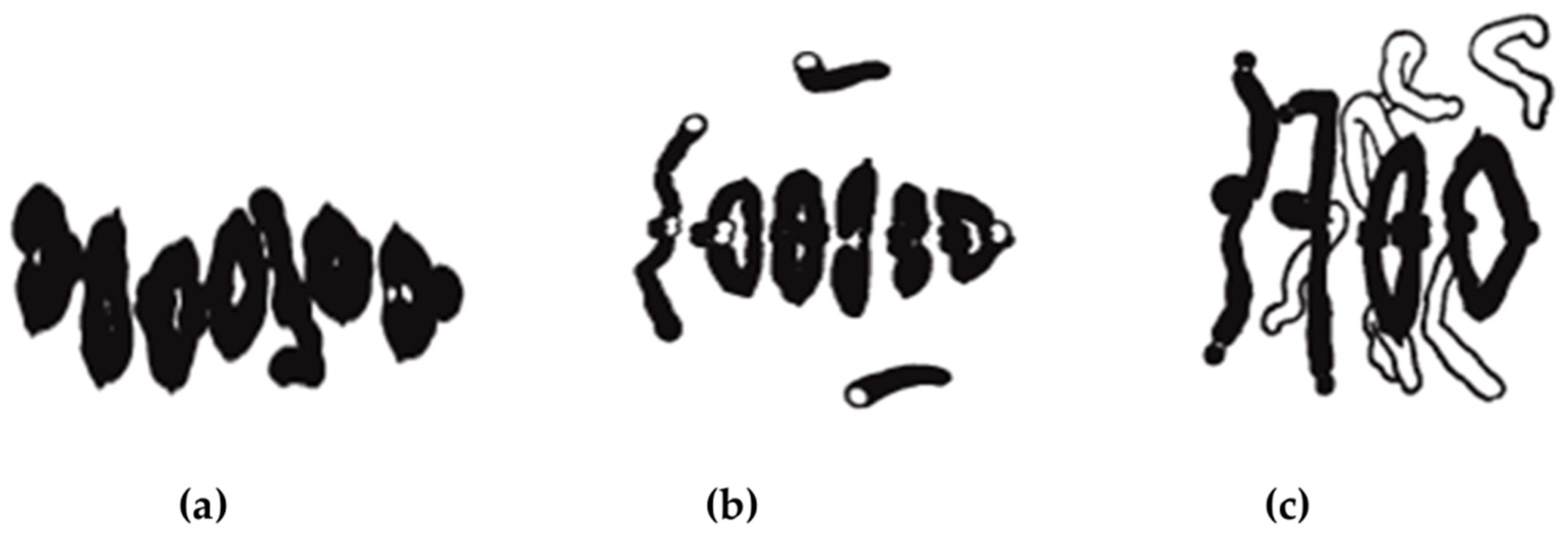
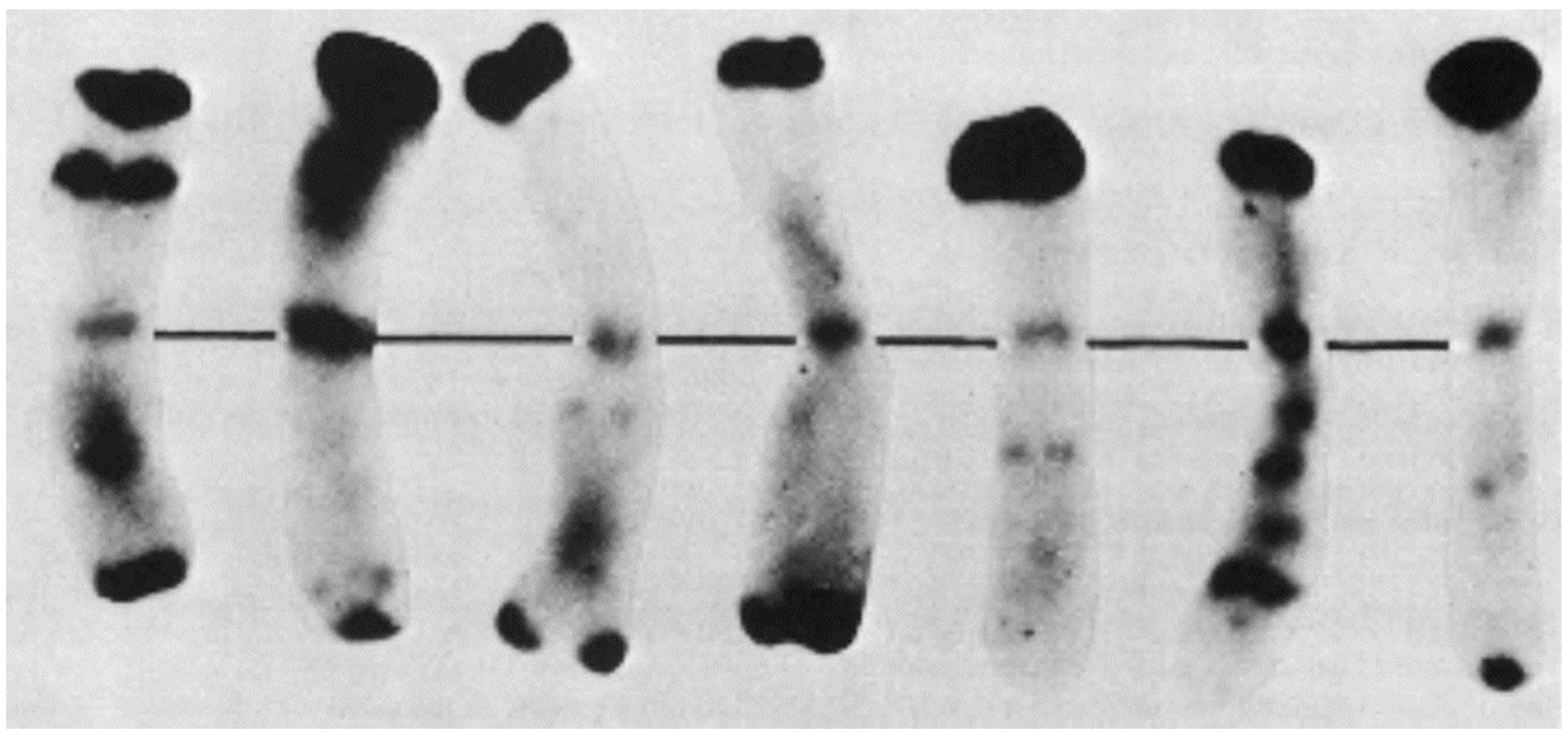
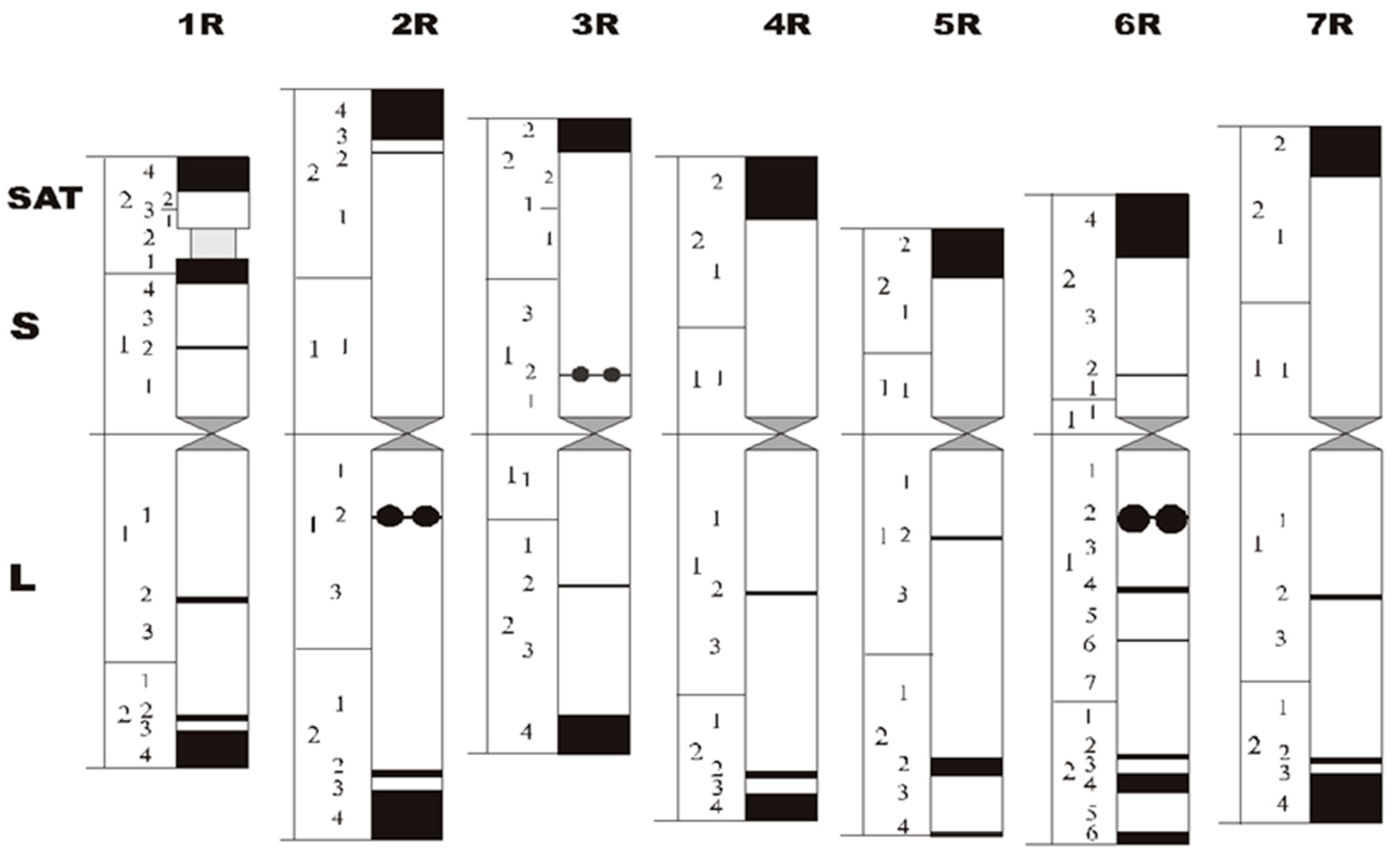
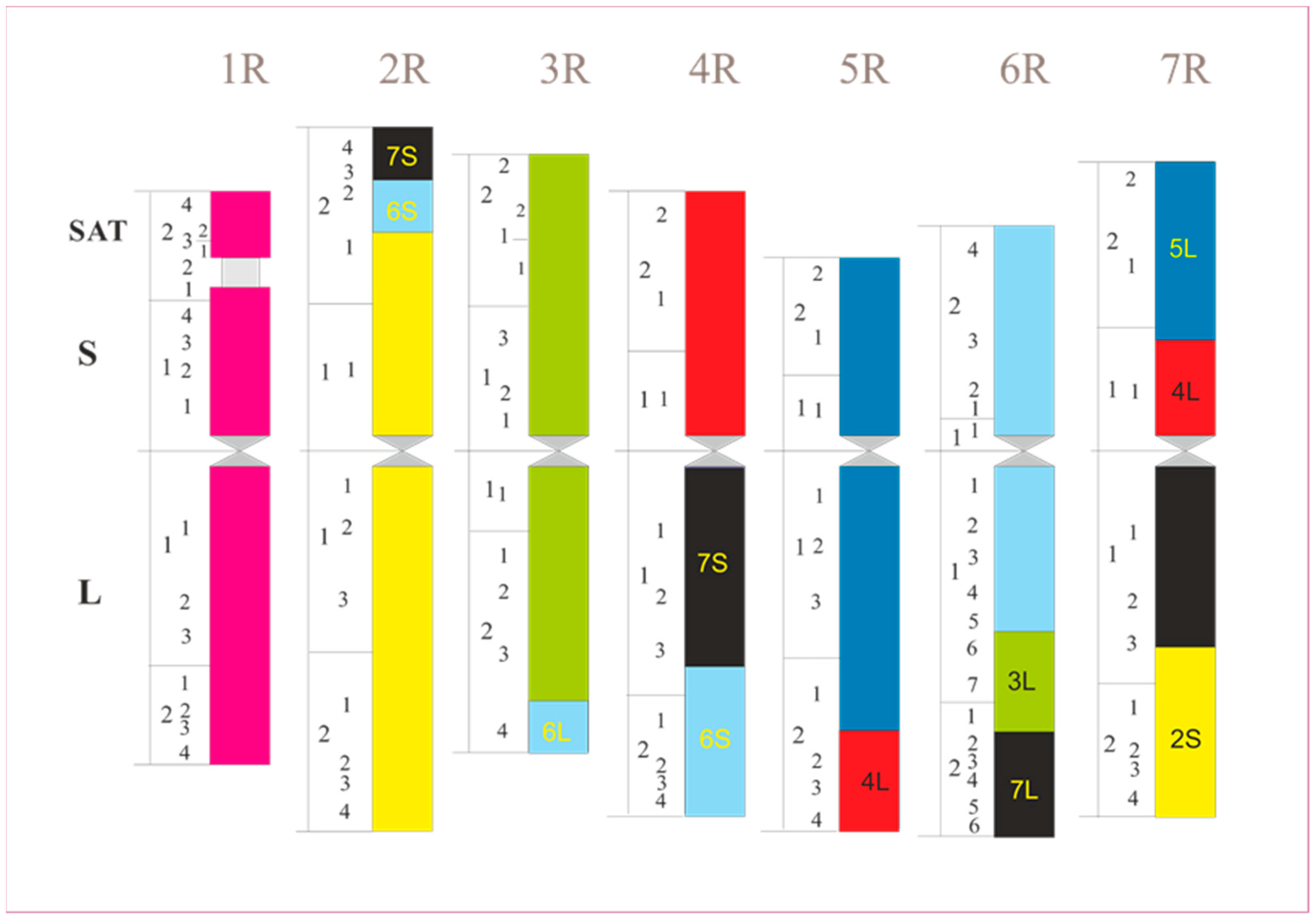
4. Chromosome Staining
4.1. Radioactive Labeling
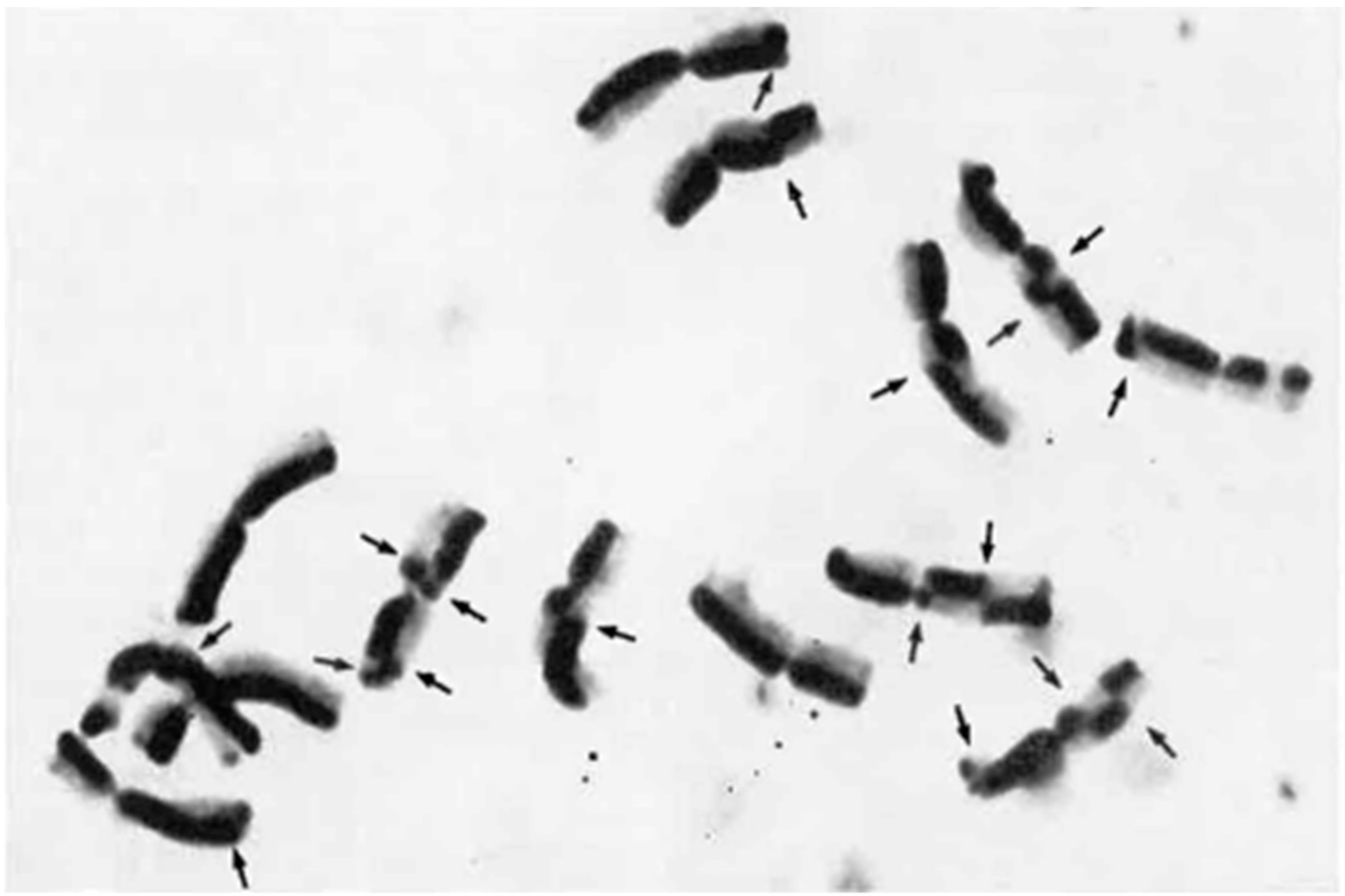
4.2. C/N-Banding
4.3. Sister Chromatid Exchange
4.4. Non-Radioactive Labeling
5. In Situ Manipulation
6. Flow Sorting
7. Outlook
Funding
Acknowledgments
Conflicts of Interest
References
- Rabanus-Wallace, M.T.; Stein, N. The Rye Genome; Springer Nature: Cham, Switzerland, 2021; p. 251. ISSN 2199-4781. [Google Scholar]
- Schlegel, R. Rye—Genetics, Breeding & Cultivation, 1st ed.; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group, Inc.: New York, NY, USA, 2013; p. 387. ISBN1 10:1466561432. ISBN2 13:978-1466561434. [Google Scholar]
- Nemec, B. Das Problem der Befruchtungsvorgänge und Andere Zytologische Fragen; Gebrüder Borntraeger: Berlin, Germany, 1910. [Google Scholar]
- Nakao, M. Cytological studies on the nuclear division of the pollen mother cells of some cereals and their hybrids. J. Coll. Agric. 1911, 4, 173–190. [Google Scholar]
- Sakamura, T.J. Kurze Mitteilung über die Chromosomenzahlen und die Verwandtschaftsverhältnisse der Triticum-Arten. Bot. Mag. Tokyo 1918, 32, 149–153. [Google Scholar] [CrossRef]
- Sakamura, T.J. Experimentelle Studien über die Zell- und Kernteilung mit besonderer Rücksicht auf Form, Größe und Zahl von Chromosomen. J. Coll. Sci. Imp. Univ. Tokyo 1920, 39, 1–221. [Google Scholar]
- Kihara, H. Cytologische Studien bei einigen Getreidearten. I. Spezies-Bastarde Weizens und Weizen-Roggen-Bastarde. Bot. Mag. Tokyo 1919, 32, 1–93. [Google Scholar]
- Gotoh, K. Über die Chromosomenzahl von Secale cereale. Bot. Mag. Tokyo 1924, 38, 135–152. [Google Scholar] [CrossRef]
- Emme, H.K. Zur Cytologie der Gattung Secale L. (Russian). Bull. Appl. Bot. Genet. Plant-Breed. 1927, 17, 73–100. [Google Scholar]
- Lewitzky, G.A. The morphology of chromosomes. Bull. Appl. Bot. Genet. Plant-Breed. 1931, 27, 19–174. [Google Scholar]
- Vavilov, N.I. On the origin of cultivated rye. Appl. Bot. 1917, 10, 561–590. [Google Scholar]
- Vavilov, N.I. Ursprungszentren der Kulturpflanzen. Bull. Appl. Bot. Sel. 1925, 16, 1–216. [Google Scholar]
- Vavilov, N.I. Geographical regularities in relation to the distribution of the genes of cultivated plants. Bull. Appl. Bot. Genet. Plant-Breed. 1927, 17, 411–428. [Google Scholar]
- Kostoff, D. Interspecific hybrids in rye (Russian). Dokl. Akad. Sci. USSR 1937, 14, 1–125. [Google Scholar]
- Nakajima, G. Cytogenetical studies on the interspecific hybrids within the genus Secale. II. Meiosis in PMC of F1 hybrids between cereal and 3 species Vavilovii, Africanum and Montanum. Jpn. J. Breed. 1956, 6, 171–176. [Google Scholar] [CrossRef]
- Schreiber, M.; Himmelbach, A.; Börner, A.; Masche, M. Genetic diversity and relationship between domesticated rye and its wild relatives as revealed through genotyping-by-sequencing. Evol. Appl. 2018, 12, 66–77. [Google Scholar] [CrossRef]
- Blakeslee, A.F.; Avery, A.G. Methods of inducing doubling of chromosomes in plants. J. Hered. 1937, 28, 393–411. [Google Scholar] [CrossRef]
- Nebel, B.R. Mechanism of polyploids through colchicine. Nature 1937, 140, 1101. [Google Scholar] [CrossRef]
- Chin, T.C. Cytology of the autotetraploid rye. Bot. Gaz. 1943, 104, 627–632. [Google Scholar] [CrossRef]
- Dorsey, E. Induced polyploidy in wheat and rye. J. Hered. 1936, 27, 155–160. [Google Scholar] [CrossRef]
- Schlegel, R. Experimentelle Untersuchungen zur Chromosomalen Stabilität und Deren Bedeutung für die Fertiltät des Autotetraploiden Roggens (Secale cereale L.). Ph.D. Thesis, Martin Luther University, Halle, Germany, 1973; pp. 1–172. [Google Scholar]
- Rees, H. Genotypic control of chromosome behaviour in rye. I. Inbred lines. Heredity 1955, 9, 93–116. [Google Scholar] [CrossRef][Green Version]
- Rees, H. Genotypic control of chromosome behaviour in rye. III. Chiasma frequency in homozygotes and heterozygotes. Heredity 1955, 10, 409–424. [Google Scholar] [CrossRef]
- Schlegel, R.; Mettin, D. Untersuchungen zur intra- und interindividuellen Variabilität der Chromosomenpaarung bei di- und tetraploidem Roggen aus Populationen (Secale cereale L). I. Intraindividuelle Variabilität. Biol. Zent. 1975, 94, 539–555. [Google Scholar]
- Schlegel, R.; Mettin, D. Untersuchungen zur intra- und interindividuellen Variabilität der Chromosomenpaarung bei di- und tetraploidem Roggen (Secale cereale L.). II. Interindividuelle Variabilität. Biol. Zent. 1975, 94, 703–715. [Google Scholar]
- Naranjo, T. Variable patterning of chromatin remodeling, telomere positioning, synapsis, and chiasma formation of individual rye chromosomes in meiosis of wheat-rye additions. Front. Plant Sci. 2018, 9, 880. [Google Scholar] [CrossRef]
- Kagawa, N.; Nagaki, K.; Tsujimoto, H. Tetrad-FISH analysis reveals recombination suppression by interstitial heterochromatin sequences in rye (Secale cereale). Mol. Genet. Genom. 2002, 267, 10–15. [Google Scholar] [CrossRef]
- Hazarika, M.H.; Rees, H. Genotypical control of chromosome behaviour in rye. X. Chromosome pairing and fertility in autotetraploids. Heredity 1967, 22, 317–321. [Google Scholar] [CrossRef]
- Schlegel, R. The relationship between meiosis and fertility in autotetraploid rye, Secale cereale L. Tag. Ber. Akad. d. Landwirtsch. 1976, 143, 31–37. [Google Scholar]
- Davies, E.D.G.; Jones, G.H. Chiasma variation and control in pollen mother cells and embryo-sac mother cells of rye. Genet. Res. 1974, 23, 185–190. [Google Scholar] [CrossRef]
- Zeller, F.J.; Herrmann, R.G.; Wanner, G. Chromosome condensation in mitosis and meiosis of rye (Secale cereale L.). Cytogenet. Genome Res. 2004, 105, 134–144. [Google Scholar] [CrossRef]
- Maestra, B.; de Jong, J.H.; Shepherd, K.; Naranjo, T. Chromosome arrangement and behaviour of two rye homologous telosomes at the onset of meiosis in disomic wheat-5RL addition lines with and without the Ph1 locus. Chromosome Res. 2002, 10, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Corredor, E.; Naranjo, T. Effect of colchicine and telocentric chromosome conformation on centromere and telomere dynamics at meiotic prophase I in wheat–rye additions. Chromosome Res. 2007, 15, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, T. Dynamics of rye telomeres in a wheat background during early meiosis. Cytogenet. Genome Res. 2014, 143, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Gillies, C.B. An electron microscope study of synaptonemal complex formation at zygotene in rye. Chromosoma 1985, 92, 165–175. [Google Scholar] [CrossRef]
- Hesse, S.; Zelkowski, M.; Mikhailova, E.I.; Keijzer, C.J.; Houben, A.; Schubert, V. Ultrastructure and dynamics of synaptonemal complex components during meiotic pairing and synapsis of standard (A) and accessory (B) rye Chromosomes. Front. Plant Sci. 2019, 10, 773. [Google Scholar] [CrossRef]
- Abirached-Darmency, M.; Zickler, D.; Cauderon, Y. Synaptonemal complex and recombination nodules in rye (Secale cereale). Chromosoma 1983, 88, 299–306. [Google Scholar] [CrossRef]
- Ahloowalia, B.S. Chromosome association and fertility in tetraploid ryegrass. Genetica 1967, 38, 471–484. [Google Scholar] [CrossRef]
- Crowley, J.G.; Rees, H. Fertility and selection in tetraploid Lolium. Chromosoma 1968, 24, 300–308. [Google Scholar] [CrossRef]
- Schlegel, R.; Mettin, D. Studies of valence crosses in rye (Secale cereale L.). IV. The relationship between meiosis and fertility in tetraploid hybrids. Biol. Zbl. 1975, 94, 295–315. [Google Scholar]
- Swami, U.B.S.; Thomas, H. Chromosome pairing in autotetraploid Avena. Z. Pflanz. 1968, 59, 163–170. [Google Scholar]
- Sybenga, J. The quantitative analysis of chromosome pairing and chiasma formation based on the relative frequencies of MI configurations. II. Primary trisomics. Genetica 1965, 36, 339–350. [Google Scholar] [CrossRef]
- Sybenga, J.; Wolters, Z.H.G. The classification of the chromosomes of rye (Secale cereale L.): A translocation tester set. Genetica 1972, 43, 453–464. [Google Scholar] [CrossRef]
- Sybenga, J.; van Eden, J.; van der Meijs, Q.G.; Roeterding, B.W. Identification of the chromosomes of the rye translocation tester set. Theor. Appl. Genet. 1985, 69, 313–316. [Google Scholar] [CrossRef]
- Augustin, C.; Schlegel, R. Chromosome manipulations. II. Production and characterization of interchromosomal translocations for genetic mapping in rye. Arch. Züchtungsforsch. 1983, 12, 180–184. [Google Scholar]
- Lili, Q.; Friebe, B.; Gill, B.S. Complex genome rearrangements reveal evolutionary dynamics of pericentromeric regions in the Triticeae. Genome 2006, 49, 1628–1639. [Google Scholar]
- Schlegel, R. Dictionary of Plant Breeding, 3rd ed.; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group, Inc.: New York, NY, USA, 2020; p. 734. ISBN 9780367494131. [Google Scholar]
- Takagi, F. Karyogenetical studies on rye. I. A trisomic plant. Cytologia 1935, 6, 496–501. [Google Scholar] [CrossRef][Green Version]
- Kamanoi, M.; Jenkins, B.C. Trisomics in common rye, Secale cereale L. Seiken Ziho 1962, 13, 118–123. [Google Scholar]
- Mettin, D.; Balkandschiewa, J.; Müller, F. Studies on trisomics of rye, Secale cereale L. German. Tag. Ber. Akad. Landwirtsch. Wiss. 1972, 119, 153–161. [Google Scholar]
- Mettin, D.; Müller, F.; Schlegel, R. Untersuchungen an Valenzkreuzungen beim Roggen (Secale cereale L.) 3. Über die Häufigkeit von 3x- und 4x-Bastarden unter verschiedenen Bestäubungsbedingungen. (Studies on ploidy crosses in rye, Secale cereale L. 3. About the frequency of triploid and tetraploid hybrids under different crossing conditions). Tag. Ber. AdL (DDR) 1972, 119, 135–152. [Google Scholar]
- Zeller, F.J.; Kimber, G.; Gill, B.S. The identification of rye trisomics by translocations and Giemsa staining. Chromosoma 1977, 62, 279–282. [Google Scholar] [CrossRef]
- Benito, C.; Romero, M.P.; Henriques-Gil, N.; Llorente, F.; Figueiras, A.M. Sex influence on recombination frequency in Secale cereale L. Theor. Appl. Genet. 1996, 93, 926–931. [Google Scholar] [CrossRef]
- Melz, G.; Schlegel, R. Identification of seven telotrisomics of rye (Secale cereale L.). Euphytica 1985, 34, 361–366. [Google Scholar] [CrossRef]
- Schlegel, R.; Sturm, W. Meiotic chromosome pairing of primary trisomics of rye, Secale cereale L. Tag. Ber. Akad. Landwirtsch. Wiss. 1982, 198, 225–243. [Google Scholar]
- Sturm, W.; Melz, G. Telotrisomics. Tag. Ber. Akad. Landwirtsch. Wiss. 1982, 198, 217–225. [Google Scholar]
- Melz, G.; Neumann, H.; Müller, H.W.; Sturm, W. Genetic analysis of rye (Secale cereale L.). I. Results of gene location on rye chromosomes using primary trisomics. Genet. Pol. 1984, 25, 111–115. [Google Scholar]
- Schlegel, R.; Korzun, V. Genes, Markers, and Linkage Data of Rye (Secale cereale L.), 12th Updated Inventory. V01.22, 1–115. 2022. Available online: http://www.rye-gene-map.de (accessed on 1 January 2020).
- Sturm, W. Identification of trisomics in the rye variety “Esto” and trisomic analysis of short straw gene Hl. Ph.D. Thesis, Akademie der Landwirtschaftswissenschaften, Berlin, Germany, 1978; pp. 1–164. [Google Scholar]
- Lima de Faria, A. Differential uptake of tritiated thymidine into hetero- and euchromatin in Melanoplus and Secale. J. Biophys. Biochem. Cytol. 1959, 6, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Sarma, N.P.; Natarajan, A.T. Identification of heterochromatic regions in the chromosomes of rye. Hereditas 1973, 74, 233–238. [Google Scholar] [CrossRef]
- Gill, B.S.; Kimber, G. The Giemsa C-banded karyotype of rye. Proc. Natl. Acad. Sci. USA 1974, 71, 1247–1249. [Google Scholar] [CrossRef]
- Lima de Faria, A. Chromomere analysis of the chromosome complement of rye. Chromosoma 1952, 5, 1–68. [Google Scholar] [CrossRef]
- Schlegel, R.; Friedrich, F. Erste Untersuchungen zum meiotischen Paarungsverhalten Giemsa-markierter Chromosomen des diploiden Roggens (Secale cereale L.) (First studies on meiotic chromosome pairing considering Giemsa stained chromosomes of diploid rye, Secale cereale L.). Biol. Rundsch. 1975, 13, 300–305. [Google Scholar]
- Heneen, W.K. Chromosome morphology in inbred rye. Hereditas 1962, 48, 182–200. [Google Scholar] [CrossRef]
- Bhattacharyya, N.K.; Jenkins, B.C. Karyotype analysis and chromosome designation for Secale cereale L. “Dakold.” Can. J. Genet. Cytol. 1960, 2, 168–277. [Google Scholar] [CrossRef]
- De Vries, J.N.; Sybenga, J. Identification of rye chromosomes: The Giemsa banding pattern and the translocation tester set. Theor. Appl. Genet. 1976, 48, 35–43. [Google Scholar] [CrossRef]
- Oinuma, T. Karyomorphology of cereals. Karyotype alteration of rye, Secale cereale L. Jpn. J. Genet. 1953, 28, 28–34. [Google Scholar] [CrossRef]
- Pathak, G.N. Studies on the cytology of cereals. J. Genet. 1940, 39, 437–467. [Google Scholar] [CrossRef]
- Schlegel, R.; Mettin, D. The present status of chromosome recognition and gene localization in rye, Secale cereale L. Tag. Ber. Akad. Landwirtsch. Wiss. 1982, 198, 131–152. [Google Scholar]
- Schlegel, R.; Melz, G.; Nestrowicz, R. A universal reference karyotype in rye, Secale cereale L. Theor. Appl. Genet. 1987, 74, 820–826. [Google Scholar] [CrossRef]
- Tijo, J.N.; Levan, A. The use of oxyquinoline in chromosome analysis. Anal. Est. Exp. Aula Dei 1950, 2, 21–64. [Google Scholar]
- Verma, S.G.; Rees, H. Giemsa staining and the distribution of heterochromatin in rye chromosomes. Heredity 1974, 32, 118–122. [Google Scholar] [CrossRef][Green Version]
- Vosa, C.G. The basic karyotype of rye (Secale cereale) analysed with Giemsa and fluorescence method. Heredity 1974, 33, 403–408. [Google Scholar] [CrossRef]
- Sybenga, J. Rye chromosome nomenclature and homoeology relationships—1st workshop on rye. Z. Pflanz. 1983, 90, 297–304. [Google Scholar]
- Bennett, M.D.; Rees, H. Natural and induced changes in chromosome size and mass in meristems. Nature 1967, 215, 93–94. [Google Scholar] [CrossRef]
- Schlegel, R.; Gill, S. N-banding analysis of rye chromosomes and the relationship between N-banded and C-banded heterochromatin. Can. J. Genet. Cytol. 1984, 26, 765–769. [Google Scholar] [CrossRef]
- Schlegel, R.; Melz, G.; Mettin, D. Rye cytology and cytogenetics—Current status. Theor. Appl. Genet. 1986, 72, 721–734. [Google Scholar] [CrossRef]
- Devos, K.M.; Atkinson, M.D.; Chinoy, C.N.; Francis, H.A.; Harcourt, R.L.; Koebner, R.M.D.; Liu, C.J.; Masojć, P.; Xie, D.X.; Gale, M.D. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 1993, 85, 673–680. [Google Scholar] [CrossRef]
- Friebe, B. Untersuchungen zum Schwesterchromatidenaustausch bei Secale cereale. Microsc. Acta 1978, 81, 159–165. [Google Scholar]
- Jones, G.H. Giemsa C-banding of rye meiotic chromosomes and the nature of “terminal” chiasmata. Chromosoma 1978, 66, 45–57. [Google Scholar] [CrossRef]
- Schwarzacher, T.; Leitch, A.R.; Bennett, M.D.; Heslop-Harrison, J.S. In situ localization of parental genomes in a wide hybrid. Ann. Bot. 1989, 64, 315–324. [Google Scholar] [CrossRef]
- Bashir, A.; Auger, J.A.; Rayburn, L.A. Flow cytometric DNA analysis of wheat-rye addition lines. Cytometry 1993, 4, 843–847. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S.; Leitch, A.R.; Schwarzacher, T.; Anamthawat-Jonsson, K. Detection and characterization of 1B/1R translocations in hexaploid wheat. Heredity 1990, 65, 385–392. [Google Scholar] [CrossRef][Green Version]
- Mikhailova, E.L.; Philipps, D.; Sosnikhina, S.P.; Lovtsyus, A.V.; Jones, R.N.; Jenkins, R. Molecular assembly of meiotic proteins Asy1 and Zyp1 and pairing promiscuity in rye (Secale cereale L.) and its synaptic mutant sy10. Genetics 2006, 174, 1247–1258. [Google Scholar] [CrossRef]
- Rabanus-Wallace, M.T.; Hackauf, B.; Mascher, M.; Lux, T.; Wicker, T.; Gundlach, H.; Baez, M.; Houben, A.; Mayer, K.F.X.; Guo, L.; et al. Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat. Genet. 2021, 53, 564–573. [Google Scholar] [CrossRef]
- Schlegel, R.; Kynast, R.; Schwarzacher, T.; Römheld, V.; Walter, A. Mapping of genes for copper efficiency in rye and the relationship between copper and iron efficiency. Plant Soil 1993, 154, 61–65. [Google Scholar] [CrossRef]
- Schlegel, R. Current List of Wheats with Rye and Alien Introgression. V01-22, 1–18. 2022. Available online: http://www.rye-gene-map.de/rye-introgression (accessed on 1 January 2020).
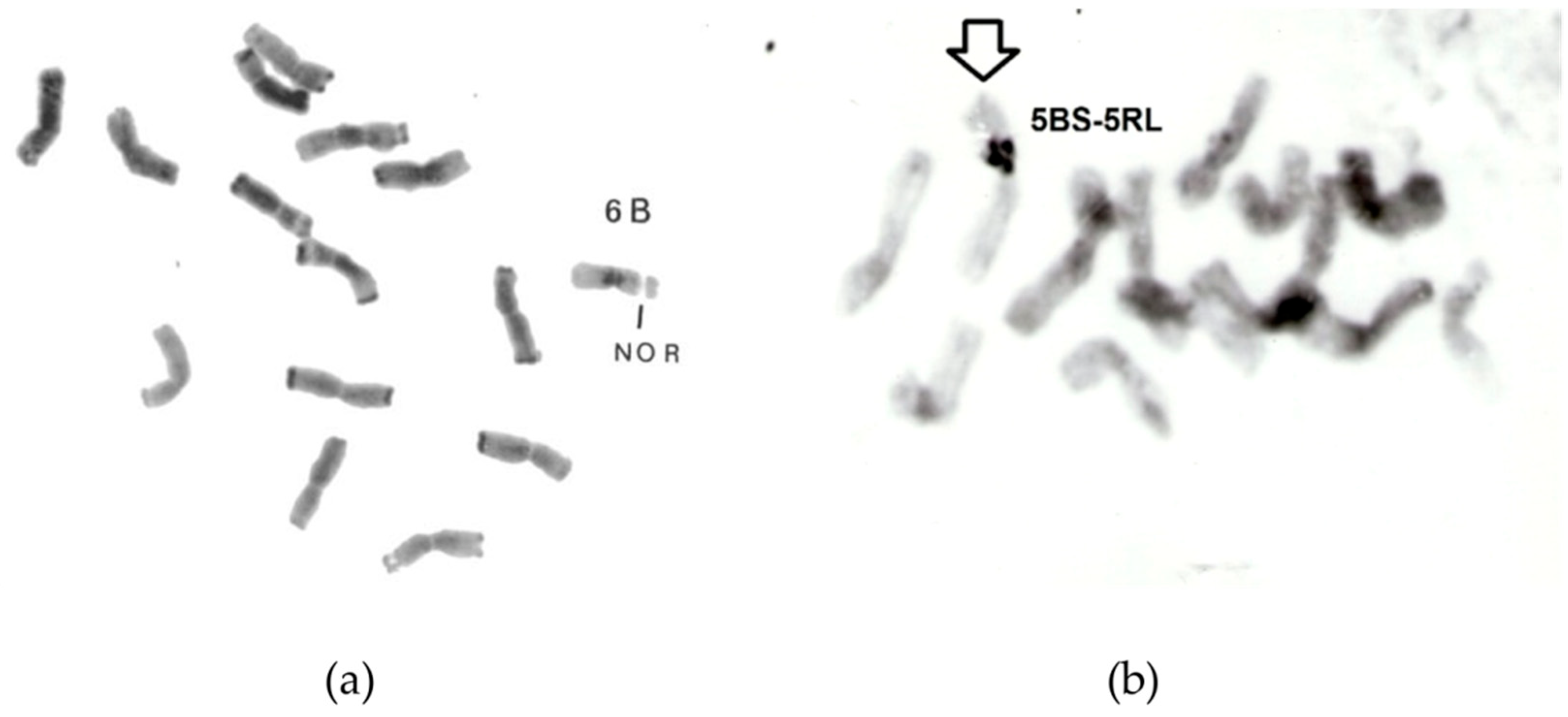
- Schlegel, R. First evidence for rye-wheat additions. Biol. Zentralbl. 1982, 101, 641–646. [Google Scholar]
- Schlegel, R.; Kynast, R.; Schmidt, J.C. Alien chromosome transfer from wheat into rye. In Genetic Manipulation in Plant Breeding; Walter de Gruyter: Berlin, Germany; New York, NY, USA, 1986; pp. 129–136. [Google Scholar]
- Schlegel, R.; Kynast, R. Wheat chromosome 6B compensates genetic information of diploid rye, Secale cereale L. In Proceedings of the 7th International Wheat Genetics Symposium, Cambridge, UK, 13–19 July 1988; pp. 421–426. [Google Scholar]
- Lukaszewski, A.J.; Kopecky, D. The Ph1 locus from wheat controls meiotic chromosome pairing in autotetraploid rye (Secale cereale L.). Cytogenet. Genome Res. 2010, 129, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Lukaszewski, A.J.; Brzezinski, W.; Klockiewicz-Kaminska, E. Transfer of the Glu-D1 locus encoding high molecular weight glutenin subunits 5+10 from bread wheat to diploid rye. Euphytica 2000, 115, 49–57. [Google Scholar] [CrossRef]
- Schlegel, R.; Börner, A.; Thiele, V.; Melz, G. The effect of the Ph1 gene in diploid rye, Secale cereale L. Genome 1991, 34, 913–917. [Google Scholar] [CrossRef]
- Xi, W.; Tang, S.; Du, H.; Luo, J.; Tang, Z.; Fu, S. ND-FISH-positive oligonucleotide probes for detecting specific segments of rye (Secale cereale L.) chromosomes and new tandem repeats in rye. Crop J. 2020, 8, 171–181. [Google Scholar] [CrossRef]
- Jones, R.N.; Rees, H. The influence of B-chromosomes upon the nuclear phenotype in rye. Chromosoma 1968, 24, 158–176. [Google Scholar] [CrossRef]
- Houben, A.; Kynast, R.G.; Heim, U.; Hermann, H.; Jones, R.N.; Forster, J.W. Molecular cytogenetic characterisation of the terminal heterochromatic segment of the B-chromosome of rye (Secale cereale). Chromosoma 1996, 105, 97–103. [Google Scholar] [CrossRef]
- Houben, A.; Ma, W.; Banaei-Moghaddam, A.M. The B chromosome of rye. In The Rye Genome; Rabanus-Wallace, M.T., Stein, N., Eds.; Springer Nature: Cham, Switzerland, 2021; pp. 63–76. ISSN 2199-4781. [Google Scholar]
- Küster, H. Am Anfang war das Korn: Eine andere Geschichte der Menschheit; C.H. Beck: Munich, Germany, 2013; p. 298. [Google Scholar]
- Langdon, T.; Seago, C.; Jones, R.N.; Ougham, H.; Thomas, H.; Forster, J.W.; Jenkins, G. De novo evolution of satellite DNA on the rye B chromosome. Genetics 2000, 154, 869–884. [Google Scholar] [CrossRef]
- Pohler, W.; Schlegel, R. A rye plant with frequent A-B chromosome pairing. Hereditas 1990, 112, 217–220. [Google Scholar] [CrossRef]
- Schlegel, R.; Pohler, W. Identification of an A-B translocation in diploid rye. Breed. Sci. 1994, 44, 279–283. [Google Scholar]
- Houben, A.; Schlegel, R. Chromosomen-Transfer bei Pflanzen. Wiss. Fortschr. 1991, 41, 358–360. [Google Scholar]
- Hülgenhof, E.; Weidhase, R.A.; Schlegel, R.; Tewes, A. Flow cytometric determination of DNA content in isolated nuclei of cereals. Genome 1988, 30, 565–569. [Google Scholar] [CrossRef]
- Pan, W.H.; Houben, A.; Schlegel, R. Highly effective cell synchronization in plant roots by hydroxyurea and amiprophos-methyl or colchicine. Genome 1993, 36, 387–390. [Google Scholar] [CrossRef]
- Hülgenhof, E.; Schlegel, R. Alterations of chromosome structure in hexaploid triticale and its effect on yield characters. 2. Experimental results of cytological behaviour in some heterochromatin-deficient triticale lines (German). Biol. Zbl. 1985, 104, 245–259. [Google Scholar]
- Kubalakova, M.; Valarik, M.; Bartos, J.; Vrana, J.; Cíhalíkova, J.; Molnar-Lang, M.; Dolezel, J. Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome 2003, 46, 893–905. [Google Scholar] [CrossRef]
- Mikhailova, E.L.; Dolezel, J. Rye cytogenetics and chromosome genomics. In The Rye Genome; Rabanus-Wallace, M.T., Stein, N., Eds.; Springer Nature: Cham, Switzerland, 2021; pp. 43–62. ISSN 2199-4781. [Google Scholar]
- Simkova, H.; Safar, J.; Suchankova, P.; Kovarova, P.; Bartos, J.; Kubalakova, M.; Janda, J.; Cíhalíkova, J.; Mago, R.; Lelley, T.; et al. A novel resource for genomics of Triticeae: BAC library specific for the short arm of rye (Secale cereale L.) chromosome 1R (1RS). BMC Genom. 2008, 9, 237–246. [Google Scholar] [CrossRef]
- Martis, M.M.; Zhou, R.; Haseneyer, G.; Schmutzer, T.; Vrána, J.; Kubaláková, M.; König, S.; Kugler, K.G.; Scholz, U.; Hackauf, B.; et al. Reticulate Evolution of the rye genome. Plant Cell 2013, 25, 3685–3698. [Google Scholar] [CrossRef]
- Nagaki, K.; Furuta, T.; Yamaji, N.; Kuniyoshi, D.; Ishihara, M.; Kishima, Y.; Murata, M.; Hoshino, A.; Takatsuka, H. Effectiveness of Create ML in microscopy image classifications: A simple and inexpensive deep learning pipeline for non-data scientists. Chromosome Res. 2021, 29, 361–371. [Google Scholar] [CrossRef]
- Smith, D.B.; Flavell, R.B. Nucleotide sequence organization in rye genome. Biochem. Biophys. Acta 1997, 474, 82–97. [Google Scholar]
- Hackauf, B.; Wehling, P. Development of microsatellite markers in rye: Map construction. Plant Breed. Seed Sci. 2003, 48, 143–151. [Google Scholar]
- Li, G.; Wang, L.; Yang, J.; He, H.; Jin, H.; Li, X.; Ren, T.; Ren, Z.; Li, F.; Han, X.; et al. A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 2021, 53, 574–584. [Google Scholar] [CrossRef]
- Camacho, M.V.; Matos, M.; Gonzales, C.; Perez-Flores, V.; Pernauta, B.; Pinto-Carnida, O.; Benito, C. Secale cereale inter-microsatellites (SCIMs): Chromosomal location and genetic inheritance. Genetica 2005, 123, 303–311. [Google Scholar] [CrossRef]
- Keilwagen, J.; Lehnert, H.; Berner, T.; Beier, S.; Scholz, U.; Himmelbach, A.; Stein, N.; Badaeva, E.D.; Lang, D.; Kilian, B.; et al. Detecting large chromosomal modifications using short read data from genotyping-by-sequencing. Front. Plant Sci. 2019, 10, 1133. [Google Scholar] [CrossRef]
- Gehrke, F.; Schindele, A.; Puchta, H. Nonhomologous end joining as key to CRISPR/Cas-mediated plant chromosome engineering. Plant Physiol. 2022, 188, 1769–1779. [Google Scholar] [CrossRef]
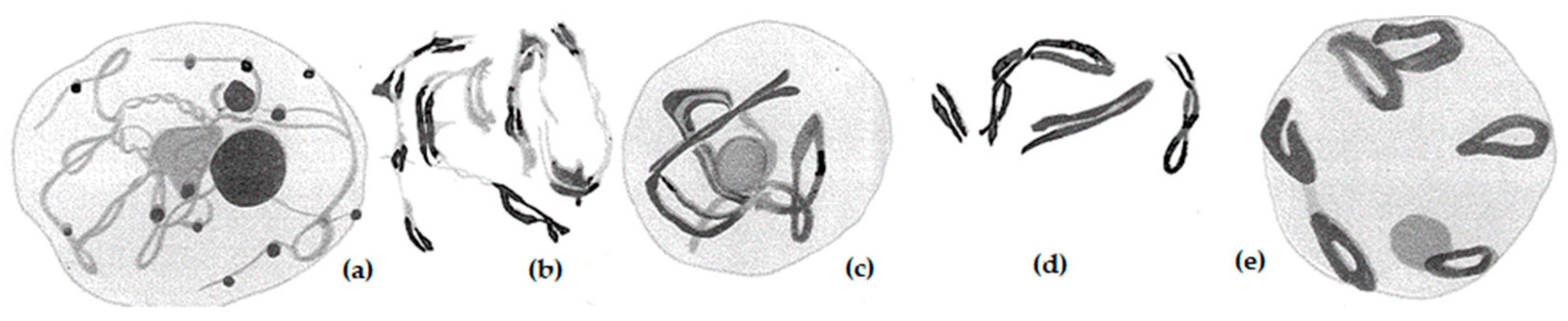
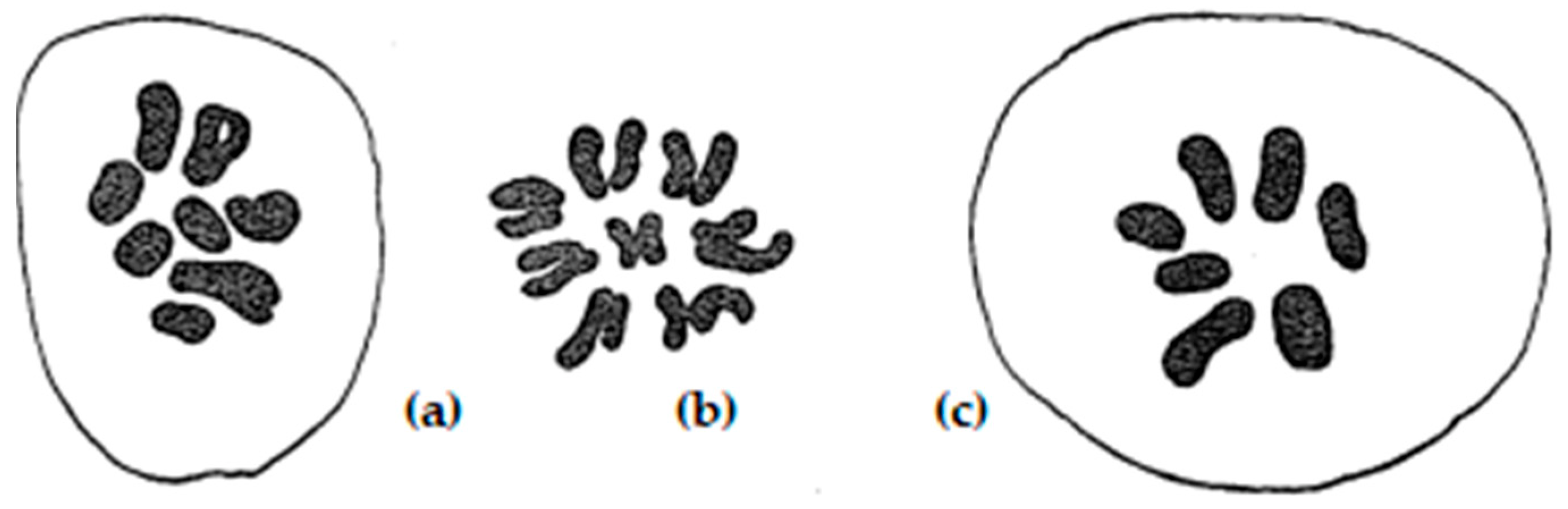

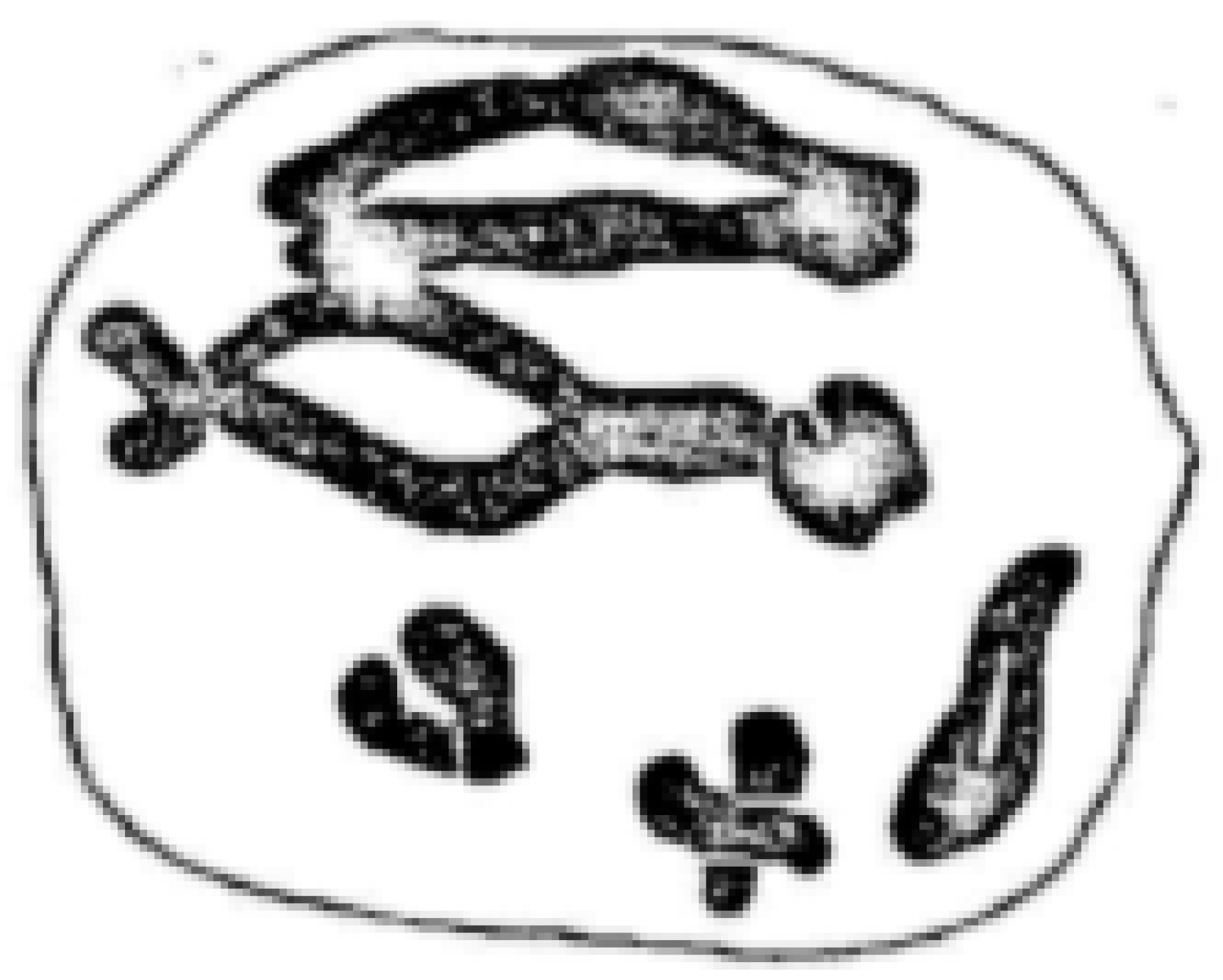







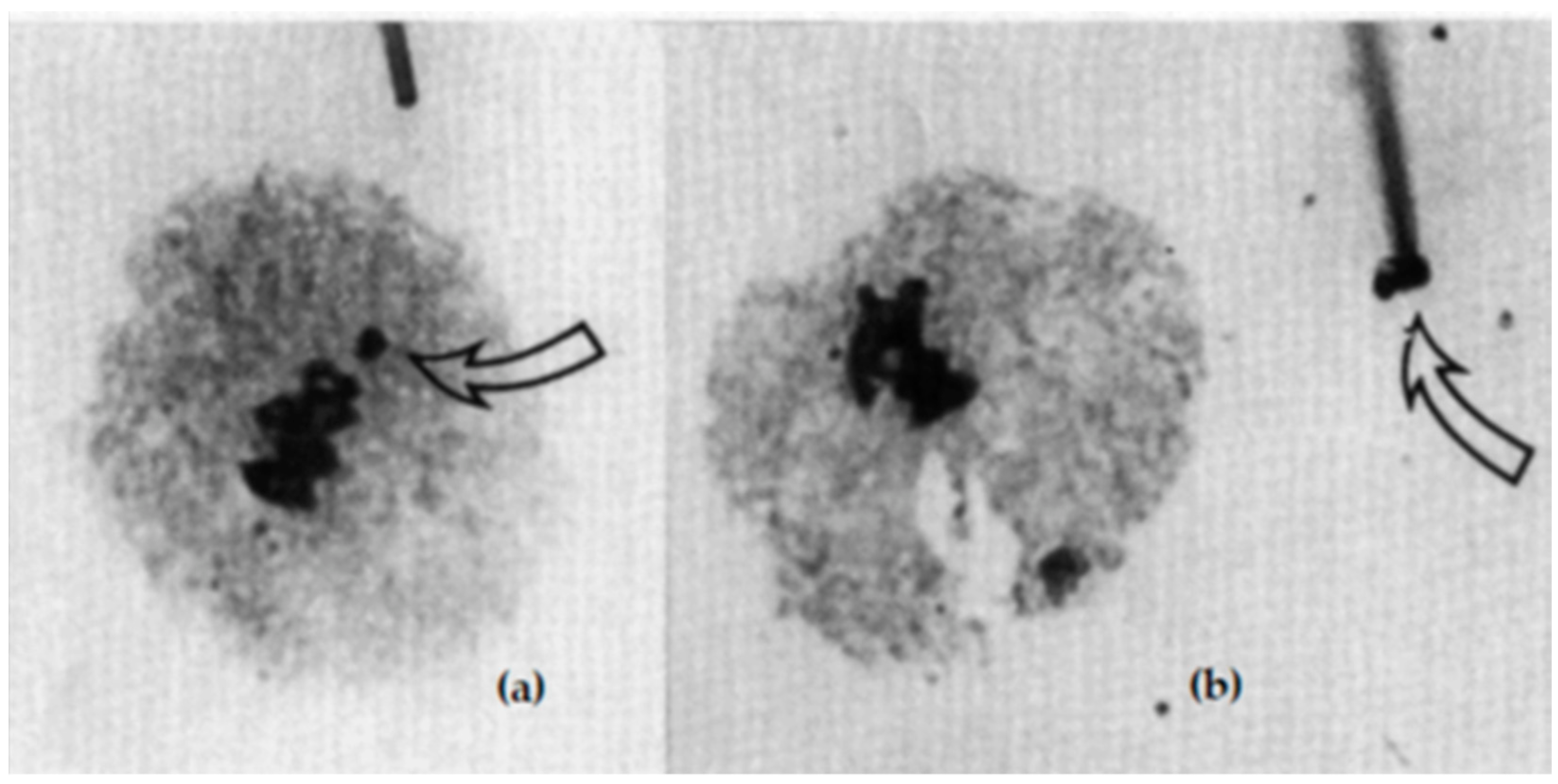
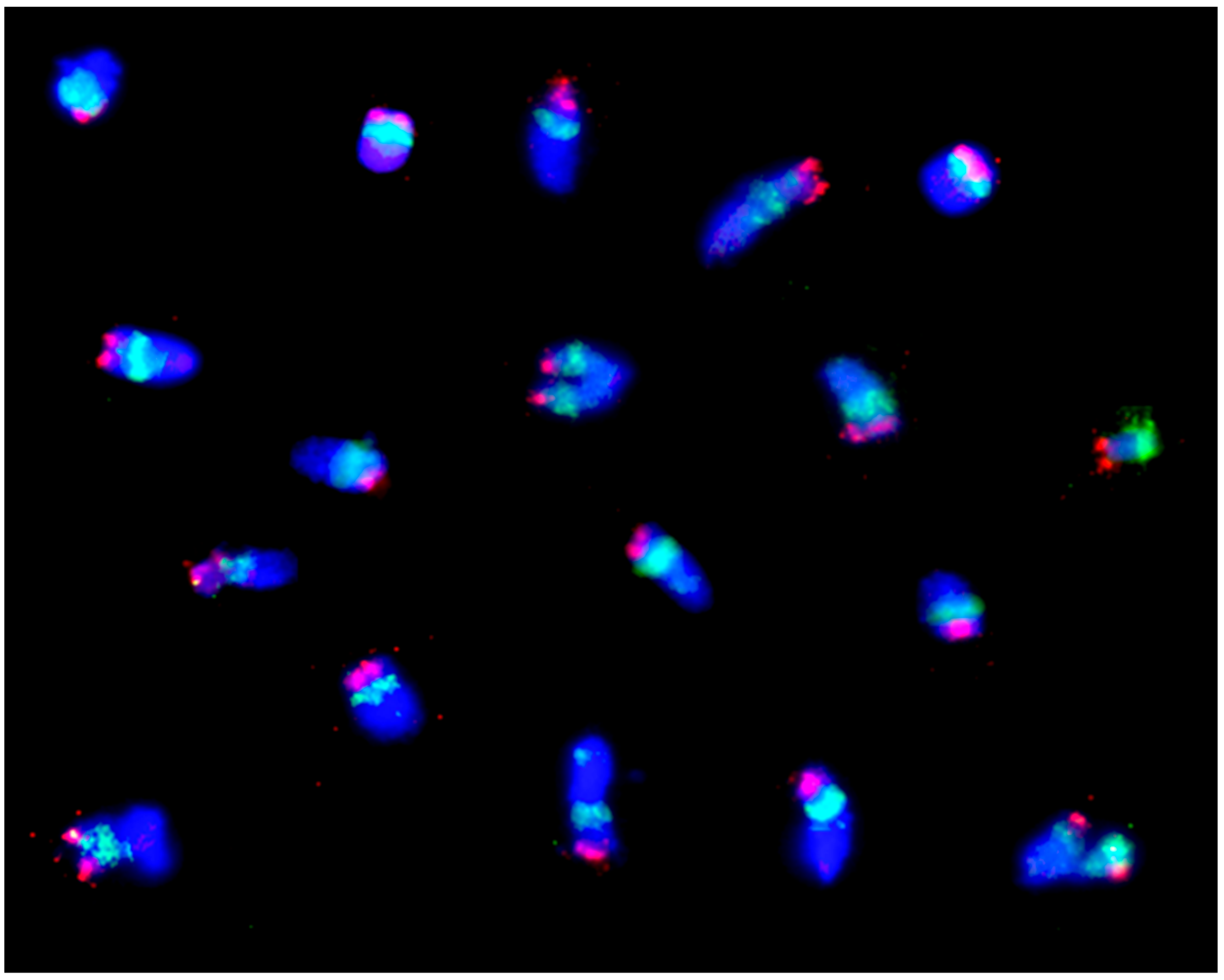
| Line No. | T273 | T248 | T305 | T242 | T240 | T501 | T282 |
|---|---|---|---|---|---|---|---|
| Chromosomes/arm sinvolved | 1RS–4RL, 1RL–5RS | 1RS–5RL, 1RS–6RS | 2RS–5RS, 2RL–5RS | 2RL–6RL | 3RS–5RS | 3RS–5RL, 4RL–5RL | 5RL–7RS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schlegel, R. 100 Years of Chromosome Research in Rye, Secale L. Plants 2022, 11, 1753. https://doi.org/10.3390/plants11131753
Schlegel R. 100 Years of Chromosome Research in Rye, Secale L. Plants. 2022; 11(13):1753. https://doi.org/10.3390/plants11131753
Chicago/Turabian StyleSchlegel, Rolf. 2022. "100 Years of Chromosome Research in Rye, Secale L." Plants 11, no. 13: 1753. https://doi.org/10.3390/plants11131753
APA StyleSchlegel, R. (2022). 100 Years of Chromosome Research in Rye, Secale L. Plants, 11(13), 1753. https://doi.org/10.3390/plants11131753






