Seed-to-Seedling Transition in Pisum sativum L.: A Transcriptomic Approach
Abstract
1. Introduction
2. Results
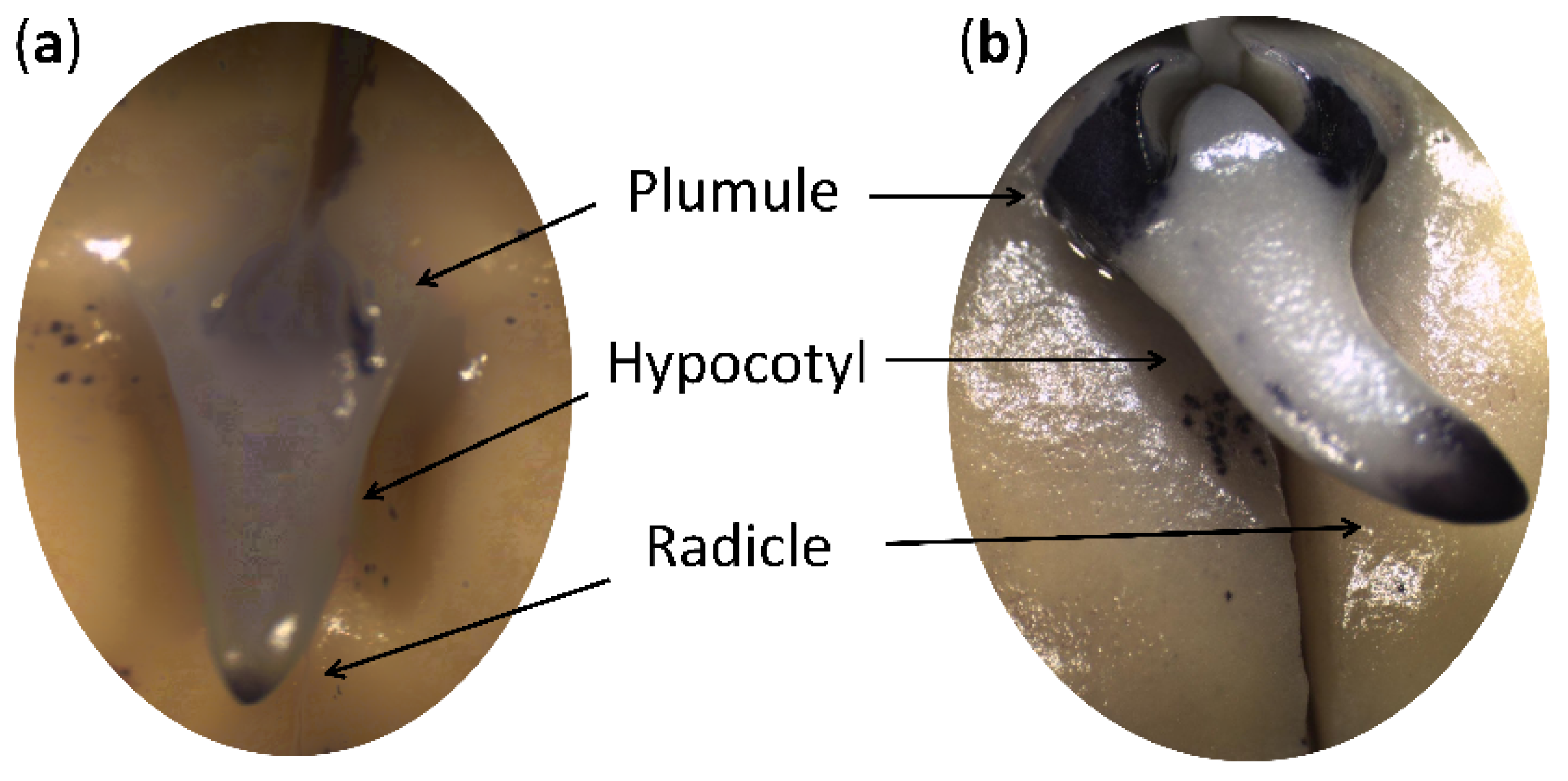
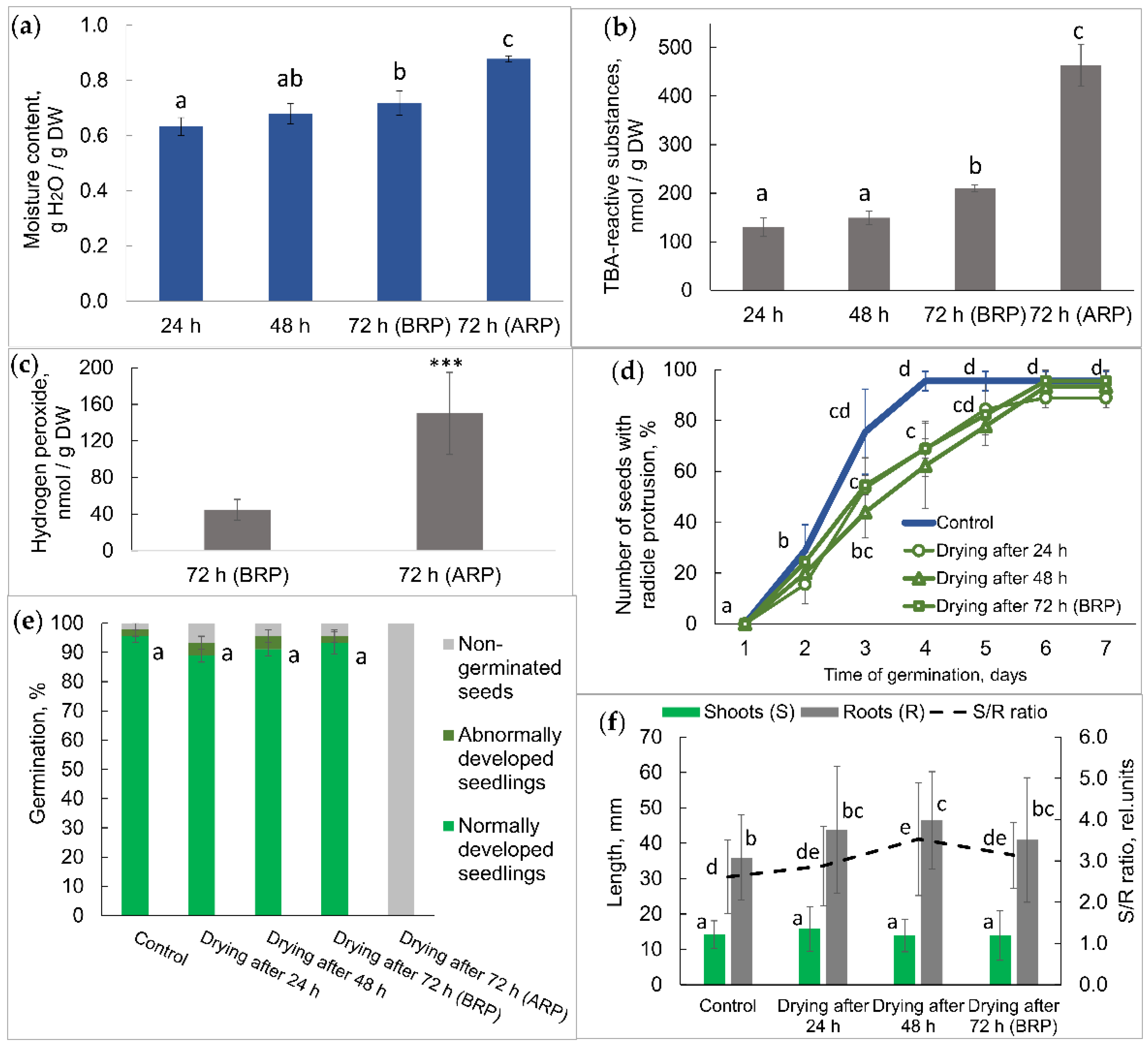
2.1. Physiological Status of Germinated Pea Seeds
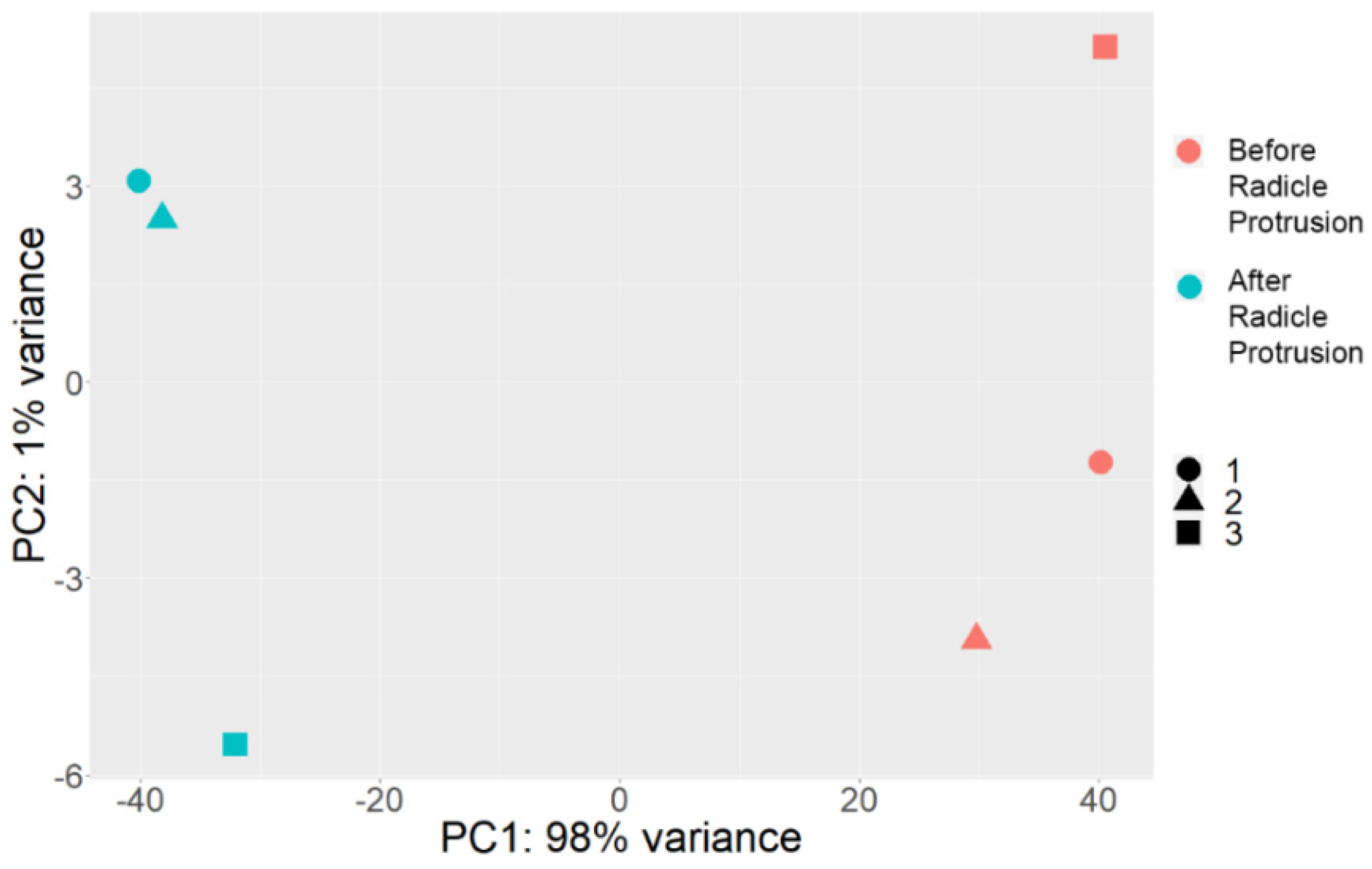
2.2. Comparative Transcriptome Profiling of Pea Seed Axes during Seed-to-Seedling Transition
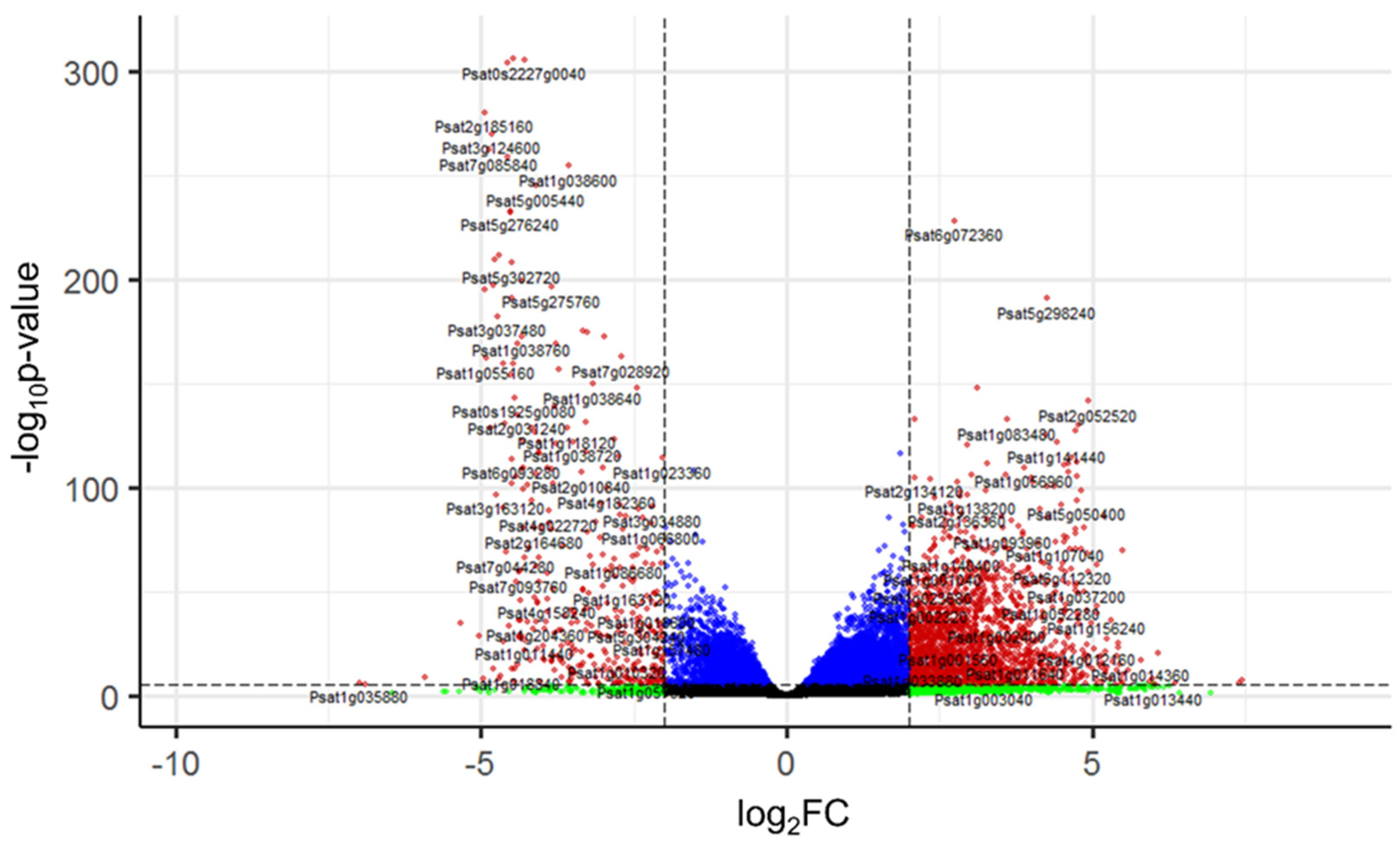
2.3. Categorization and Functional Annotation of the DEGs in Pea Seed Axes during Seed-to-Seedling Transition
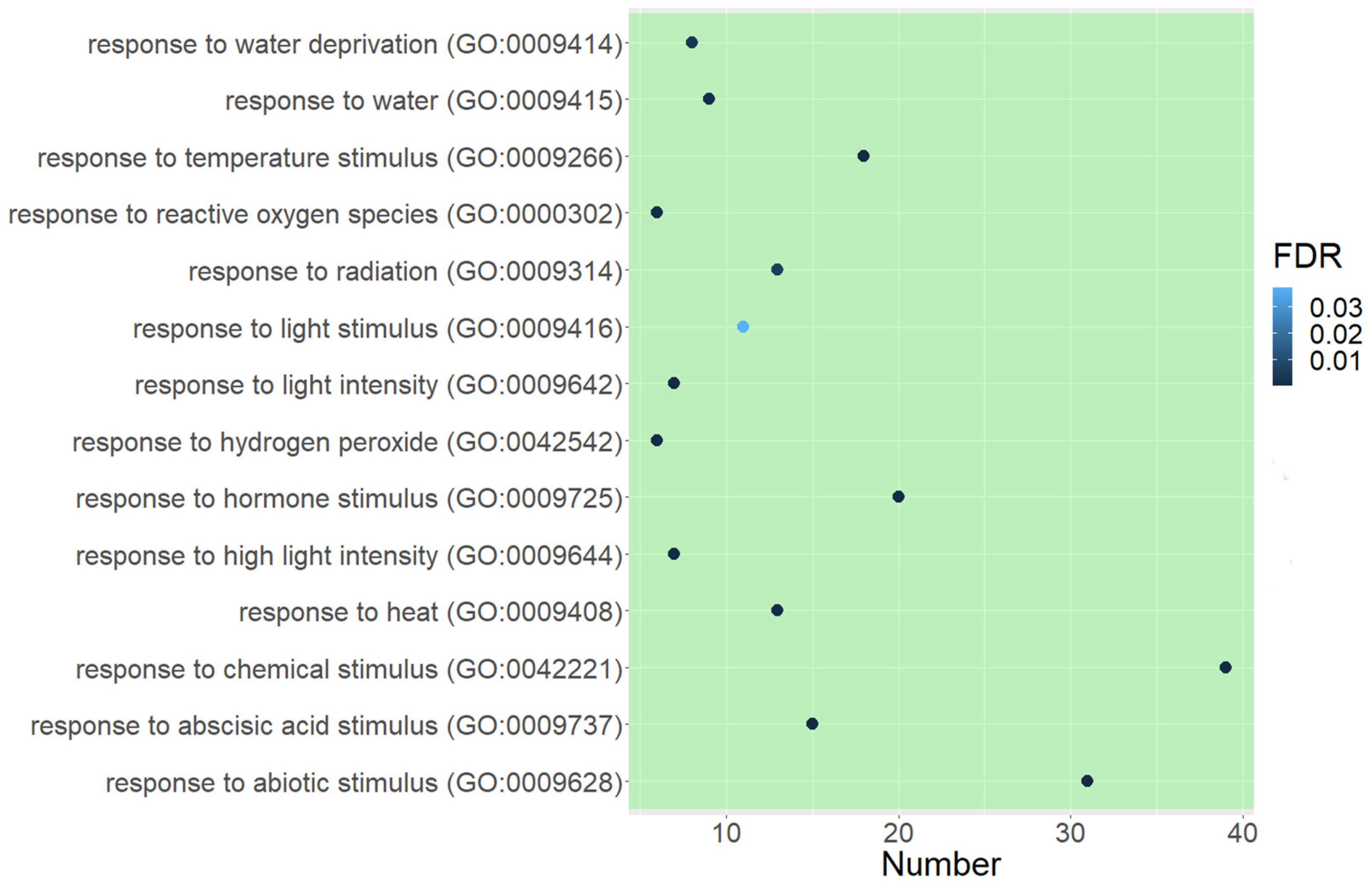
2.3.1. GO Functional Enrichment Analysis
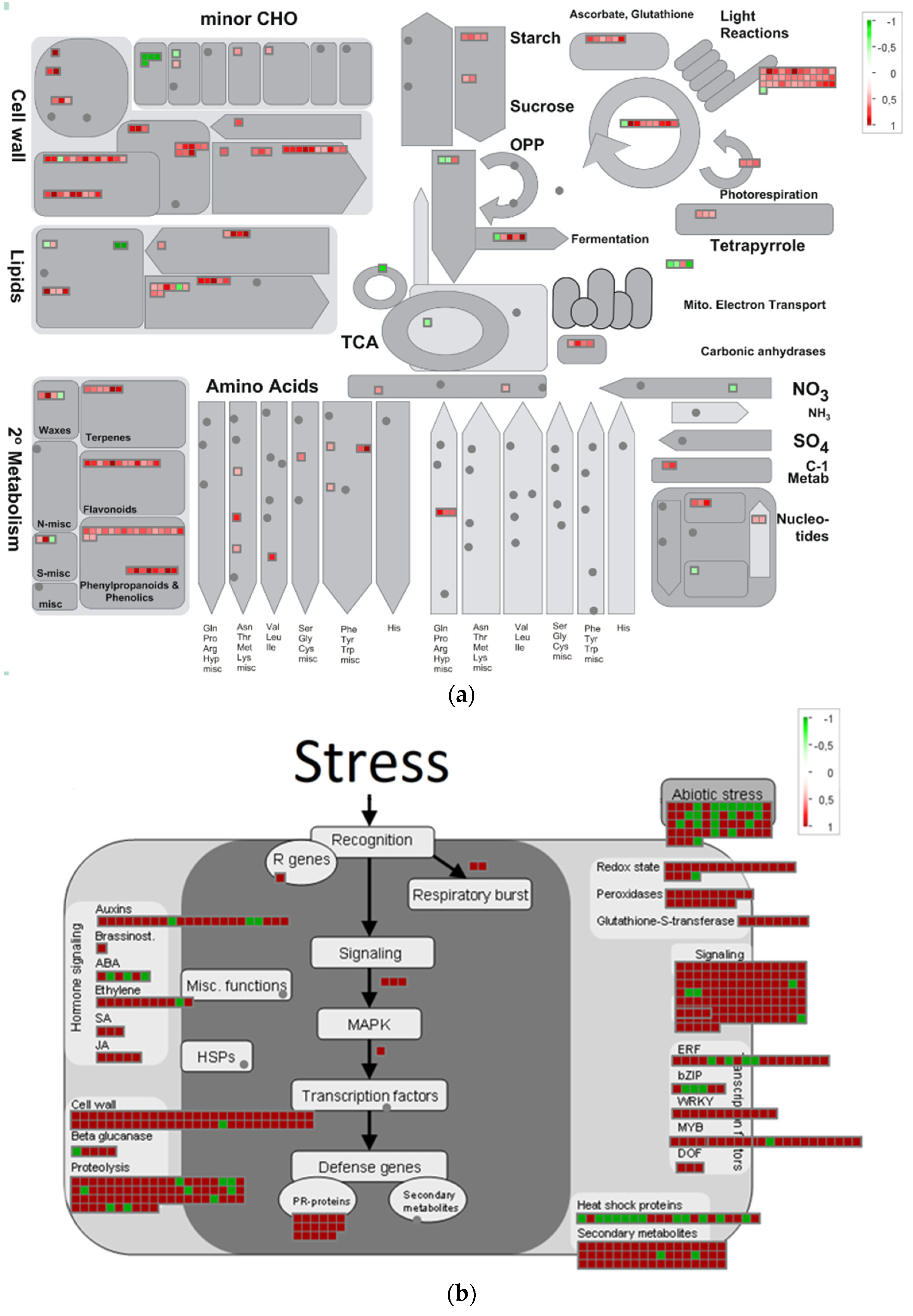
2.3.2. Overview of Metabolism
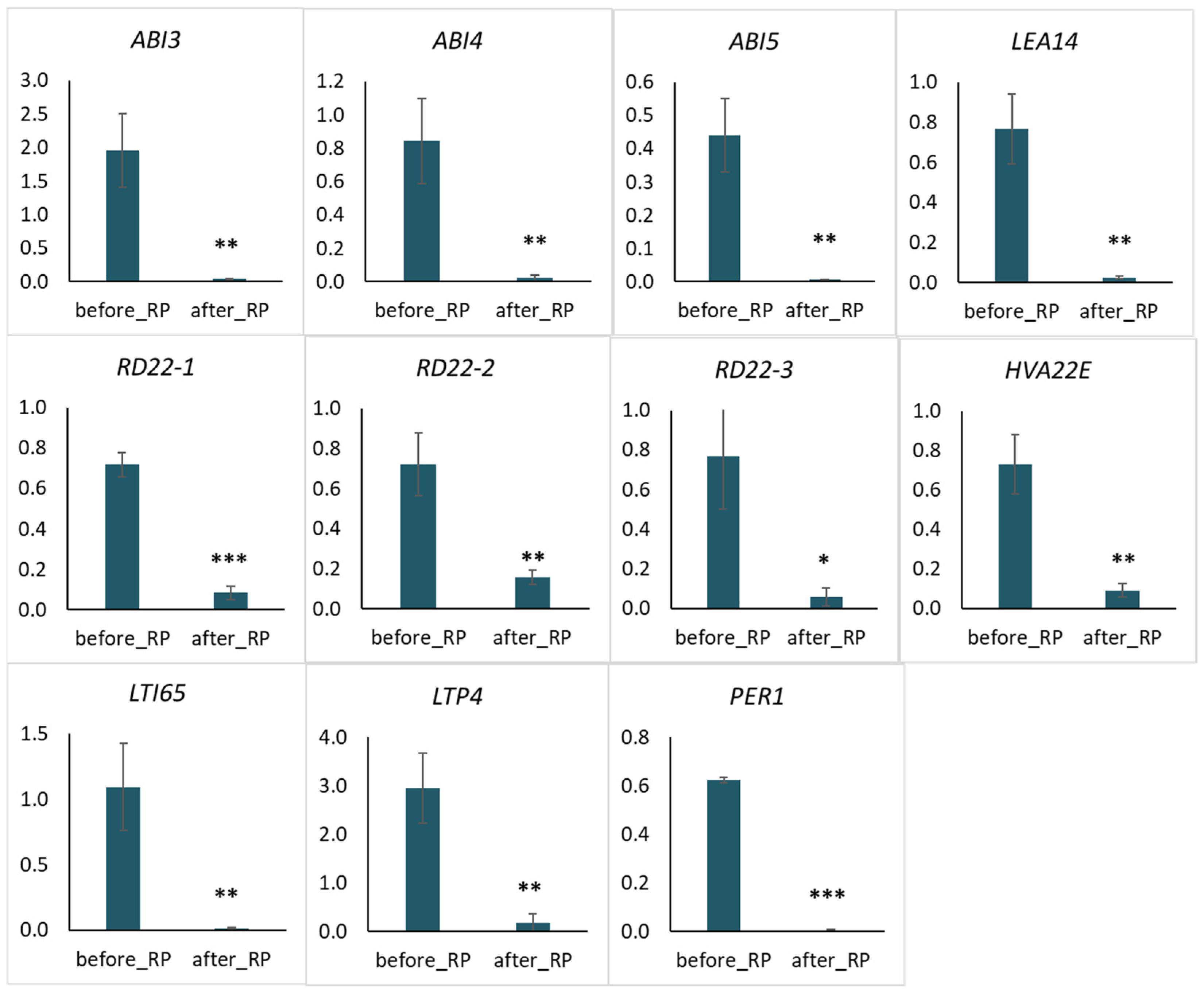
2.4. Quantitative PCR Analysis of Selected Genes Related to Water Deprivation
3. Discussion
3.1. Key Genes of the Response to Drought Tolerance of Pea Seeds
3.2. The LAFL Network and the Developmental Program
4. Conclusions
5. Materials and Methods
5.1. Preparation of Plant Material
5.2. Physiological Evaluation
5.3. Isolation of Total DNA and RNA
5.4. RNA Library Preparation
5.5. Reads Preprocessing, Mapping and Differential Expression Analysis
5.6. Differentially Expressed Genes (DEGs) Annotation
5.7. Quantitative Reverse-Transcription PCR (qRT-PCR) and Sequence Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed Germination and Vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D.; Bradford, K.J.; Hilhorst, H.W.M.; Nonogaki, H. Seeds: Physiology of Development, Germination and Dormancy, 3rd ed.; Springer: New York, NY, USA, 2013; ISBN 978-1-4614-4692-7. [Google Scholar]
- Née, G.; Xiang, Y.; Soppe, W.J. The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 2017, 35, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Carrera-Castaño, G.; Calleja-Cabrera, J.; Pernas, M.; Gómez, L.; Oñate-Sánchez, L. An Updated Overview on the Regulation of Seed Germination. Plants 2020, 9, 703. [Google Scholar] [CrossRef]
- Bentsink, L.; Koornneef, M. Seed dormancy and germination. Arab. Book 2008, 6, e0119. [Google Scholar] [CrossRef]
- Penfield, S. Seed dormancy and germination. Curr. Biol. 2017, 27, R874–R878. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Leprince, O.; Buitink, J. Desiccation tolerance: From genomics to the field. Plant Sci. 2010, 179, 554–564. [Google Scholar] [CrossRef]
- Smolikova, G.; Leonova, T.; Vashurina, N.; Frolov, A.; Medvedev, S. Desiccation tolerance as the basis of long-term seed viability. Int. J. Mol. Sci. 2021, 22, 101. [Google Scholar] [CrossRef]
- Ballesteros, D.; Pritchard, H.W.; Walters, C. Dry architecture: Towards the understanding of the variation of longevity in desiccation-tolerant germplasm. Seed Sci. Res. 2020, 30, 142–155. [Google Scholar] [CrossRef]
- Leprince, O.; Pellizzaro, A.; Berriri, S.; Buitink, J. Late seed maturation: Drying without dying. J. Exp. Bot. 2016, 68, erw363. [Google Scholar] [CrossRef]
- Oliver, M.J.; Farrant, J.M.; Hilhorst, H.W.M.; Mundree, S.; Williams, B.; Bewley, J.D. Desiccation Tolerance: Avoiding Cellular Damage During Drying and Rehydration. Annu. Rev. Plant Biol. 2020, 71, 435–460. [Google Scholar] [CrossRef] [PubMed]
- Matilla, A.J. The Orthodox Dry Seeds Are Alive: A Clear Example of Desiccation Tolerance. Plants 2021, 11, 20. [Google Scholar] [CrossRef]
- Bizouerne, E.; Buitink, J.; Vu, B.L.; Vu, J.L.; Esteban, E.; Pasha, A.; Provart, N.; Verdier, J.; Leprince, O. Gene co-expression analysis of tomato seed maturation reveals tissue-specific regulatory networks and hubs associated with the acquisition of desiccation tolerance and seed vigour. BMC Plant Biol. 2021, 21, 124. [Google Scholar] [CrossRef]
- Jia, H.; Suzuki, M.; McCarty, D.R. Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Wiley Interdiscip. Rev. Dev. Biol. 2014, 3, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Devic, M.; Roscoe, T.J.; Bouyer, D.; Zhou, D.-X.; Boulard, C.; Baud, S.; Dubreucq, B. Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod. 2018, 31, 291–307. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Custard, R.D.; Brown, R.C.; Lemmon, B.E.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. DNA methylation is critical for Arabidopsis embroyogenesis and seed viability. Plant Cell 2006, 18, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Wang, H.H.Y.; McCarty, D.R. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 2007, 143, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; McCarty, D.R. Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 2008, 11, 548–553. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Nery, J.R.; Castanon, R.; Ecker, J.R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017, 18, 171. [Google Scholar] [CrossRef]
- Nonogaki, H.; Bassel, G.W.; Bewley, J.D. Germination—Still a mystery. Plant Sci. 2010, 179, 574–581. [Google Scholar] [CrossRef]
- Weitbrecht, K.; Müller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef] [PubMed]
- Leprince, O.; Harren, F.J.M.; Buitink, J.; Alberda, M.; Hoekstra, F.A. Metabolic Dysfunction and Unabated Respiration Precede the Loss of Membrane Integrity during Dehydration of Germinating Radicles. Plant Physiol. 2000, 122, 597–608. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buitink, J.; Ly Vu, B.; Satour, P.; Leprince, O. The re-establishment of desiccation tolerance in germinated radicles of Medicago truncatula Gaertn. seeds. Seed Sci. Res. 2003, 13, 273–286. [Google Scholar] [CrossRef]
- Rocha Faria, J.M.; van Lammeren, A.A.M.; Hilhorst, H.W.M. Desiccation sensitivity and cell cycle aspects in seeds of Inga vera subsp. affinis. Seed Sci. Res. 2004, 14, 165–178. [Google Scholar] [CrossRef][Green Version]
- Maia, J.; Dekkers, B.J.W.; Provart, N.J.; Ligterink, W.; Hilhorst, H.W.M. The re-establishment of desiccation tolerance in germinated Arabidopsis thaliana seeds and its associated transcriptome. PLoS ONE 2011, 6, e29123. [Google Scholar] [CrossRef] [PubMed]
- Smolikova, G.N. Loss of tolerance to desiccation during germination of Brasicca oleraceae L. seeds with different content of residual chlorophylls. Trans. Karelian Res. Cent. Russ. Acad. Sci. 2011, 3, 105–111. [Google Scholar]
- Dekkers, B.J.W.; Costa, M.C.D.; Maia, J.; Bentsink, L.; Ligterink, W.; Hilhorst, H.W.M. Acquisition and loss of desiccation tolerance in seeds: From experimental model to biological relevance. Planta 2015, 241, 563–577. [Google Scholar] [CrossRef]
- Costa, M.C.D. Desiccation Toletance in Seeds and Plants. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2016. [Google Scholar]
- Faria, J.M.R.; Buitink, J.; van Lammeren, A.A.M.; Hilhorst, H.W.M. Changes in DNA and microtubules during loss and re-establishment of desiccation tolerance in germinating Medicago truncatula seeds. J. Exp. Bot. 2005, 56, 2119–2130. [Google Scholar] [CrossRef]
- Śliwińska, E. Cell cycle and germination of fresh, dried and deteriorated sugarbeet seeds as indicators of optimal harvest time. Seed Sci. Res. 2003, 13, 131–138. [Google Scholar] [CrossRef]
- Warpeha, K.M.; Montgomery, B.L. Light and hormone interactions in the seed-to-seedling transition. Environ. Exp. Bot. 2016, 121, 56–65. [Google Scholar] [CrossRef]
- Boccaccini, A.; Lorrai, R.; Ruta, V.; Frey, A.; Mercey-Boutet, S.; Marion-Poll, A.; Tarkowská, D.; Strnad, M.; Costantino, P.; Vittorioso, P. The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels. BMC Plant Biol. 2016, 16, 198. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Tuan, P.A.; Izydorczyk, M.S.; Ayele, B.T. Ethylene regulates post-germination seedling growth in wheat through spatial and temporal modulation of ABA/GA balance. J. Exp. Bot. 2020, 71, 1985–2004. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Chen, Z. The Control of Seed Dormancy and Germination by Temperature, Light and Nitrate. Bot. Rev. 2020, 86, 39–75. [Google Scholar] [CrossRef]
- Marczak, M.; Cieśla, A.; Janicki, M.; Kasprowicz-Maluśki, A.; Kubiak, P.; Ludwików, A. Protein Phosphatases Type 2C Group A Interact with and Regulate the Stability of ACC Synthase 7 in Arabidopsis. Cells 2020, 9, 978. [Google Scholar] [CrossRef]
- Liu, S.; Yang, L.; Li, J.; Tang, W.; Li, J.; Lin, R. FHY3 interacts with phytochrome B and regulates seed dormancy and germination. Plant Physiol. 2021, 187, 289–302. [Google Scholar] [CrossRef]
- Ruta, V.; Longo, C.; Boccaccini, A.; Madia, V.N.; Saccoliti, F.; Tudino, V.; Di Santo, R.; Lorrai, R.; Dello Ioio, R.; Sabatini, S.; et al. Inhibition of Polycomb Repressive Complex 2 activity reduces trimethylation of H3K27 and affects development in Arabidopsis seedlings. BMC Plant Biol. 2019, 19, 429. [Google Scholar] [CrossRef]
- Nonogaki, H. A repressor complex silencing ABA signaling in seeds? J. Exp. Bot. 2020, 71, 2847–2853. [Google Scholar] [CrossRef]
- Narsai, R.; Gouil, Q.; Secco, D.; Srivastava, A.; Karpievitch, Y.V.; Liew, L.C.; Lister, R.; Lewsey, M.G.; Whelan, J. Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol. 2017, 18, 172. [Google Scholar] [CrossRef]
- Smolikova, G.; Strygina, K.; Krylova, E.; Leonova, T.; Frolov, A.; Khlestkina, E.; Medvedev, S. Transition from Seeds to Seedlings: Hormonal and Epigenetic Aspects. Plants 2021, 10, 1884. [Google Scholar] [CrossRef]
- Luján-Soto, E.; Dinkova, T.D. Time to wake up: Epigenetic and small-RNA-mediated regulation during seed germination. Plants 2021, 10, 236. [Google Scholar] [CrossRef]
- Molitor, A.M.; Bu, Z.; Yu, Y.; Shen, W.-H. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 2014, 10, e1004091. [Google Scholar] [CrossRef]
- Mozgova, I.; Köhler, C.; Hennig, L. Keeping the gate closed: Functions of the polycomb repressive complex PRC2 in development. Plant J. 2015, 83, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Makarevich, G.; Leroy, O.; Akinci, U.; Schubert, D.; Clarenz, O.; Goodrich, J.; Grossniklaus, U.; Köhler, C. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006, 7, 947–952. [Google Scholar] [CrossRef] [PubMed]
- Bratzel, F.; López-Torrejón, G.; Koch, M.; Del Pozo, J.C.; Calonje, M. Keeping cell identity in arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 2010, 20, 1853–1859. [Google Scholar] [CrossRef] [PubMed]
- Bouyer, D.; Roudier, F.; Heese, M.; Andersen, E.D.; Gey, D.; Nowack, M.K.; Goodrich, J.; Renou, J.-P.; Grini, P.E.; Colot, V.; et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 2011, 7, e1002014. [Google Scholar] [CrossRef] [PubMed]
- Kreplak, J.; Madoui, M.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed germination and dormancy: The classic story, new puzzles, and evolution. J. Integr. Plant Biol. 2019, 61, 541–563. [Google Scholar] [CrossRef]
- Silva, A.T.; Ligterink, W.; Hilhorst, H.W.M. Metabolite profiling and associated gene expression reveal two metabolic shifts during the seed-to-seedling transition in Arabidopsis thaliana. Plant Mol. Biol. 2017, 95, 481–496. [Google Scholar] [CrossRef]
- Silva, A.T.; Ribone, P.A.; Chan, R.L.; Ligterink, W.; Hilhorst, H.W.M. A predictive coexpression network identifies novel genes controlling the seed-to-seedling phase transition in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2218–2231. [Google Scholar] [CrossRef]
- Bailly, C. The signalling role of ROS in the regulation of seed germination and dormancy. Biochem. J. 2019, 476, 3019–3032. [Google Scholar] [CrossRef]
- Matilla, A.J. Exploring Breakthroughs in Three Traits Belonging to Seed Life. Plants 2022, 11, 490. [Google Scholar] [CrossRef] [PubMed]
- Cairns, N.G.; Pasternak, M.; Wachter, A.; Cobbett, C.S.; Meyer, A.J. Maturation of arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 2006, 141, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Li, Y.; Fang, T.; Shi, X.; Chen, X. Specific roles of tocopherols and tocotrienols in seed longevity and germination tolerance to abiotic stress in transgenic rice. Plant Sci. 2016, 244, 31–39. [Google Scholar] [CrossRef]
- Smolikova, G.N.; Medvedev, S.S. Seed carotenoids:synthesis, diversity, and functions. Russ. J. Plant Physiol. 2015, 62, 1–13. [Google Scholar] [CrossRef]
- Hoekstra, F.A.; Golovina, E.A.; Buitink, J. Mechanisms of plant desiccation tolerance. Trends Plant Sci. 2001, 6, 431–438. [Google Scholar] [CrossRef]
- Buitink, J.; Leprince, O. Intracellular glasses and seed survival in the dry state. C. R. Biol. 2008, 331, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Walters, C. Orthodoxy, recalcitrance and in-between: Describing variation in seed storage characteristics using threshold responses to water loss. Planta 2015, 242, 397–406. [Google Scholar] [CrossRef]
- Sano, N.; Rajjou, L.; North, H.M.; Debeaujon, I.; Marion-Poll, A.; Seo, M. Staying alive: Molecular aspects of seed longevity. Plant Cell Physiol. 2016, 57, 660–674. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of an Arabidopsis Transcription Factor, DREB2A, Involved in Drought-Responsive Gene Expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
- Rathor, P.; Borza, T.; Stone, S.; Tonon, T.; Yurgel, S.; Potin, P.; Prithiviraj, B. A Novel Protein from Ectocarpus sp. Improves Salinity and High Temperature Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 1971. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Guo, W.; Feng, W.; Liu, L.; Song, X.; Chen, J.; Hou, W.; Zhu, H.; Tang, S.; Hu, J. LTP3 contributes to disease susceptibility in A rabidopsis by enhancing abscisic acid (ABA) biosynthesis. Mol. Plant Pathol. 2016, 17, 412–426. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P.; et al. Acyl-Lipid Metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; De Coninck, B.M.A.; Cammue, B.P.A.; De Bolle, M.F.C. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- Hu, J.; Barlet, X.; Deslandes, L.; Hirsch, J.; Feng, D.X.; Somssich, I.; Marco, Y. Transcriptional Responses of Arabidopsis thaliana during Wilt Disease Caused by the Soil-Borne Phytopathogenic Bacterium, Ralstonia solanacearum. PLoS ONE 2008, 3, e2589. [Google Scholar] [CrossRef]
- Shen, Q.; Uknes, S.J.; Ho, T.H. Hormone response complex in a novel abscisic acid and cycloheximide-inducible barley gene. J. Biol. Chem. 1993, 268, 23652–23660. [Google Scholar] [CrossRef]
- Chen, C.N.; Chu, C.C.; Zentella, R.; Pan, S.M.; Ho, T.H.D. AtHVA22 gene family in Arabidopsis: Phylogenetic relationship, ABA and stress regulation, and tissue-specific expression. Plant Mol. Biol. 2002, 49, 633–644. [Google Scholar] [CrossRef]
- Guo, W.-J.; David Ho, T.-H. An Abscisic Acid-Induced Protein, HVA22, Inhibits Gibberellin-Mediated Programmed Cell Death in Cereal Aleurone Cells. Plant Physiol. 2008, 147, 1710–1722. [Google Scholar] [CrossRef]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Abdul Aziz, M.; Sabeem, M.; Mullath, S.K.; Brini, F.; Masmoudi, K. Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses. Biomolecules 2021, 11, 1662. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Cornilescu, C.C.; Tyler, R.C.; Cornilescu, G.; Tonelli, M.; Lee, M.S.; Markley, J.L. Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein. Protein Sci. 2005, 14, 2601–2609. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Qi, S.; Li, H.; Liu, P.; Li, P.; Wu, C.; Zheng, C.; Huang, J. Overexpression of Late Embryogenesis Abundant 14 enhances Arabidopsis salt stress tolerance. Biochem. Biophys. Res. Commun. 2014, 454, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Dunaeva, M.; Adamska, I. Identification of genes expressed in response to light stress in leaves of Arabidopsis thaliana using RNA differential display. Eur. J. Biochem. 2001, 268, 5521–5529. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, T.; Itai, R.; Maeda, M.; Kitashiba, H.; Isuzugawa, K.; Kato, K.; Kanayama, Y. Characterization of PcLEA14, a Group 5 Late Embryogenesis Abundant Protein Gene from Pear (Pyrus communis). Plants 2020, 9, 1138. [Google Scholar] [CrossRef] [PubMed]
- Hattori, J.; Boutilier, K.A.; Campagne, M.M.L.; Miki, B.L. A conserved BURP domain defines a novel group of plant proteins with unusual primary structures. Mol. Gen. Genet. MGG 1998, 259, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Harshavardhan, V.T.; Van Son, L.; Seiler, C.; Junker, A.; Weigelt-Fischer, K.; Klukas, C.; Altmann, T.; Sreenivasulu, N.; Bäumlein, H.; Kuhlmann, M. AtRD22 and AtUSPL1, Members of the Plant-Specific BURP Domain Family Involved in Arabidopsis thaliana Drought Tolerance. PLoS ONE 2014, 9, e110065. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol. Gen. Genet. MGG 1993, 238–238, 17–25. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Lindemose, S.; O’Shea, C.; Jensen, M.; Skriver, K. Structure, Function and Networks of Transcription Factors Involved in Abiotic Stress Responses. Int. J. Mol. Sci. 2013, 14, 5842–5878. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, L.; Fu, Y.; Cheung, M.-Y.; Wong, F.-L.; Phang, T.-H.; Sun, Z.; Lam, H.-M. Expression of an apoplast-localized BURP-domain protein from soybean (GmRD22) enhances tolerance towards abiotic stress. Plant. Cell Environ. 2012, 35, 1932–1947. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.-J. Plant Peroxiredoxins. Annu. Rev. Plant Biol. 2003, 54, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.J. Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal. 2011, 15, 1129–1159. [Google Scholar] [CrossRef]
- Chen, H.; Chu, P.; Zhou, Y.; Ding, Y.; Li, Y.; Liu, J.; Jiang, L.; Huang, S. Ectopic expression of NnPER1, a Nelumbo nucifera 1-cysteine peroxiredoxin antioxidant, enhances seed longevity and stress tolerance in Arabidopsis. Plant J. 2016, 88, 608–619. [Google Scholar] [CrossRef]
- Stacy, R.A.P.; Munthe, E.; Steinum, T.; Sharma, B.; Aalen, R.B. A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol. Biol. 1996, 31, 1205–1216. [Google Scholar] [CrossRef]
- Haslekås, C.; Viken, M.K.; Grini, P.E.; Nygaard, V.; Nordgard, S.H.; Meza, T.J.; Aalen, R.B. Seed 1-Cysteine Peroxiredoxin Antioxidants Are Not Involved in Dormancy, But Contribute to Inhibition of Germination during Stress. Plant Physiol. 2003, 133, 1148–1157. [Google Scholar] [CrossRef]
- Kim, S.Y.; Paeng, S.K.; Nawkar, G.M.; Maibam, P.; Lee, E.S.; Kim, K.-S.; Lee, D.H.; Park, D.-J.; Kang, S.B.; Kim, M.R.; et al. The 1-Cys peroxiredoxin, a regulator of seed dormancy, functions as a molecular chaperone under oxidative stress conditions. Plant Sci. 2011, 181, 119–124. [Google Scholar] [CrossRef]
- Chen, H.; Ruan, J.; Chu, P.; Fu, W.; Liang, Z.; Li, Y.; Tong, J.; Xiao, L.; Liu, J.; Li, C.; et al. AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant J. 2020, 101, 310–323. [Google Scholar] [CrossRef]
- Lalanne, D.; Malabarba, J.; Ly Vu, J.; Hundertmark, M.; Delahaie, J.; Leprince, O.; Buitink, J.; Verdier, J. Medicago ABI3 Splicing Isoforms Regulate the Expression of Different Gene Clusters to Orchestrate Seed Maturation. Plants 2021, 10, 1710. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular Basis of the Core Regulatory Network in ABA Responses: Sensing, Signaling and Transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef] [PubMed]
- Söderman, E.M.; Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and Function of the Arabidopsis ABA-insensitive4 Gene in Seed and Abscisic Acid Response Signaling Networks. Plant Physiol. 2000, 124, 1752–1765. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.-H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Reeves, W.M.; Lynch, T.J.; Mobin, R.; Finkelstein, R.R. Direct targets of the transcription factors ABA-Insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol. Biol. 2011, 75, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 Regulates Primary Seed Dormancy by Regulating the Biogenesis of Abscisic Acid and Gibberellins in Arabidopsis. PLoS Genet. 2013, 9, e1003577. [Google Scholar] [CrossRef]
- Feng, C.; Chen, Y.; Wang, C.; Kong, Y.; Wu, W.; Chen, Y. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and earl. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef]
- Li, X.; Zhong, M.; Qu, L.; Yang, J.; Liu, X.; Zhao, Q.; Liu, X.; Zhao, X. AtMYB32 regulates the ABA response by targeting ABI3, ABI4 and ABI5 and the drought response by targeting CBF4 in Arabidopsis. Plant Sci. 2021, 310, 110983. [Google Scholar] [CrossRef]
- Wind, J.J.; Peviani, A.; Snel, B.; Hanson, J.; Smeekens, S.C. ABI4: Versatile activator and repressor. Trends Plant Sci. 2013, 18, 125–132. [Google Scholar] [CrossRef]
- Dekkers, B.J.W.; He, H.; Hanson, J.; Willems, L.A.J.; Jamar, D.C.L.; Cueff, G.; Rajjou, L.; Hilhorst, H.W.M.; Bentsink, L. The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J. 2016, 85, 451–465. [Google Scholar] [CrossRef]
- Zinsmeister, J.; Lalanne, D.; Terrasson, E.; Chatelain, E.; Vandecasteele, C.; Vu, B.L.; Dubois-Laurent, C.; Geoffriau, E.; Le Signor, C.; Dalmais, M.; et al. ABI5 is a regulator of seed maturation and longevity in legumes. Plant Cell 2016, 28, 2735–2754. [Google Scholar] [CrossRef] [PubMed]
- Smolikova, G.; Dolgikh, E.; Vikhnina, M.; Frolov, A.; Medvedev, S. Genetic and hormonal regulation of chlorophyll degradation during maturation of seeds with green embryos. Int. J. Mol. Sci. 2017, 18, 1993. [Google Scholar] [CrossRef] [PubMed]
- Smolikova, G.; Shiroglazova, O.; Vinogradova, G.; Leppyanen, I.; Dinastiya, E.; Yakovleva, O.; Dolgikh, E.; Titova, G.; Frolov, A.; Medvedev, S. Comparative analysis of the plastid conversion, photochemical activity and chlorophyll degradation in developing embryos of green-seeded and yellow-seeded pea (Pisum sativum) cultivars. Funct. Plant Biol. 2020, 47, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Han, X.; Yang, M.; Zhang, M.; Pan, J.; Yu, D. The Transcription Factor INDUCER OF CBF EXPRESSION1 Interacts with ABSCISIC ACID INSENSITIVE5 and DELLA Proteins to Fine-Tune Abscisic Acid Signaling during Seed Germination in Arabidopsis. Plant Cell 2019, 31, 1520–1538. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.-H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef]
- Chandrasekaran, U.; Luo, X.; Zhou, W.; Shu, K. Multifaceted Signaling Networks Mediated by Abscisic Acid Insensitive 4. Plant Commun. 2020, 1, 100040. [Google Scholar] [CrossRef]
- Chen, C.; Twito, S.; Miller, G. New cross talk between ROS, ABA and auxin controlling seed maturation and germination unraveled in APX6 deficient Arabidopsis seeds. Plant Signal. Behav. 2014, 9, e976489. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, X.; Gong, Z.; Yang, S.; Shi, Y. ABI4 represses the expression of type-A ARRs to inhibit seed germination in Arabidopsis. Plant J. 2017, 89, 354–365. [Google Scholar] [CrossRef]
- Oracz, K.; Bouteau, H.E.M.; Farrant, J.M.; Cooper, K.; Belghazi, M.; Job, C.; Job, D.; Corbineau, F.; Bailly, C.; El-Maarouf Bouteau, H.; et al. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 2007, 50, 452–465. [Google Scholar] [CrossRef]
- Beyer, W.F.; Fridovich, I. Assaying for superoxide dismutase activity: Some large consequences of minor changes in conditions. Anal. Biochem. 1987, 161, 559–566. [Google Scholar] [CrossRef]
- Chantseva, V.; Bilova, T.; Smolikova, G.; Frolov, A.; Medvedev, S. 3D-clinorotation induces specific alterations in metabolite profiles of germinating Brassica napus L. seeds. Biol. Commun. 2019, 64, 55–74. [Google Scholar] [CrossRef]
- Gay, C.; Collins, J.; Gebicki, J.M. Hydroperoxide assay with the ferric–xylenol orange complex. Anal. Biochem. 1999, 273, 149–155. [Google Scholar] [CrossRef]
- Paudel, G.; Bilova, T.; Schmidt, R.; Greifenhagen, U.; Berger, R.; Tarakhovskaya, E.; Stöckhardt, S.; Balcke, G.U.; Humbeck, K.; Brandt, W.; et al. Osmotic stress is accompanied by protein glycation in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6283–6295. [Google Scholar] [CrossRef]
- Don, R. ISTA Handbook on Seedling Evaluation, 3rd ed.; The International Seed Testing Association: Basserdorf, Switzerland, 2003; ISBN 3-906549-39-9. [Google Scholar]
- Andrews, S. FASTQC: A quality control tool for high throughput sequence data. In Transcriptomics in Entomological Research; Babraham Institute: Cambridge, UK, 2015. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. R Package Version 1.10.0. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 19 June 2021).
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Die, J.V.; Román, B.; Nadal, S.; González-Verdejo, C.I. Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 2010, 232, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smolikova, G.; Strygina, K.; Krylova, E.; Vikhorev, A.; Bilova, T.; Frolov, A.; Khlestkina, E.; Medvedev, S. Seed-to-Seedling Transition in Pisum sativum L.: A Transcriptomic Approach. Plants 2022, 11, 1686. https://doi.org/10.3390/plants11131686
Smolikova G, Strygina K, Krylova E, Vikhorev A, Bilova T, Frolov A, Khlestkina E, Medvedev S. Seed-to-Seedling Transition in Pisum sativum L.: A Transcriptomic Approach. Plants. 2022; 11(13):1686. https://doi.org/10.3390/plants11131686
Chicago/Turabian StyleSmolikova, Galina, Ksenia Strygina, Ekaterina Krylova, Aleksander Vikhorev, Tatiana Bilova, Andrej Frolov, Elena Khlestkina, and Sergei Medvedev. 2022. "Seed-to-Seedling Transition in Pisum sativum L.: A Transcriptomic Approach" Plants 11, no. 13: 1686. https://doi.org/10.3390/plants11131686
APA StyleSmolikova, G., Strygina, K., Krylova, E., Vikhorev, A., Bilova, T., Frolov, A., Khlestkina, E., & Medvedev, S. (2022). Seed-to-Seedling Transition in Pisum sativum L.: A Transcriptomic Approach. Plants, 11(13), 1686. https://doi.org/10.3390/plants11131686









