Genome-Wide Identification of MAPKK and MAPKKK Gene Family Members and Transcriptional Profiling Analysis during Bud Dormancy in Pear (Pyrus x bretschneideri)
Abstract
:1. Introduction
2. Results
2.1. Identification and Sequence Analysis of Pyrus MAPKKs and MAPKKKs
2.2. Phylogenetic Analysis and Multiple Alignment of MAPKK and MAPKKK Genes in Pyrus
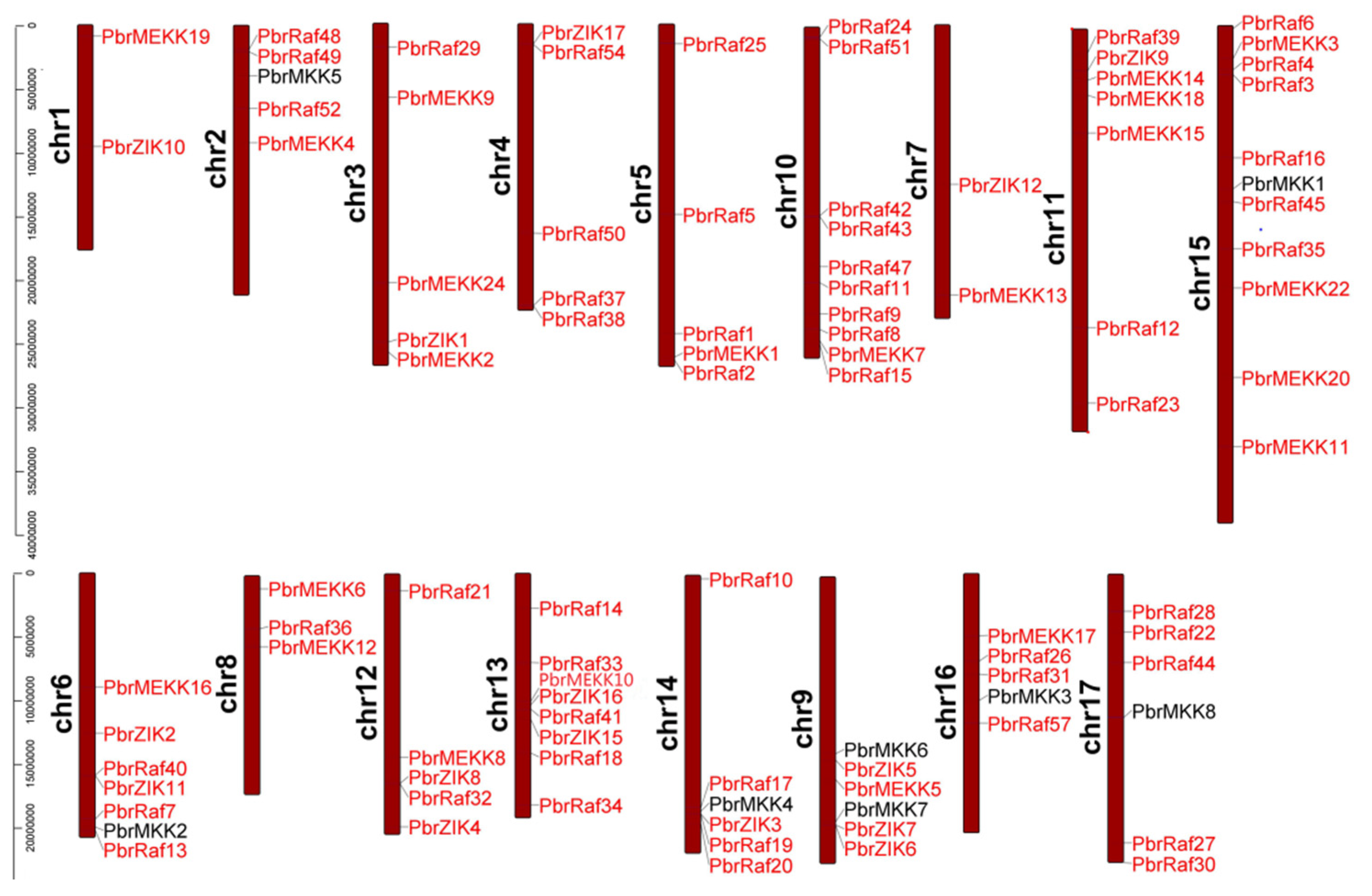
2.3. Gene Structure, Conserved Motifs and Chromosomal Distribution Analysis of Pyrus MAPKKs and MAPKKKs
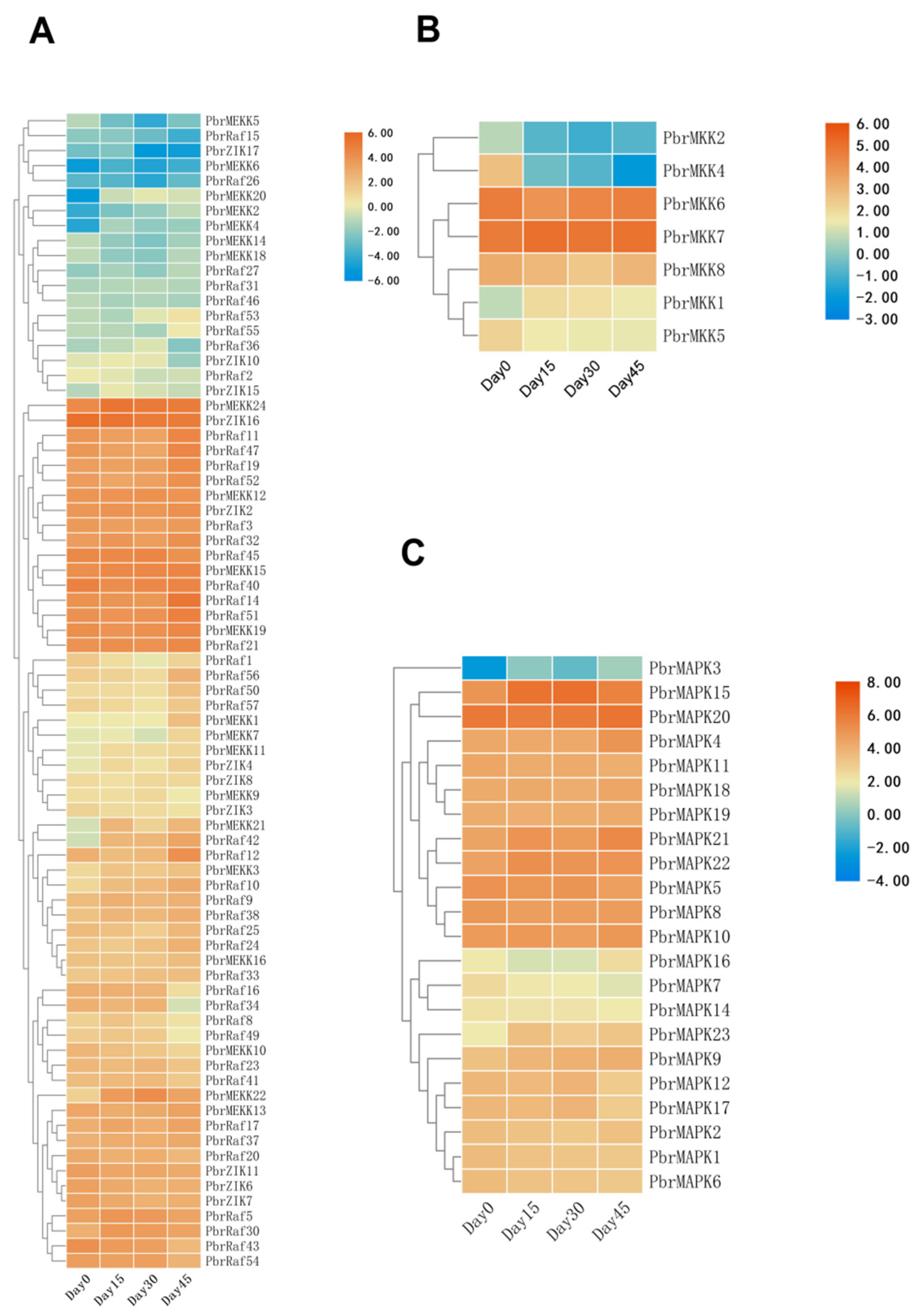
2.4. Expression Profiles of MAPK Cascade Genes in Different Stages of Dormancy in Pear
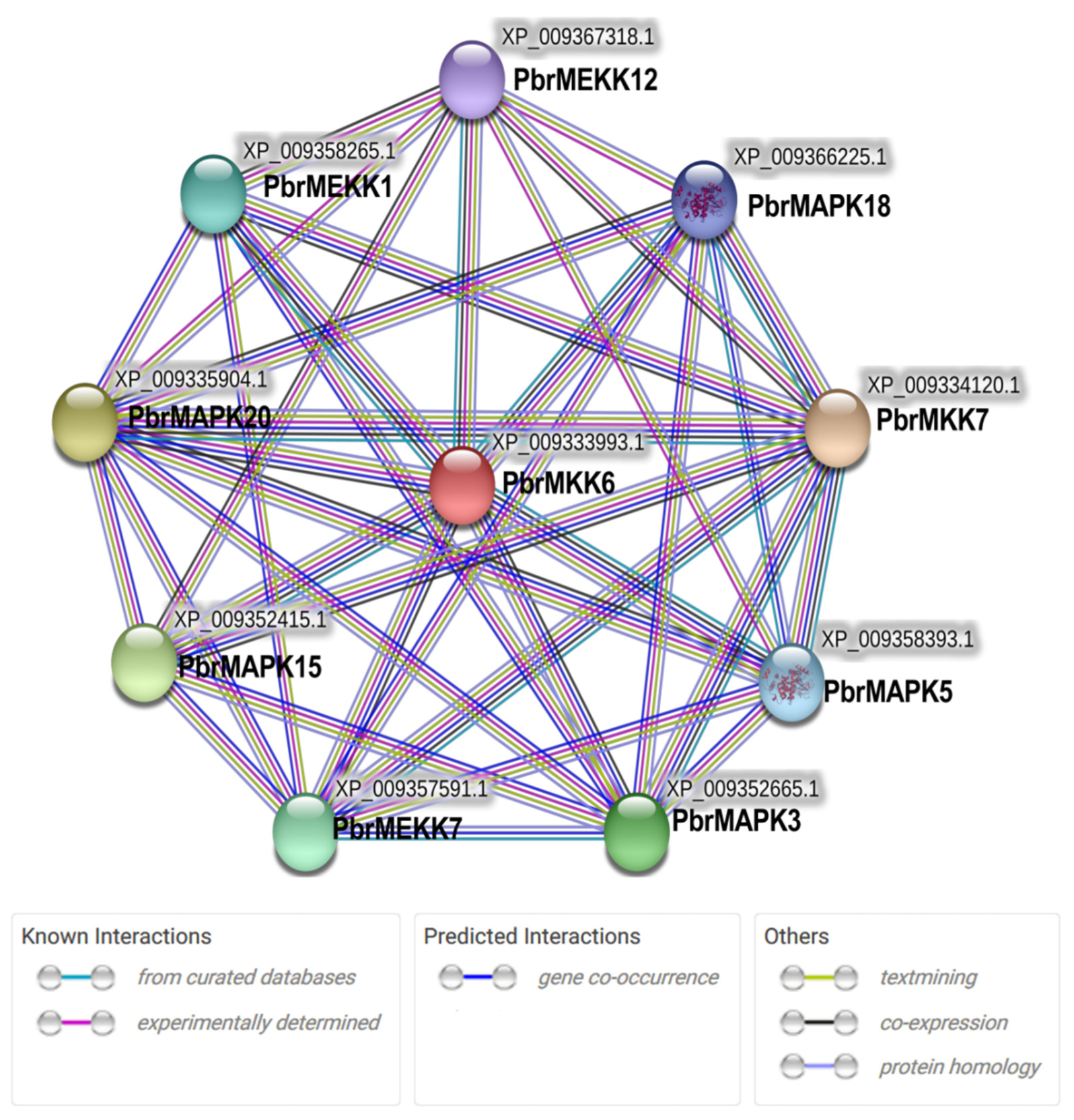
2.5. MAPK Cascade Networks and Their Co-Expression during Bud Dormancy
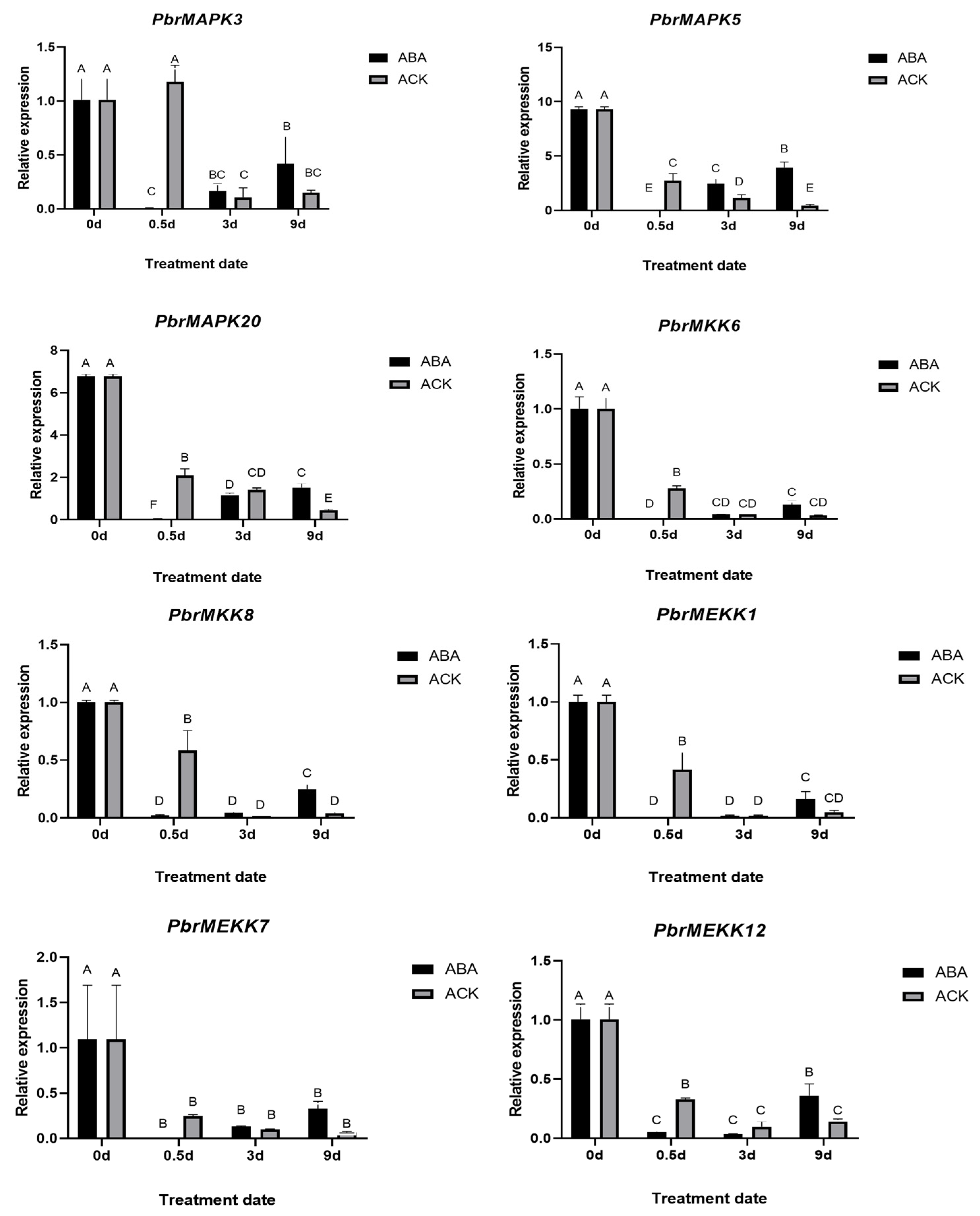
2.6. Validation of PbrMAPKKK-PbrMAPKK-PbrMAPK Cascade Gene Function in Pear Dormancy by qRT–PCR Analysis
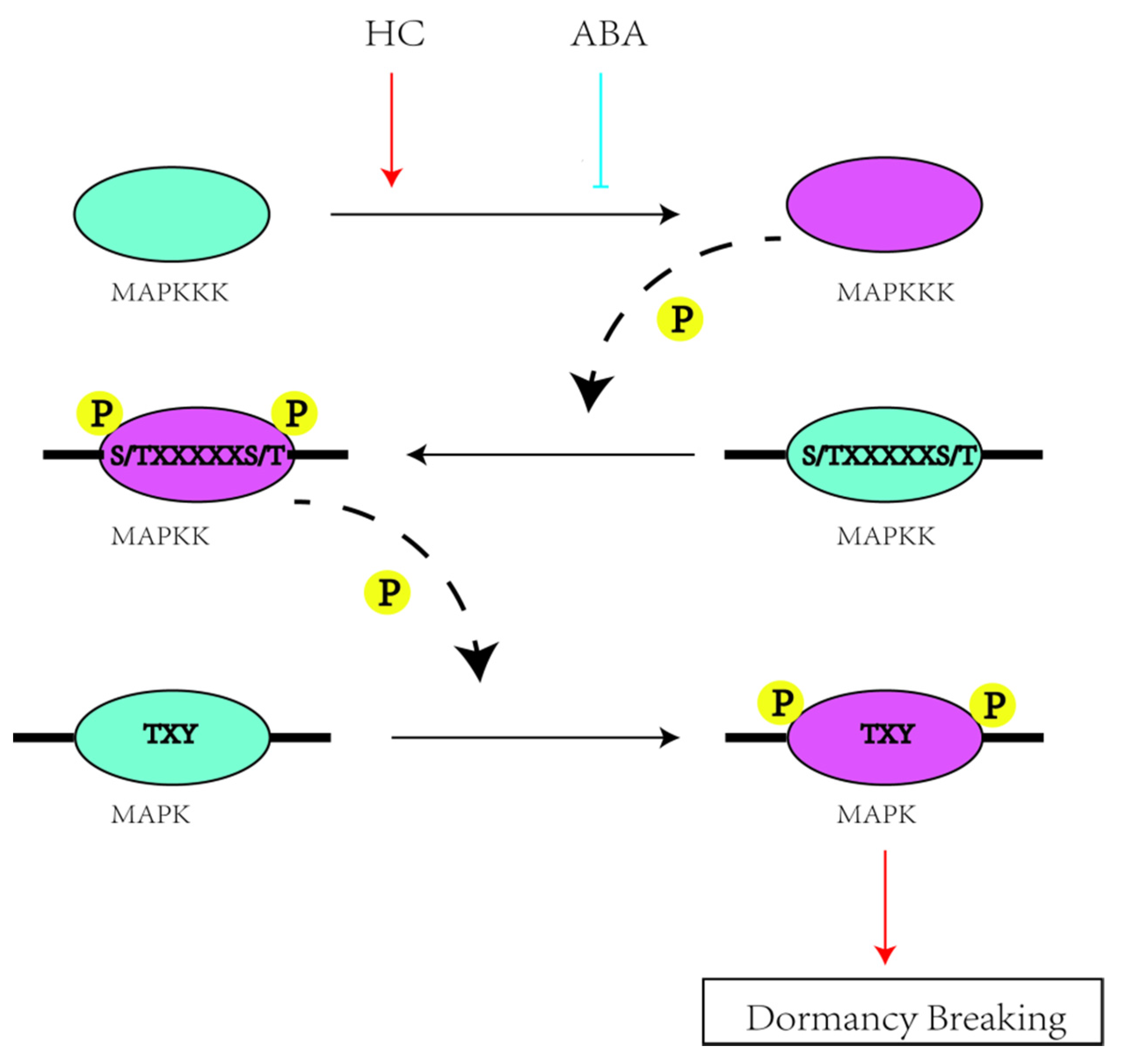
3. Discussion
4. Materials and Methods
4.1. Identification of the MAPKK and MAPKKK Gene Families in Pyrus
4.2. Multiple Sequence Alignments and Phylogenetic Analysis
4.3. Sequence, Protein Property and Chromosome Location Analyses
4.4. Differential Expression Analysis of MAPK Cascade Genes during Dormancy Using RNA-Seq
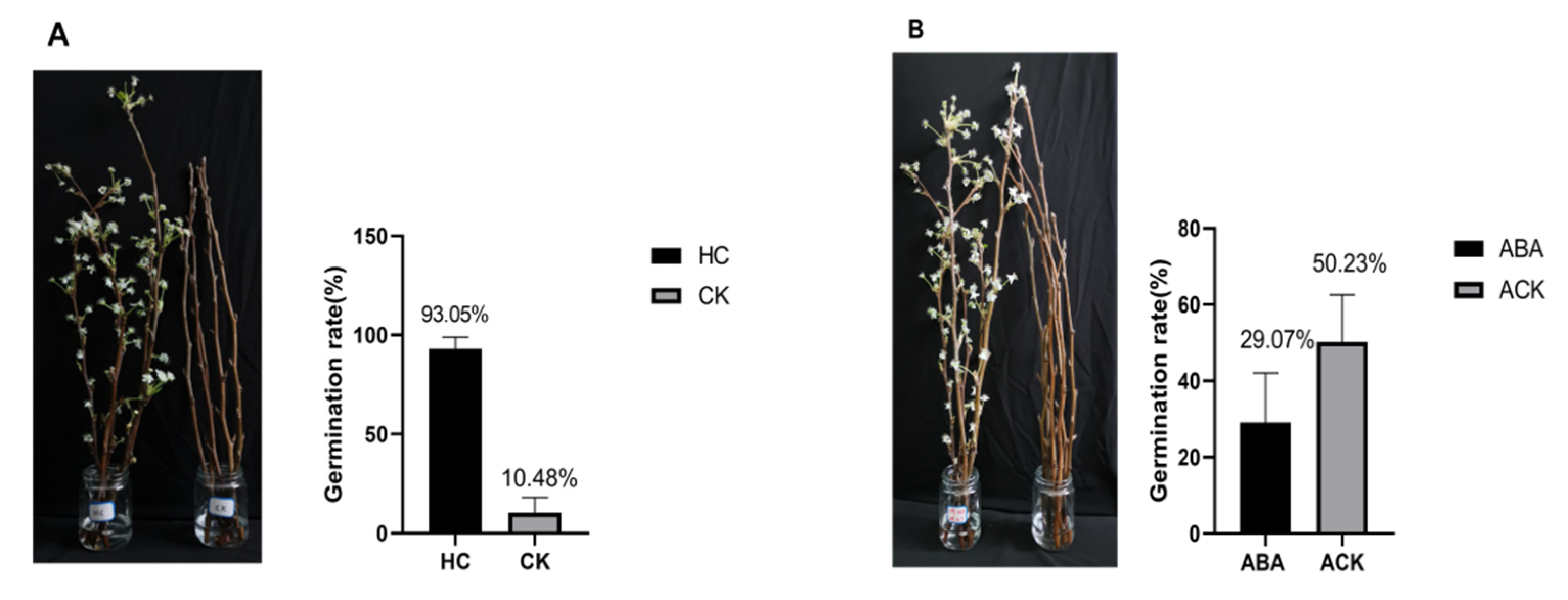
4.5. Plant Materials and Treatments
4.6. MAPK Signalling Cascade Co-Expression Analysis during Pear Dormancy
4.7. RNA Isolation and qRT–PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rohde, A.; Bhalerao, R.P. Plant dormancy in the perennial context. Trends Plant Sci. 2007, 12, 217–223. [Google Scholar] [CrossRef]
- Erez, A.; Norman, H.A.; Rowland, L.J.; Faust, M.; Wang, S.Y. Bud Dormancy in Perennial Fruit Trees: Physiological Basis for Dormancy Induction, Maintenance, and Release. HortScience 1997, 32, 623–629. [Google Scholar]
- Hänninen, H. Modelling bud dormancy release in trees from cool and temperate regions. Acta For. Fenn. 1990, 213, 7660. [Google Scholar] [CrossRef] [Green Version]
- Kilany, A.E.; Hegazi, E.S.; Kilany, O.A. Ethylene evolution, CO2 production and some endogenous compounds in relation to bud dormancy of pear trees [Egypt]. Bull. Fac. Agric. 1987, 75, 1584–1588. [Google Scholar]
- Jimenez, C.; Cossio, B.R.; Rivard, C.J.; Berl, T.; Capasso, J.M. Cell division in the unicellular microalga Dunaliella viridis depends on phosphorylation of extracellular signal-regulated kinases (ERKs). J. Exp. Bot. 2007, 58, 1001–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Nath, O.; Singh, S.; Kumar, S.; Singh, I.K. Corrigendum to “Genome-Wide Identification of the MAPK Gene Family in Chickpea and Expression Analysis during Development and Stress Response”. Plant Gene 2017, 17, 100138, Erratum in Plant Gene 2018, 13, 25–35. [Google Scholar] [CrossRef]
- Chavez Suarez, L.; Ramirez Fernandez, R. Signalling pathway in plants affected by salinity and drought. Itea-Inf. Tec. Econ. Agrar. 2010, 106, 157–169. [Google Scholar]
- Jonak, C.; Ökrész, L.; Bögre, L.; Hirt, H. Complexity, Cross Talk and Integration of Plant MAP Kinase Signalling. Curr. Opin. Plant Biol. 2002, 5, 415–424. [Google Scholar] [CrossRef]
- Meng, W.; Hong, Y.; Kewei, F.; Pingchuan, D.; Weining, S.; Xiaojun, N. Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.). BMC Genom. 2016, 17, 668. [Google Scholar]
- Ichimura, K.; Mizoguchi, T.; Irie, K.; Morris, P.; Giraudat, J.; Matsumoto, K.; Shinozaki, K. Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem. Biophys. Res. Commun. 1998, 253, 532–543. [Google Scholar] [CrossRef]
- Smekalova, V.; Doskocilova, A.; Komis, G.; Samaj, J. Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol. Adv. 2014, 32, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Colcombet, J.; Hirt, H. Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Teige, M.; Scheikl, E.; Eulgem, T.; Doczi, F.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Rohila, J.S.; Yang, Y. Rice mitogen-activated protein kinase gene family and its role in biotic and abiotic stress response. J. Integr. Plant Biol. 2007, 49, 751–759. [Google Scholar] [CrossRef]
- Wankhede, D.P.; Misra, M.; Singh, P.; Sinha, A.K. Rice Mitogen Activated Protein Kinase Kinase and Mitogen Activated Protein Kinase Interaction Network Revealed by In-Silico Docking and Yeast Two-Hybrid Approaches. PLoS ONE 2013, 8, e65011. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Xu, R.; Luo, X.; Jiang, Z.; Shu, H. Genome-wide identification and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene 2013, 531, 377–387. [Google Scholar] [CrossRef]
- Jiao, J.; Liu, X.; Wu, J.; Xu, G.; Zhang, S. Characterization of the MAPK Gene Family and PbrMAPK13 Response to Hormone and Temperature Stresses via Different Expression Pattern in Pyrus x bretschneideri Pollen. J. Am. Soc. Hortic. Sci. 2017, 142, 163–174. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Jia, W.; Zhang, J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008, 54, 440–451. [Google Scholar] [CrossRef]
- Beck, M.; Komis, G.; Ziemann, A.; Menzel, D.; Samaj, J. Mitogen-activated protein kinase 4 is involved in the regulation of mitotic and cytokinetic microtubule transitions in Arabidopsis thaliana. New Phytol. 2011, 189, 1069–1083. [Google Scholar] [CrossRef]
- Takahashi, Y.; Soyano, T.; Kosetsu, K.; Sasabe, M.; Machida, Y. HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 1766–1776. [Google Scholar] [CrossRef] [Green Version]
- Du, P.; Wang, D.; Tan, Q.; Liu, L.; Fu, X.; Chen, X.; Xiao, W.; Li, D.; Zhu, C.; Li, L.; et al. Expression analysis of MAPK genes involved in nectarine floral buds during dormancy release. Plant Physiol. J. 2016, 52, 216–224. [Google Scholar]
- Mao, X.; Zhang, J.; Liu, W.; Yan, S.; Liu, Q.; Fu, H.; Zhao, J.; Huang, W.; Dong, J.; Zhang, S.; et al. The MKKK62-MKK3-MAPK7/14 module negatively regulates seed dormancy in rice. Rice 2019, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Li, W.; Zheng, P.; Xu, T.; Chen, L.; Liu, D.; Hussain, S.; Teng, Y. Transcriptomic analysis of ‘Suli’ pear (Pyrus pyrifolia white pear group) buds during the dormancy by RNA-Seq. BMC Genom. 2012, 13, 700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Wang, J.; Pan, C.; Guan, X.; Wang, Y.; Liu, S.; He, Y.; Chen, J.; Chen, L.; Lu, G. Genome-Wide Identification of MAPKK and MAPKKK Gene Families in Tomato and Transcriptional Profiling Analysis during Development and Stress Response. PLoS ONE 2014, 9, e103032. [Google Scholar] [CrossRef]
- Rao, K.P.; Richa, T.; Kumar, K.; Raghuram, B.; Sinha, A.K. In Silico Analysis Reveals 75 Members of Mitogen-Activated Protein Kinase Kinase Kinase Gene Family in Rice. DNA Res. 2010, 17, 139–153. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Gong, M.; Guo, J.; Xin, H.; Gao, Y.; Liu, C.; Dai, D.; Tang, L. Genome-wide Identification of Jatropha curcas MAPK, MAPKK, and MAPKKK Gene Families and Their Expression Profile Under Cold Stress. Sci. Rep. 2018, 8, 16163. [Google Scholar] [CrossRef] [Green Version]
- Lang, G.A. Dormancy: A new universal terminology. HortScience 1987, 22, 817–820. [Google Scholar]
- Xu, J.; Zhang, S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef]
- Wang, J.; Pan, C.; Wang, Y.; Ye, L.; Wu, J.; Chen, L.; Zou, T.; Lu, G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genom. 2015, 16, 386. [Google Scholar] [CrossRef] [Green Version]
- Lang, G.A.; Early, J.D.; Martin, G.C.; Darnell, R.L. Endo-, para-, and ecodormancy: Physiological terminology and classification for dormancy research. HortScience 1987, 22, 371–377. [Google Scholar]
- Fadon, E.; Herrero, M.; Rodrigo, J. Dormant Flower Buds Actively Accumulate Starch over Winter in Sweet Cherry. Front. Plant Sci. 2018, 9, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heide, O.M.; Prestrud, A.K. Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol. 2005, 25, 109–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.; Hirt, H.; Wilson, C.; et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Hass, R. Hormone receptors, MAPKs and transcription factors. Signal Transduct. Recept. Mediat. Genes Off. J. Signal Transduct. Soc. 2006, 6, 3–4. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Lu, W.; Wang, X.; Wu, C.-A.; Guo, X. Overexpression of cotton GhMKK4 enhances disease susceptibility and affects abscisic acid, gibberellin and hydrogen peroxide signalling in transgenic Nicotiana benthamiana. Mol. Plant Pathol. 2014, 15, 94–108. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Niu, Q.; He, L.; Teng, Y.; Bai, S. Abscisic Acid (ABA) Promotes the Induction and Maintenance of Pear (Pyrus pyrifolia White Pear Group) Flower Bud Endodormancy. Int. J. Mol. Sci. 2018, 19, 310. [Google Scholar] [CrossRef] [Green Version]
- Tamura, F.; Tanabe, K.; Ikeda, T. Relationship between Intensity of Bud Dormancy and Level of ABA in Japanese Pear ‘Nijisseiki’. J. Jpn. Soc. Hortic. Sci. 1993, 62, 75–81. [Google Scholar] [CrossRef]
- Li, K.; Wang, X.; Yang, F.; Xu, S. MAPK cascade pathway is involved in plant growth and development regulated by ABA signal transduction. J. Plant Sci. 2014, 32, 531–539. [Google Scholar]
- Wang, H.; Huang, T.; Li, Y.; Li, L.; Wu, S. Study on floral organ development and endogenous hormones during bud dormancy of Pear. China Sci. Online. 2015. Available online: http://www.paper.edu.cn/releasepaper/content/201501-358 (accessed on 19 May 2022).
- Perez, F.J.; Vergara, R.; Rubio, S. H2O2 is involved in the dormancy-breaking effect of hydrogen cyanamide in grapevine buds. Plant Growth Regul. 2008, 55, 149–155. [Google Scholar] [CrossRef]
- Son, Y.; Cheong, Y.-K.; Kim, N.-H.; Chung, H.-T.; Kang, D.G.; Pae, H.-O. Mitogen-Activated Protein Kinases and Reactive Oxygen Species: How Can ROS Activate MAPK Pathways? J. Signal Transduct. 2011, 2011, 792639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Dong, F.; Gao, J.; Galbraith, D.W.; Song, C.P. Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 2001, 126, 1438–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittler, R.; Blumwald, E. The Roles of ROS and ABA in Systemic Acquired Acclimation. Plant Cell 2015, 27, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Mori, I.C.; Muto, S. Abscisic Acid Activates a 48-Kilodalton Protein Kinase in Guard Cell Protoplasts. Plant Physiol. 1997, 113, 833–839. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Hou, S. Role of Post-Translational Modification of Proteins in ABA Signaling Transduction. Chin. Bull. Bot. 2019, 54, 300–315. [Google Scholar]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comp. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Feng, T.; Zhang, D.; Zhuo, R.; Liu, M. Identification and comprehensive analysis of the characteristics and roles of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in Sedum alfredii Hance responding to cadmium stress. Ecotoxicol. Environ. Saf. 2019, 167, 95–106. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, R.X.; Zhong, X.J.; Wang, T.J.; Dong, L.X. A study advance on effect of temperature on fruit trees bud dormancy. J. Hubei Agric. Univ. 2002, 0z1, 101–103. [Google Scholar]
- Liu, D.; Chen, L.; Li, G.; Wang, T.; Ten, Y. Effect of cyanogen on dormancy release and endogenous hormone and carbohydrate content of Pear flower buds of “Emerald crown”. J. Zhejiang Univ. (Agric. Life Sci.) 2013, 39, 69–74. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, 45. [Google Scholar] [CrossRef] [PubMed]

| Differentially Expressed Gene Set | Differentially Expressed Gene Number | Up Regulated | Down Regulated |
|---|---|---|---|
| Day 0 vs. Day 15 | 66 | 32 | 34 |
| Day 15 vs. Day 30 | 22 | 9 | 13 |
| Day 30 vs. Day 45 | 66 | 47 | 19 |
| Process | Temperature | Time | Cycle Number |
|---|---|---|---|
| Initial Denaturation | 94 °C | 30 s | 1 |
| Denaturation | 94 °C | 5 s | 40 |
| Annealing | 60 °C | 30 s | |
| Extension/Elongation | 72 °C | 10 s |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Q.; Lin, X.; Liu, J.; Feng, Y.; Niu, X.; Wang, C.; Song, K.; Yang, C.; Li, L.; Li, Y. Genome-Wide Identification of MAPKK and MAPKKK Gene Family Members and Transcriptional Profiling Analysis during Bud Dormancy in Pear (Pyrus x bretschneideri). Plants 2022, 11, 1731. https://doi.org/10.3390/plants11131731
Liang Q, Lin X, Liu J, Feng Y, Niu X, Wang C, Song K, Yang C, Li L, Li Y. Genome-Wide Identification of MAPKK and MAPKKK Gene Family Members and Transcriptional Profiling Analysis during Bud Dormancy in Pear (Pyrus x bretschneideri). Plants. 2022; 11(13):1731. https://doi.org/10.3390/plants11131731
Chicago/Turabian StyleLiang, Qin, Xiaojie Lin, Jinhang Liu, Yu Feng, Xianqian Niu, Chao Wang, Keke Song, Chao Yang, Liang Li, and Yongyu Li. 2022. "Genome-Wide Identification of MAPKK and MAPKKK Gene Family Members and Transcriptional Profiling Analysis during Bud Dormancy in Pear (Pyrus x bretschneideri)" Plants 11, no. 13: 1731. https://doi.org/10.3390/plants11131731
APA StyleLiang, Q., Lin, X., Liu, J., Feng, Y., Niu, X., Wang, C., Song, K., Yang, C., Li, L., & Li, Y. (2022). Genome-Wide Identification of MAPKK and MAPKKK Gene Family Members and Transcriptional Profiling Analysis during Bud Dormancy in Pear (Pyrus x bretschneideri). Plants, 11(13), 1731. https://doi.org/10.3390/plants11131731





