From Soil Amendments to Controlling Autophagy: Supporting Plant Metabolism under Conditions of Water Shortage and Salinity
Abstract
:1. Introduction
2. Plant Performance in the Field
| Type of Stress | Application/Duration | Example | Reference |
|---|---|---|---|
| Eustress | whole plant priming | taking advantage of cross tolerance to different types of stress | Villagómez-Arande et al., 2022 [34] |
| Distress | fluctuating environmental conditions | day–night cycles; rain–sunshine cycles | Lichtenthaler, 1996 [15] |
| short time, hours to 4 days | laboratory experiments to identify stress-responsive traits (genes) | Miller et al., 2015 [35] | |
| series of adverse environmental conditions (weeks to months) | drought periods | Fahad et al., 2017 [36] | |
| poor growth conditions | soil salinity lack of nutrients deserts | Zhao et al., 2020 [37] | |
| transfer to a different environment | cultivation of plants not native to an area | Geppert et al., 2021 [38] |
3. Photosynthetic Performance under Stress
4. Improving Soil Quality by the Addition of Biochar
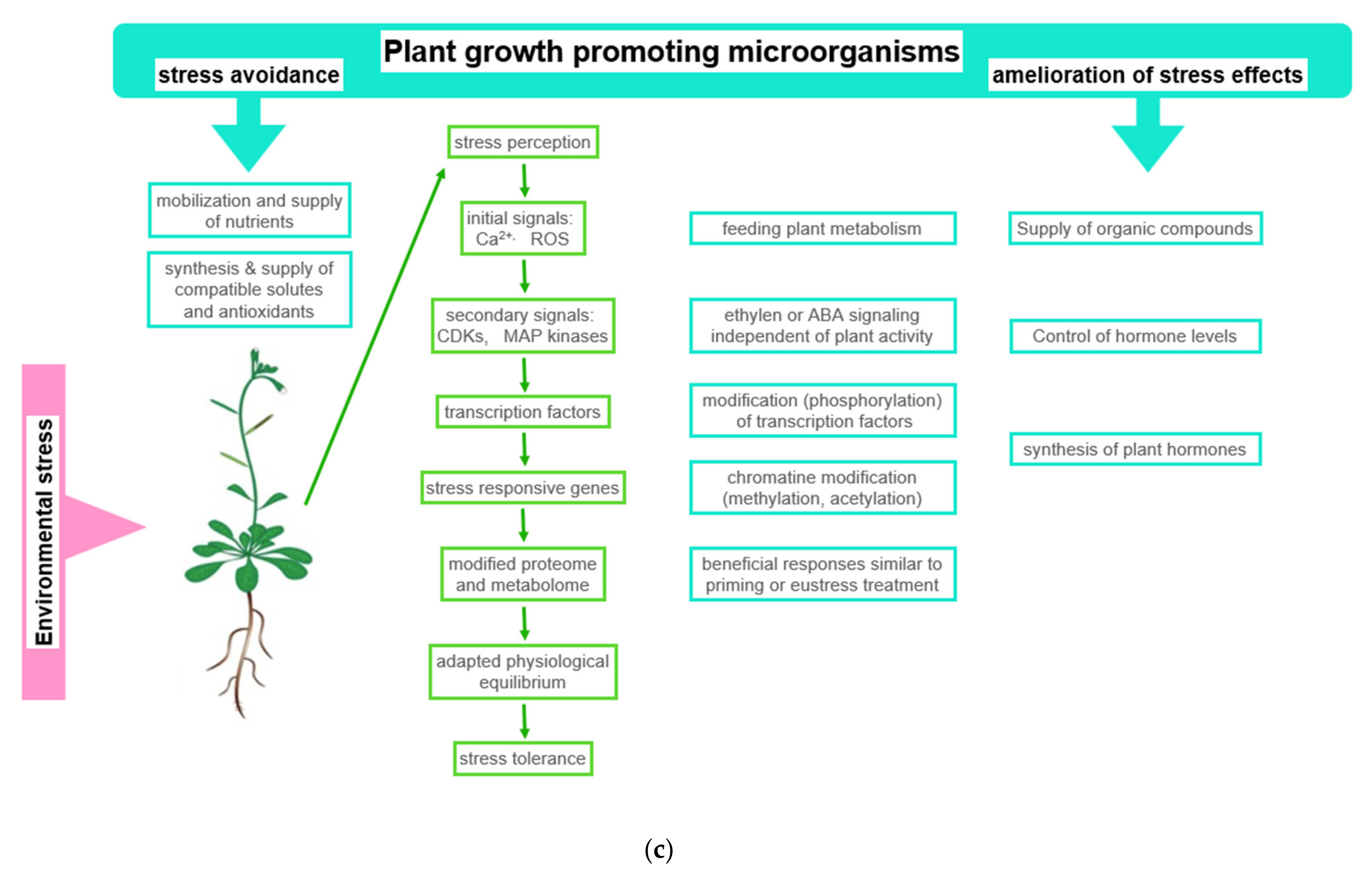
5. Interaction of Plants and Microorganisms
5.1. General Observations
5.2. Acquisition of Symbionts
5.3. Two Examples of Improved Nutrient Supply Provided by Plant Growth Promoting Microorganisms
5.3.1. Mobilization of Plant-Inaccessible Phosphate
5.3.2. Support of Nitrogen Uptake and Fixation
6. Response of Plants to Environmental Stress in Dependence of the External Application of Plant Stress Reducing and Growth Promoting Compounds
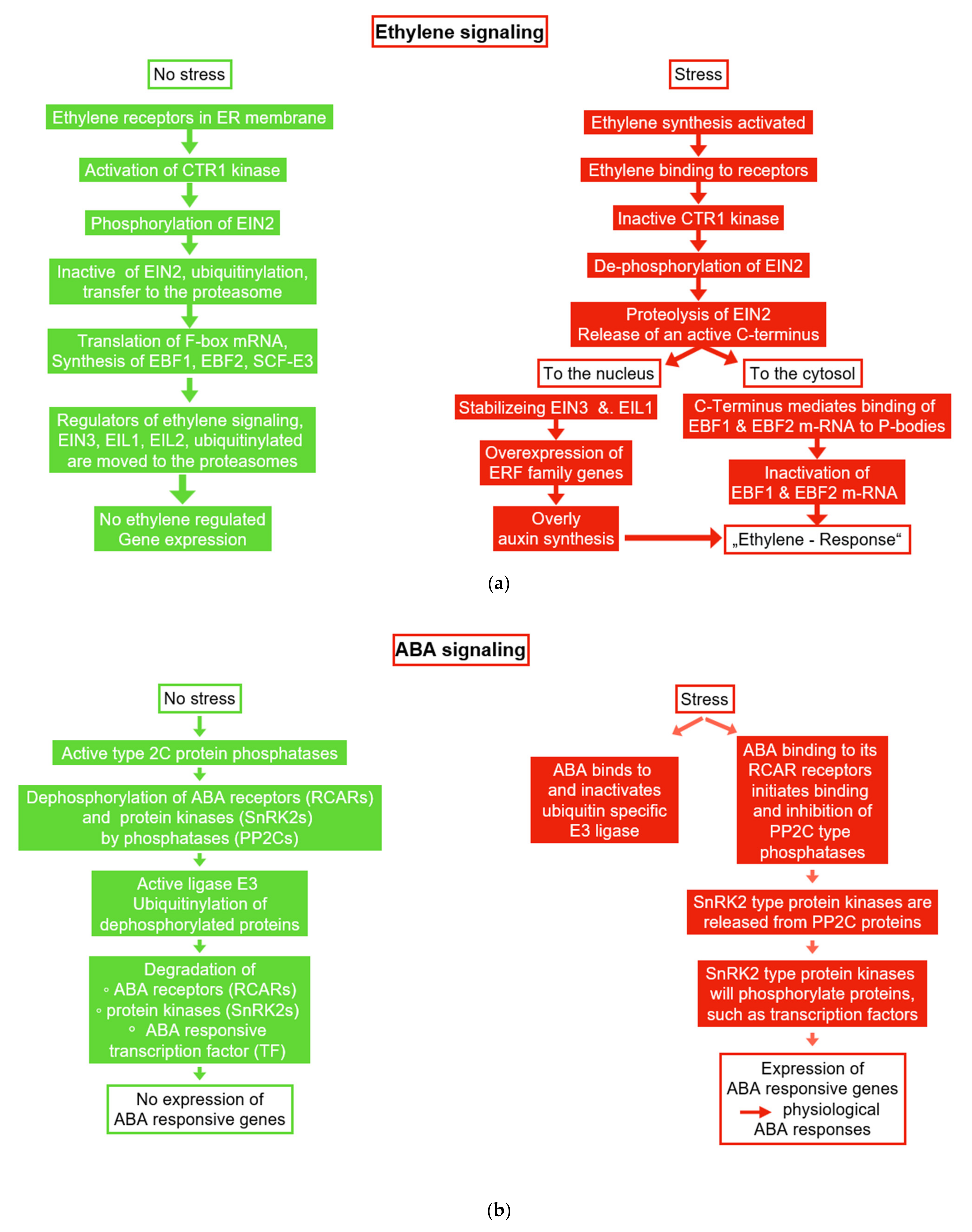
6.1. Ethylene and ACC
6.2. Abscisic Acid (ABA)
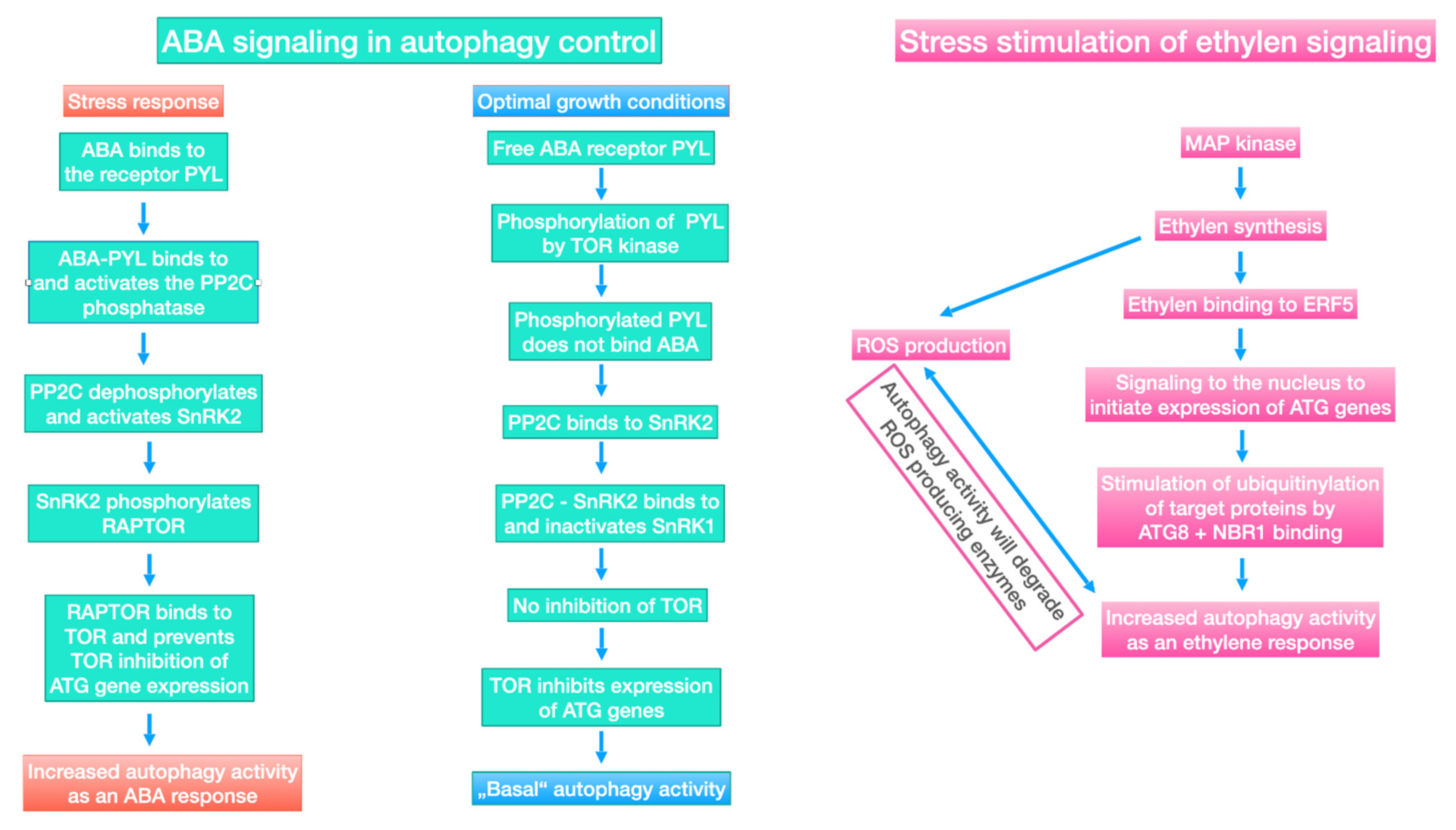
6.3. Regulation of Autophagy by Endophyte and Plant-Induced Signaling under Environmental Stress
6.4. Control of Autophagy by ABA and Ethylene Signaling
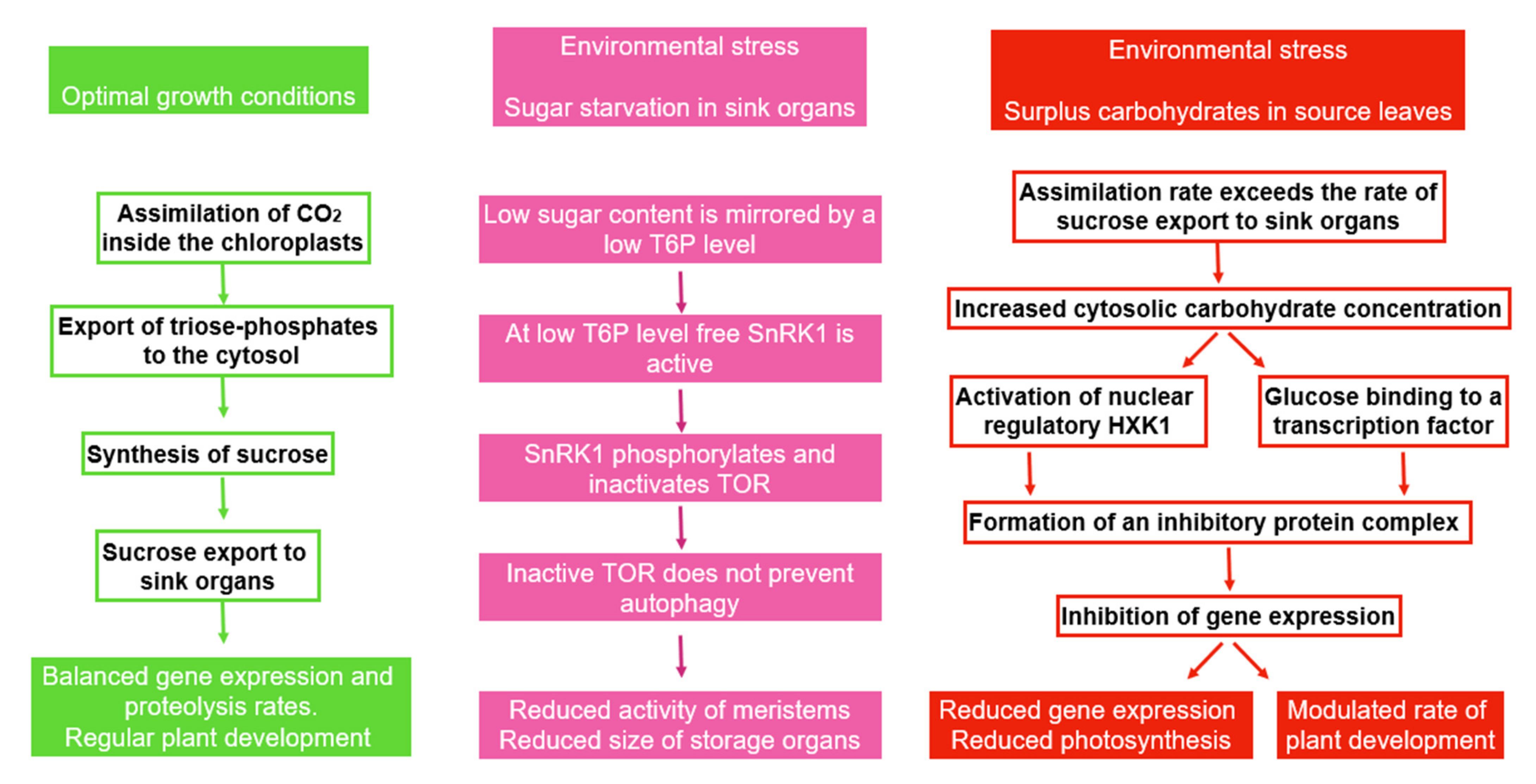
6.5. Autophagy in Source and Sink Tissues
7. Summary
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Muscat, A.; de Olde, E.M.; de Boer, I.J.M.; Ripoll-Bosch, R. The battle for biomass: A systematic review of food-feed-fuel competition. Glob. Food Secur. 2020, 25, 100330. [Google Scholar] [CrossRef]
- Shahbaz, M.; Ashraf, M. Improving Salinity Tolerance in Cereals. Crit. Rev. Plant Sci. 2013, 32, 237–249. [Google Scholar] [CrossRef]
- Daryanto, S.; Wang, L.; Jacinthe, P.A. Global synthesis of drought effects on maize and wheat production. PLoS ONE 2016, 11, e0156362. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, P.; Singh, P.K.; Chakraborty, D.; Mishra, S.; Pattnaik, R. Insight into the Role of PGPR in Sustainable Agriculture and Environment. Front. Sustain. Food Syst. 2021, 5, 667150. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2014, 22, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Jägermeyr, J.; Frieler, K. Spatial variations in crop growing seasons pivotal to reproduce global fluctuations in maize and wheat yields. Sci. Adv. 2018, 4, eaat4517. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Schillaci, M.; Walker, R.; Smith, P.M.C.; Watt, M.; Roessner, U. Alleviation of salinity stress in plants by endophytic plant-fungal symbiosis: Current knowledge, perspectives and future directions. Plant Soil 2021, 461, 219–244. [Google Scholar] [CrossRef]
- Tilman, D.; Cassman, K.G.; Matson, P.A.; Naylor, R.; Polasky, S. Agricultural sustainability and intensive production practices. Nature 2002, 418, 671–677. [Google Scholar] [CrossRef]
- Verma, J.P.; Jaiswal, D.K.; Meena, V.S.; Kumar, A.; Meena, R.S. Issues and challenges about sustainable agriculture production for management of natural resources to sustain soil fertility and health. J. Clean. Prod. 2015, 107, 793–794. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [Green Version]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Lichtenthaler, H. The stress concept in plants: An introduction. Ann. N. Y. Acad. Sci. 2006, 851, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Anumalla, M.; Roychowdhury, R.; Geda, C.K.; Bharathkumar, S.; Goutam, K.D.; Dev, T.S.S.M. Mechanism of stress signal transduction and involvement of stress inducible transcription factors and genes in response to abiotic stresses in plants. Int. J. Recent Sci. Res. 2016, 7, 12754–12771. [Google Scholar]
- Agegnehu, G.; Amede, T. Integrated soil fertility and plant nutrient management in tropical agro-ecosystems: A review. Pedosphere 2017, 27, 662–680. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Vegetation stress: An introduction to the stress concept in plants. J. Plant Physiol. 1996, 148, 4–14. [Google Scholar] [CrossRef]
- Fischer, K.S.; Fukai, S.; Kumar, A.; Leung, H.; Jongdee, B. Field phenotyping strategies and breeding for adaptation of rice to drought. Front. Physiol. 2012, 3, 282. [Google Scholar] [CrossRef] [Green Version]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef]
- Senapati, N.; Semenov, M.A. Large genetic yield potential and genetic yield gap estimated for wheat in europe. Glob. Food Secur. 2020, 24, 100340. [Google Scholar] [CrossRef]
- Lobell, D.B.; Roberts, M.J.; Schlenker, W.; Braun, N.; Little, B.B.; Rejesus, R.M.; Hammer, G.L. Greater sensitivity to drought accompanies maize yield increase in the u.s. midwest. Science 2014, 344, 516–519. [Google Scholar] [CrossRef]
- Tollenaar, M.; Lee, E.A. Yield potential, yield stability and stress tolerance in maize. Field Crops Res. 2002, 75, 161–169. [Google Scholar] [CrossRef]
- Witcombe, J.R.; Hollington, P.A.; Howarth, C.J.; Reader, S.; Steele, K.A. Breeding for abiotic stresses for sustainable agriculture. Philos. Trans. R. Soc. B 2008, 363, 703–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Li, Y.; Zhu, J.-K. Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 2018, 4, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Godoy, F.; Olivos-Hernández, K.; Stange, C.; Handford, M. Abiotic stress in crop species: Improving tolerance by applying plant metabolites. Plants 2021, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Sallam, A.; Sehgal, D.; Sukumaran, S.; Farhad, M.d.; Krishnan, J.N.; Kumar, U.; Biswal, A. Advances in breeding for abiotic stress tolerance in wheat. In Genomic Designing for Abiotic Stress Resistant Cereal Crops; Kole, C., Ed.; Springer: Cham, Switzerland, 2021; Volume 2, pp. 71–103. [Google Scholar] [CrossRef]
- Lebaudy, A.; Vavasseur, A.; Hosy, E.; Dreyer, I.; Leonhardt, N.; Thibaud, J.-B.; Very, A.-A.; Simonneau, T.; Sentenac, H. Plant adaptation to fluctuating environment and biomass production are strongly dependent on guard cell potassiu channels. Proc. Natl. Acad. Sci. USA 2008, 105, 5271–5276. [Google Scholar] [CrossRef] [Green Version]
- Larcher, W. Physiological Plant Ecology: Ecophysiology and Stress Physiology of Functional Groups; Springer: Berlin/Heidelberg, Germany, 2003; ISBN 978-3-540-43516-7. [Google Scholar]
- Huang, J.; Rozelle, S. Environmental stress and grain yields in china. Am. J. Agric. Econ. 1995, 77, 853–864. [Google Scholar] [CrossRef]
- Yadav, S.; Modi, P.; Dave, A.; Vijapura, A.; Patel, D.; Patel, M. Effect of abiotic stress on crops. In Sustainable Crop Production; Hasanuzzaman, M., Teixeira Filho, M.C.M., Masayuki Fujita, M., Rodrigues Nogueira, T.A., Eds.; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Ashraf, M.; Wu, L. Breeding for salinity tolerance in plants. Crit. Rev. Plant Sci. 1994, 13, 17–42. [Google Scholar] [CrossRef]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Guerrier, G. Fluxes of Na+. K+ and Cl− and osmotic adjustment in Lycopersicon pimpinellifolium and L. esculentum during short- and long-term exposures to NaCl. Physiol. Plant. 1996, 97, 583–591. [Google Scholar] [CrossRef]
- Yousfi, N.; Slama, I.; Ghnaya, T.; Savoure, A.; Abdelly, C. Effects of water deficit stress on growth, water relations and osmolyte accumulation in Medicago truncatula and M. laciniata populations. Comptes Rendus Biol. 2010, 333, 205–213. [Google Scholar] [CrossRef]
- Slama, S.; Abdelly, C.; Bouchereau, A.; Flowers, T.; Savouré, A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 2015, 115, 433–447. [Google Scholar] [CrossRef] [Green Version]
- Villagómez-Aranda, A.L.; Feregrino-Pérez, A.A.; García-Ortega, L.F.; González-Chavira, M.M.; Torres-Pacheco, I.; Guevara-González, R.G. Activating stress memory: Eustressors as potential tools for plant breeding. Plant Cell Rep. 2022. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Qingxin Song, Q.; Shi, X.; Juenger, T.E.; Chen, Z.J. Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat. Commun. 2015, 6, 7453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.-K.; Shabala, S. Mechanisms of plant responses and adaptation to soil alinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef]
- Geppert, C.; Boscutti, F.; La Bella, G.; De Marchi, V.; Corcos, D.; Filippi, A.; Marini, L. Contrasting response of native and non-native plants to disturbance and herbivory in mountain environments. Front. Plant Sci. 2021, 8, 1147. [Google Scholar] [CrossRef]
- Smirnoff, N.; Cumbes, Q.J. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 1989, 28, 1057–1060. [Google Scholar] [CrossRef]
- Stocker, O. Das Halophytenproblem. Ergeb. Biol. 1928, 3, 265–353. [Google Scholar] [CrossRef]
- Byrt, C.S.; Munns, R. Living with salinity. New Phytol. 2008, 179, 903–905. [Google Scholar] [CrossRef]
- Huchzermeyer, B.; Flowers, T. Putting halophytes to work—Genetics, biochemistry and physiology. Funct. Plant Biol. 2013, 40, 5–8. [Google Scholar] [CrossRef] [Green Version]
- Cornelissen, J.H.C.; Lavorel, S.; Garnier, E.; Diaz, S.; Buchmann, N.; Gurvich, D.E.; Reich, P.B.; ter Steege, H.; Morgan, H.D.; van der Heijden, M.G.A.; et al. A handbook of protocols for standardized and easy measurements of plant functional traits worldwide. Aust. J. Bot. 2003, 51, 335–380. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Harguindeguy, N.; Díaz, S.; Garnier, E.; Lavorel, S.; Poorter, H.; Jaureguiberry, P.; Bret-Harte, M.S.; Cornwell, W.K.; Craine, J.M.; Gurvich, D.E.; et al. New handbook for standardised measurement of plant functional traits worldwide. Aust. J. Bot. 2013, 61, 167–234. [Google Scholar] [CrossRef]
- Ahmed, M.Z.; Shimazaki, T.; Gulzar, S.; Kikuchi, A.B.; Khan, M.A.; Koyro, H.-W.; Huchzermeyer, B.; Watanabe, K.N. The influence of genes regulating transmembrane transport of Na+ on the salt resistance of Aeluropus lagopoides. Funct. Plant Biol. 2013, 40, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Kosová, K.; Prášil, I.T.; Vítámvás, P. Protein contribution to plant salinity response and tolerance acquisition. Int. J. Mol. Sci. 2013, 14, 6757–6789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyro, H.-W.; Zörb, C.; Debez, A.; Huchzermeyer, B. The effect of hyper-osmotic salinity on protein pattern and enzyme activities of halophytes. Funct. Plant Biol. 2013, 40, 787–804. [Google Scholar] [CrossRef] [PubMed]
- Bartels, D.; Dinakar, C. Balancing salinity stress responses in halophytes and non-halophytes: A comparison between Thellungiella and Arabidopsis thaliana. Funct. Plant Biol. 2013, 40, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Kranner, I.; Seal, C. Salt stress, signalling and redox control in seeds. Funct. Plant Biol. 2013, 40, 848–859. [Google Scholar] [CrossRef] [PubMed]
- Ozgur, R.; Turkan, I.; Uzilday, B.; Sekmen, A.H. Endoplasmic reticulum stress triggers ROS signalling, changes the redox state, and regulates the antioxidant defence of Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 1377–1390. [Google Scholar] [CrossRef]
- Ben Amora, N.; Jiménez, A.; Megdiche, W.; Lundqvist, M.; Sevilla, F.; Abdelly, C. Response of antioxidant systems to NaCl stress in the halophyte Cakile maritima. Physiol. Plant. 2006, 126, 446–457. [Google Scholar] [CrossRef]
- Lämke, J.; Bäuerle, I. Epigenetik and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124. [Google Scholar] [CrossRef]
- Turgut-Kara, N.; Arikan, B.; Celik, H. Epigenetic memory and priming in plants. Genetica 2020, 148, 47–54. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agronomy 2009, 29, 185–212. [Google Scholar] [CrossRef] [Green Version]
- Latef, A.A.H.A.; Kordrostami, M.; Zakir, A.; Zaki, H.; Saleh, O.M. Eustress with H2O2 facilitates plant growth by improving tolerance to salt stress in two wheat cultivars. Plants 2019, 8, 303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amritha, M.S.; Sridharan, K.; Puthur, J.T.; Dhankher, O.P. Priming with nanoscale materials for boosting abiotic stress tolerance in crop plants. J. Agric. Food Chem. 2021, 69, 10017–10035. [Google Scholar]
- Barickman, T.C.; Adhikari, B.; Sehgal, A.; Walne, C.H.; Reddy, K.R.; Gao, W. Drought and elevated carbon dioxide impact the morphophysiological profile of basil (Ocimum basilicum L.). Crops 2021, 1, 118–128. [Google Scholar] [CrossRef]
- Lataef, A.A.A.; Hasanuzzaman, M.; Tahjib-ul-Arif, M. Mitigation of salinity stress by exogenous application of cytokinin in faba bean (Vicia faba L.). Not. Bot. Horti Agrobot. 2021, 49, 12192. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Tie, D.; Quan, J.; Yue, M.; Liu, X. Epigenetic memory and growth responses of the clonal plant Glechoma longituba to parental recurrent UV-B stress. Funct. Plant Biol. 2021, 48, 827–838. [Google Scholar] [CrossRef]
- Zheng, Y.; Xia, Z.; Wu, J.; Ma, H. Effects of repeated drought stress on the physiological characteristics and lipid metabolism of Bombax ceiba L. during subsequent drought and heat stresses. BMC Plant Biol. 2021, 21, 467. [Google Scholar] [CrossRef]
- Roach, T.; Stöggl, W.; Baur, T.; Kranner, I. Distress and eustress of reactive electrophiles and relevance to light stress acclimation via stimulation of thiol/disulphide-based redox defences. Free Radic. Biol. Med. 2018, 122, 65–73. [Google Scholar] [CrossRef]
- Powles, S.B. Photoinhibition of photosynthesis induced by visible light. Annu. Rev. Plant Physiol. 1984, 35, 15–44. [Google Scholar] [CrossRef]
- Björkman, O.; Holmgren, P. Adaptability of the photosynthetic apparatus to light intensity in ecotypes from exposed and shaded habitats. Physiol. Plant. 1963, 16, 889–914. [Google Scholar] [CrossRef]
- Krause, G.H. Photoinhibition of photosynthesis. An evaluation of damaging and protective mechanisms. Physiol. Plant. 1988, 74, 566–574. [Google Scholar] [CrossRef]
- Nishiyama, Y.; Yamamoto, H.; Allakhverdiev, S.I.; Masami Inaba, M.; Yokota, A.; Murata, N. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 2001, 20, 5587–5594. [Google Scholar] [CrossRef] [PubMed]
- Goh, C.-H.; Ko, S.-M.; Koh, S.; Kim, Y.-J.; Bae, H.-J. Photosynthesis and environments: Photoinhibition and repair mechanisms in plants. J. Plant Biol. 2012, 55, 93–101. [Google Scholar] [CrossRef]
- Foyer, C.H.; Trebst, A.; Noctor, G. Protective and signaling functions of ascorbate, glutathione and tocopherol in chloroplasts. In Advances in Photosynthesis and Respiration: Photoprotection, Photoinhibition, Gene Regulation, and Environment; Demmig-Adams, B., Adams, W.W., Eds.; Springer Science Publishers: Dordrecht, The Netherlands, 2005; pp. 241–268. [Google Scholar]
- Paul, M.J.; Foyer, C.H. Sink regulation of photosynthesis. J. Exp. Bot. 2001, 52, 1383–1400. [Google Scholar] [CrossRef] [PubMed]
- McCormick, A.J.; Cramer, M.D.; Watt, D.A. Sink strength regulates photosynthesis in sugarcane. New Phytol. 2006, 171, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Koyro, H.-W.; Geissler, N.; Seenivasan, R.; Huchzermeyer, B. Plant stress physiology: Physiological and biochemical strategies allowing plants/crops to thrive under ionic stress. In Handbook of Plant and Crop Stress, 3rd ed.; Pessarakli, M., Ed.; CRC Press: Boca Raton, FL, USA, 2011; Volume 42, pp. 1051–1093. [Google Scholar]
- Huchzermeyer, B.; Koyro, H.-W. Salt and drought stress effects on photosynthesis. Enzyme cohesion and high turnover metabolite shuttling, essential for functioning of pathways, is impaired by changes in cytosolic water potential. In Handbook of Photosynthesis, 2nd ed.; Pessarakli, M., Ed.; CRC Press: Boca Raton, FL, USA, 2005; Volume 39, pp. 751–777. [Google Scholar]
- Foyer, C.H. Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ. Exp. Bot. 2018, 154, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. abiotic stress and reactive oxygen species: Generation, signaling, and defense mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef]
- Pospíšil, P. Production of reactive oxygen species by photosystem ii as a response to light and temperature stress. Front. Plant Sci. 2016, 7, 1950. [Google Scholar] [CrossRef]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Eckardt, N.A. Photorespiration revisited. Plant Cell 2005, 17, 2139–2141. [Google Scholar] [CrossRef]
- Peterhänsel, C.; Horst, I.; Niessen, N.; Blume, C.; Kebeish, R.; Kürkcüoglu, S.; Kreuzaler, F. Photorespiration. Arab. Book 2010, 8, e0130. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; VanLoocke, A.; Bernacchi, C.J.; Ort, D.R. The costs of photorespiration to food production now and in the future. Annu. Rev. Plant Biol. 2016, 67, 107–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharkey, T.D. Estimating the rate of photorespiration in leaves. Physiol. Plant. 1988, 73, 147–152. [Google Scholar] [CrossRef]
- Claeyssen, E.; Dorion, S.; Clendenning, A.; He, J.Z.; Wally, O.; Chen, J.; Auslender, E.L.; Moisan, M.C.; Jolicoeur, M.; Rivoal, J. The futile cycling of hexose phosphates could account for the fact that hexokinase exerts a high control on glucose phosphorylation but not on glycolytic rate in transgenic potato (Solanum tuberosum) roots. PLoS ONE 2013, 8, e53898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomeo, N.J.; Rosenthal, D.M. Photorespiration differs among Arabidopsis thaliana ecotypes and is correlated with photosynthesis. J. Exp. Bot. 2018, 69, 5191–5204. [Google Scholar] [CrossRef]
- Bandehagh, A.; Taylor, N.L. Can alternative metabolic pathways and shunts overcome salinity induced inhibition of central carbon metabolism in crops? Front. Plant Sci. 2020, 11, 1072. [Google Scholar] [CrossRef]
- Kebeish, R.; Niessen, M.; Thiruveedhi, K.; Bari, R.; Hirsch, H.-J.; Rosenkranz, R.; Stäbler, N.; Schönfeld, B.; Kreuzaler, F.; Peterhänsel, C. Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat. Biotechnol. 2007, 25, 593–599. [Google Scholar] [CrossRef]
- Von Caemmerer, S.; Evans, J.R. Enhancing C3 photosynthesis. Plant Physiol. 2010, 154, 589–592. [Google Scholar] [CrossRef] [Green Version]
- Farquhar, G.D.; von Caemmerer, S.; Berry, J.A. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 1980, 149, 78–90. [Google Scholar] [CrossRef] [Green Version]
- Numes-Nesi, A.; Fermie, A.R.; Stitt, M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 2010, 3, 973–996. [Google Scholar] [CrossRef]
- Geigenberger, P.; Fernie, A.R. Metabolic control of redox and redox control of metabolism in plants. Antioxid. Redox Signal. 2014, 21, 1389–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misra, J.B. Integrated operation of the photorespiratory cycle and cytosolic metabolism in the modulation of primary nitrogen assimilation and export of organic N-transport compounds from leaves: A hypothesis. J. Plant Physiol. 2014, 171, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chun, J.A.; Fleisher, D.; Reddy, V.; Timlin, D.; Resop, J. Parameter estimation of the Farquhar—Von Caemmerer—Berry biochemical model from photosynthetic carbon dioxide response curves. Sustainability 2017, 9, 1288. [Google Scholar] [CrossRef] [Green Version]
- Voss, I.; Sunil, B.; Scheibe, R.; Raghavendra, A.S. Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol. 2013, 15, 713–722. [Google Scholar] [CrossRef]
- Scheibe, R. Maintaining homeostasis by controlled alternatives for energy distribution in plant cells under changing conditions of supply and demand. Photosynth. Res. 2019, 139, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Selinski, J.; Scheibe, R. Malate valves: Old shuttles with new perspectives. Plant Biol. 2019, 21, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Kim, C. Chloroplast ROS and stress signaling. Plant Commun. 2021, 3, 100264. [Google Scholar] [CrossRef]
- Rumeau, D.; Peltier, G.; Cournac, L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 2007, 30, 1041–1051. [Google Scholar] [CrossRef]
- Poolman, M.G.; Fell, D.A.; Thomas, S. Modelling photosynthesis and its control. J. Exp. Bot. 2000, 51, 319–328. [Google Scholar] [CrossRef] [Green Version]
- Rhoads, D.M.; Umbach, A.L.; Subbaiah, C.C.; Siedow, J.N. Mitochondrial reactive oxygen species. contribution to oxidative stress and interorganellar signaling. Plant Physiol. 2006, 141, 357–366. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Van Aken, O.; Schwarzländer, M.; Belt, K.; Millar, A.H. The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol. 2016, 171, 1551–1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahal, K.; Li, X.Q.; Tau, H.; Creelaman, A.; Bizimungu, B. Improving potato stress tolerance and tuber yield under a climate change scenario—A current overview. Front. Plant Sci. 2019, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Leister, D. Piecing the Puzzle Together: The central role of reactive oxygen species and redox hubs in chloroplast retrgrade signaling. Antioxid. Redox Signal. 2019, 30, 1206–1219. [Google Scholar] [CrossRef]
- Kanai, S.; Ohkura, K.; Adu-Gyamfi, J.J.; Mohapatra, P.K.; Nguyen, N.T.; Saneoka, H.; Fujita, K. Depression of sink activity precedes the inhibition of biomass production in tomato plants subjected to potassium deficiency stress. J. Exp. Bot. 2007, 58, 2917–2928. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, A.; Sita, K.; Siddique, K.H.M.; Kumar, R.; Bhogireddy, S.; Varshney, R.K.; Hanumantha Rao, B.; Nait, R.M.; Prasad, P.V.V.; Nayyar, H. Drought or/and heat-stress effects on seed filling in food crops: Impacts on functional biochemistry, seed yields, and nutritional quality. Front. Plant Sci. 2018, 9, 1705. [Google Scholar] [CrossRef] [Green Version]
- Huchzermeyer, B.; Menghani, E.; Khardia, P.; Shilu, A. Metabolic pathway of natural antioxidants, antioxidant enzymes, and ROS providence. Antioxidants 2022, 11, 761. [Google Scholar] [CrossRef] [PubMed]
- Heber, U.; Neimanis, S.; Dietz, K.J.; Viil, J. Assimilatory power as a driving force in photosynthesis. Biochim. Biophys. Acta 1986, 852, 144–155. [Google Scholar] [CrossRef]
- Scheibe, R. Malate valves to balance cellular energy supply. Physiol. Plant. 2004, 120, 21–26. [Google Scholar] [CrossRef]
- Sage, R.F.; Kubien, D.S. The temperature response of C3 and C4 photosynthesis. Plant Cell Environ. 2007, 30, 1086–1106. [Google Scholar] [CrossRef]
- Kromdijk, J.; Griffiths, H.; Schepers, H.E. Can the progressive increase of C4 bundle sheath leakiness at low PFD be explained by incomplete suppression of photorespiration? Plant Cell Environ. 2010, 33, 1935–1948. [Google Scholar] [CrossRef]
- Long, S.P.; Marshall-Colon, A.; Zhu, X.-G. Meeting the global food demand of the future by engineering Crop Photosynthesis and Yield Potential. Cell 2015, 161, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flexas, J.; Díaz-Espejo, A.; Conesa, M.A.; Coopman, R.E.; Douthe, C.; Gago, J.; Gallé, A.; Galmés, J.; Medrano, H.; Ribas-Carbo, M.; et al. Mesophyll conductance to CO2 and Rubisco as targets for improving intrinsic water use efficiency in C3 plants. Plant Cell Environ. 2016, 39, 965–982. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, M.; Danila, F.R.; Furbankand, R.T.; von Caemmere, S. On the road to C4 rice: Advances and perspectives. Plant J. 2020, 101, 940–950. [Google Scholar] [CrossRef] [Green Version]
- Carmo-Silva, A.E.; Powers, S.J.; Keys, A.J.; Parry, M.A.J. Photorespiration in C4 grasses remains slow under drought conditions. Plant Cell Environ. 2008, 31, 925–940. [Google Scholar] [CrossRef]
- Christin, P.-A. Traces of strong selective pressures in the genomes of C4 grasses. J. Exp. Bot. 2017, 68, 103–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellasio, C. A generalized stoichiometric model of C3, C2, C2+C4, and C4 photosynthetic metabolism. J. Exp. Bot. 2017, 68, 269–282. [Google Scholar] [CrossRef] [Green Version]
- Offermann, S.; Friso, G.; Doroshenk, K.A.; Sun, Q.; Sharpe, R.M.; Okita, T.W.; Wimmer, D.; Edwards, G.E.; van Wijk, K.J. developmental and subcellular organization of single-cell C4 photosynthesis in Bienertia sinuspersici determined by large-scale proteomics and cDNA assembly from 454 DNA sequencing. J. Proteome Res. 2015, 14, 2090–2108. [Google Scholar] [CrossRef] [PubMed]
- Von Caemmerer, S.; Furbank, R.T. Strategies for improving C4 photosynthesis. Curr. Opin. Plant Biol. 2016, 31, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Von Caemmerer, S.; Ghannoum, O.; Robert, T.; Furbank, R.T. C4 photosynthesis: 50 years of discovery and innovation. J. Exp. Bot. 2017, 68, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Ma, Y.; Manevski, K.; Neumann Andersen, M.; Li, Y.; Wei, Z.; Liu, F. Biochar and alternate wetting-drying cycles improving rhizosphere soil nutrients availability and tobacco growth by altering root growth strategy in ferralsol and anthrosol. Sci. Total Environ. 2022, 806, 150513. [Google Scholar] [CrossRef]
- El-Dakak, R.; El-Aggan, W.; Badr, G.; Helaly, A.; Tammam, A. positive salt tolerance modulation via vermicompost regulation of SOS1 gene expression and antioxidant homeostasis in Vicia faba Plant. Plants 2021, 10, 2477. [Google Scholar] [CrossRef] [PubMed]
- Abideen, Z.; Koyro, H.-W.; Huchzermeyer, B.; Ansari, R. Ameliorating effects of biochar on photosynthetic efficiency and antioxidant defense of Phragmites karka under drought stress. Plant Biol. 2020, 22, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Wirth, S.; Behrendt, U.; Abd Allah, E.F.; Berg, G. Biochar treatment resulted in a combined effect on soybean growth promotion and a shift in plant growth promoting rhizobacteria. Front. Microbiol. 2016, 7, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, A.; Zimmerman, A.R. Organic carbon and nutrient release from a range of laboratory-produced biochars and biochar–soil mixtures. Geoderma 2013, 193–194, 122–130. [Google Scholar] [CrossRef]
- Ren, H.; Huang, B.; Fernández-García, V.; Miesel, J.; Yan, L.; Lv, C. Biochar and rhizobacteria amendments improve several soil properties and bacterial diversity. Microorganisms 2020, 8, 502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Ding, J.; Wang, H.; Su, L.; Zhao, C. Biochar addition alleviate the negative effects of drought and salinity stress on soybean productivity and water use efficiency. BMC Plant Biol. 2020, 20, 288. [Google Scholar] [CrossRef]
- Hafez, E.M.; Gowayed, S.M.; Nehela, Y.; Sakran, R.M.; Rady, A.M.S.; Awadalla, A.; Omara, A.E.-D.; Alowaiesh, B.F. Incorporated biochar-based soil amendment and exogenous glycine betaine foliar application ameliorate rice (Oryza sativa L.) tolerance and resilience to osmotic stress. Plants 2021, 10, 1930. [Google Scholar] [CrossRef]
- Yin, S.; Suo, F.; Kong, Q.; You, X.; Zhang, X.; Yuan, Y.; Yu, X.; Cheng, Y.; Sun, R.; Zheng, H.; et al. Biochar Enhanced Growth and Biological Nitrogen Fixation of Wild Soybean (Glycine max subsp. soja Siebold & Zucc.) in a Coastal Soil of China. Agriculture 2021, 11, 1246. [Google Scholar] [CrossRef]
- Razzaq, M.K.; Aleem, M.; Mansoor, S.; Khan, M.A.; Rauf, S.; Iqbal, S.; Siddique, K.H.M. Omics and CRISPR-Cas9 Approaches for molecular insight, functional gene analysis, and stress tolerance development in crops. Int. J. Mol. Sci. 2021, 22, 1292. [Google Scholar] [CrossRef]
- Sarfraz, R.; Hussain, A.; Sabir, A.; Ben Fekih, I.; Ditta, A.; Xing, S. Role of biochar and plant growth promoting rhizobacteria to enhance soil carbon sequestration. Environ. Monit. Assess. 2019, 191, 251. [Google Scholar] [CrossRef]
- Nehela, Y.; Mazrou, Y.S.A.; Alshaal, T.; Rady, A.M.S.; El-Sherif, A.M.A.; Omara, A.E.-D.; Abd El-Monem, A.M.; Hafez, E.M. The integrated amendment of sodic-saline soils using biochar and plant growth-promoting rhizobacteria enhances maize (Zea mays L.) resilience to water salinity. Plants 2021, 10, 1960. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.S.; Neumann Andersen, M.; Naveed, M.; Zahir, Z.A.; Liu, F. Interactive effect of biochar and plant growth-promoting bacterial endophytes on ameliorating salinity stress in maize. Funct. Plant Biol. 2015, 42, 770–781. [Google Scholar] [CrossRef] [PubMed]
- Rasool, M.; Akhter, A.; Soja, G.; Haider, M.S. Role of biochar, compost and plant growth promoting rhizobacteria in the management of tomato early blight disease. Sci. Rep. 2021, 11, 6092. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [Green Version]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.L.; Gravel, V.; Yergeau, E. Editorial: Signaling in the phytomicrobiome. Front. Plant Sci. 2017, 8, 611. [Google Scholar] [CrossRef] [Green Version]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.-S.; Patra, J.K. Endophytes: A treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 2016, 7, 1538. [Google Scholar] [CrossRef] [Green Version]
- Nadarajah, K.; Abdul Rahman, N.S.N. Plant–Microbe Interaction: Above ground to below ground, from the good to the bad. Int. J. Mol. Sci. 2021, 22, 10388. [Google Scholar] [CrossRef]
- Epstein, S. General model of microbial uncultivability. In Uncultivated Microorganisms; Epstein, S., Ed.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 131–160. [Google Scholar]
- Berdy, B.; Spoering, A.; Ling, L.; Epstein, S. In situ cultivation of previously uncultivable microorganisms using the ichip. Nat. Protoc. 2017, 12, 2232–2242. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, P.R.; Mauchline, T.H. Who’s who in plant root microbiome? Nat. Biotechnol. 2012, 30, 961–962. [Google Scholar] [CrossRef] [PubMed]
- Delaplace, P.; Delory, B.M.; Baudson, C.; Mendaluk-Saunier de Cazenave, M.; Spaepen, S.; Varin, S.; Brostaux, Y.; du Jardin, P. Influence of rhizobacterial volatiles on the root system architecture and the production and allocation of biomass in the model grass Brachypodium distachyon (L.) P. Beauv. BMC Plant Biol. 2015, 15, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simard, S.W.; Durall, D.M. Mycorrhizal networks: A review of their extent, function, and importance. Can. J. Bot. 2004, 82, 1140–1165. [Google Scholar] [CrossRef]
- Gray, E.J.; Smith, D.L. Intracellular and extracellular PGPR: Commonalities and distinctions in the plant–bacterium signaling processes. Soil Biol. Biochem. 2005, 37, 395–412. [Google Scholar] [CrossRef]
- Nelson, E.B. The seed microbiome: Origins, interactions, and impacts. Plant Soil 2017, 422, 7–34. [Google Scholar] [CrossRef]
- Canellas, N.O.A.; Olivares, F.L.; Canellas, L.P. Metabolite fingerprints of maize and sugarcane seedlings: Searching for markers after inoculation with plant growth-promoting bacteria in humic acids. Chem. Biol. Technol. Agric. 2019, 6, 14. [Google Scholar] [CrossRef]
- Canellas, L.P.; Silva, S.F.; Olk, D.; Olivares, F.L. Foliar application of Herbaspirillum seropedicae and humic acid increase maize yields. J. Food Agric. Environ. 2015, 13, 146–153. [Google Scholar]
- Olivares, F.L.; Busato, J.G.; Paula, A.M.; Lima, L.S.; Aguiar, N.O.; Canellas, L.P. Plant growth promoting bacteria and humic substances: Crop promotion and mechanisms of action. Chem. Biol. Technol. Agric. 2017, 4, 30. [Google Scholar] [CrossRef] [Green Version]
- Nelson, L.M. Plant growth promoting rhizobacteria (PGPR): Prospects for new inoculants. Crop Manag. 2004, 3, 301–305. [Google Scholar] [CrossRef]
- Sabki, M.H.; Ong, P.Y.; Ibrahim, N.; Lee, C.T.; Klemeš, J.J.; Li, C.; Gao, Y. A Review on abiotic stress tolerance and plant growth metabolite framework by plant growth-promoting bacteria for sustainable agriculture. Chem. Eng. Trans. 2021, 83, 367–372. [Google Scholar] [CrossRef]
- Dempsey, D.A.; Klessig, D.F. How does the multifaceted plant hormone sali-cylic acid combat disease in plants and are similar mechanisms utilized in humans? BMC Biol. 2017, 15, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaparro, J.M.; Dayakar, V.; Badri, D.V.; Matthew, G.; Bakker, M.G.; Akifumi Sugiyama, A.; Daniel, K.; Manter, D.K.; Jorge, M.; Vivanco, J.M. Root exudation of phytochemicals in arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2013, 8, 1371. [Google Scholar] [CrossRef]
- Uroz, S.; Courty, P.E.; Oger, P. Plant symbionts are engineers of the plant-associated microbiome. Trends Plant Sci. 2019, 24, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Brettel, L.E.; Qiu, Z.; Sing, B.K. Microbiome-Mediated Stress Resistance in Plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef]
- Cueva-Yesquén, L.G.; Goulart, M.C.; Attili de Angelis, D.; Nopper Alves, M.; Fantinatti-Garboggini, F. Multiple plant growth-promotion traits in endophytic bacteria retrieved in the vegetative stage from passionflower. Front. Plant Sci. 2021, 11, 621740. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Nelson, M.S.; Sadowsky, M.J. Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 2015, 6, 491. [Google Scholar] [CrossRef] [Green Version]
- Massalha, H.; Korenblum, E.; Malitsky, S.; Shapiro, O.H.; Aharon, A. Live imaging of root–bacteria interactions in a microfluidics setup. Proc. Natl. Acad. Sci. USA 2017, 114, 4549–4554. [Google Scholar] [CrossRef] [Green Version]
- Nelson, E.B. Microbial dynamics and interactions in the spermosphere. Annu. Rev. Phytopathol. 2004, 42, 271–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pausch, J.; Kuzyakov, Y. Carbon input by roots into the soil: Quantification of rhizodeposition from root to ecosystem scale. Glob. Chang. Biol. 2018, 24, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rovira, A.D. Interactions between plant roots and soil microorganisms. Annu. Rev. Microbiol. 1965, 19, 241–266. [Google Scholar] [CrossRef] [PubMed]
- Lemanceau, P.; Corberand, T.; Gardan, L.; Latour, X.; Laguerre, G.; Boeufgras, J.-M.; Alabouvette, C. Effect of two plant species, flax (Linum usitatissnum L.) and tomato (Lycopersicon esculentum Mill.) on the diversity of soilborne populations of fluorescent Pseudomonas. Appl. Environ. Microbiol. 1995, 61, 1004–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grayston, S.J.; Wang, S.; Campbell, C.D.; Edwards, A.C. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 1998, 30, 369–378. [Google Scholar] [CrossRef]
- Miethling, R.; Wieland, G.; Backhaus, H. Variation of microbial rhizosphere communities in response to crop species, soil origin, and inoculation with Sinorhizobium meliloti L33. Microb. Ecol. 2000, 41, 43–56. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Gaiero, J.R.; Mc Cali, C.M.; Thompson, K.A.; Day, N.J. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef] [Green Version]
- Rout, M.E.; Southworth, A.S. The root microbiome influences scales from molecules to ecosystems: The unseen majority. Am. J. Bot. 2013, 100, 1689–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leach, J.E.; Triplett, L.R.; Argueso, C.T.; Trivedi, P. Communication in the phytobiome. Cell 2017, 169, 587–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, H.; Bagyaraj, D.; Selvakumar, G.; Sundaram, S. Novel plant growth promoting rhizobacteria—Prospects and potential. Appl. Soil Ecol. 2015, 95, 38–53. [Google Scholar] [CrossRef]
- Etesami, H.; Alikhani, H.A. Rhizosphere and endorhiza of oilseed rape (Brassica napus L.) plant harbor bacteria with multifaceted beneficial effects. Biol. Control 2016, 94, 11–24. [Google Scholar] [CrossRef]
- Khalifa, A.Y.Z.; Alsyeeh, A.-M.; Almalki, M.A.; Saleh, F.A. Characterization of the plant growth promoting bacterium, Enterobacter cloacae MSR1, isolated from roots of non-nodulating Medicago sativa. Saudi J. Biol. Sci. 2016, 23, 79–86. [Google Scholar] [CrossRef]
- Devi, K.A.; Pandey, G.; Rawat, A.K.S.; Sharma, G.D.; Pandey, P. The endophytic symbiont—Pseudomonas aeruginosa stimulates the antioxidant activity and growth of Achyranthes aspera L. Front. Microbiol. 2017, 8, 1897. [Google Scholar] [CrossRef] [Green Version]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 2006, 97, 883–893. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.L.; Touraine, B.; Moenne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyè, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 19. [Google Scholar] [CrossRef] [Green Version]
- Mahmood, A.; Kataoka, R. Metabolite profiling reveals a complex response of plants to application of plant growth-promoting endophytic bacteria. Microbiol. Res. 2020, 234, 126421. [Google Scholar] [CrossRef]
- Zaidi, A.; Khan, M.S.; Ahemad, M.; Oves, M. Plant growth promotion by phosphate solubilizing bacteria. Acta Microbiol. Immunol. Hung. 2009, 56, 263–284. [Google Scholar] [CrossRef]
- Campos, P.; Borie, F.; Cornejo, P.; Ráez, J.A.L.; López-Garciá, A.; Seguel, A. Phosphorus acquisition efficiency related to root traits: Is mycorrhizal symbiosis a key factor to wheat and barley cropping? Front. Plant Sci. 2018, 9, 752. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Khan, M.S.; Zaidi, A.; Wani, P.A. Role of phosphate-solubilizing microorganisms in sustainable agriculture—A review. Agron. Sustain. Dev. 2007, 27, 29–43. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Hao, T.; Chen, S. Colonization and maize growth promotion induced by phosphate solubilizing bacterial isolates. Int. J. Mol. Sci. 2017, 18, 1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goswami, D.; Dhandhukia, P.; Patel, P.; Thakker, J.N. Screening of PGPR from saline desert of Kutch: Growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol. Res. 2014, 169, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Oteino, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Liu, S. Identification and characterization of the phosphate-solubilizing bacterium Pantoea sp. S32 in Reclamation Soil in Shanxi, China. Front. Microbiol. 2019, 10, 21. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.T.; Vail, S.; Arcand, M.M. Nitrogen use efficiency in parent vs. hybrid canola under varying nitrogen availabilities. Plants 2021, 10, 2364. [Google Scholar] [CrossRef]
- Paungfoo-Lonhienne, C.; Lonhienne, T.G.A.; Yeoh, Y.K.; Donose, B.C.; Webb, R.I.; Parsons, J.; Liao, W.; Sagulenko, E.; Lakshmanan, P.; Hugenholtz, P.; et al. Crosstalk between sugarcane and a plant-growth promoting Burkholderia species. Sci. Rep. 2016, 6, 37389. [Google Scholar] [CrossRef]
- Xie, S.-S.; Wu, H.-J.; Zang, H.-Y.; Wu, L.-M.; Zhu, Q.-Q.; Gao, X.-W. Plant growth promotion by spermidine-producing Bacillus subtilis OKB105. Mol. Plant-Microbe Interact. 2014, 27, 655–663. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.Y.; Roessner, U.; Eickmeier, I.; Genc, Y.; Callahan, D.L.; Shirley, N.; Langridge, P.; Bacic, A. Metabolite profiling reveals distinct changes in carbon and nitrogen metabolism in phosphate-deficient barley plants (Hordeum vulgare L.). Plant Cell Physiol. 2008, 49, 691–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, H.; Ishiga, Y.; Watanabe, S.; Konishi, T.; Egusa, M.; Akiyoshi, N.; Matsuura, T.; Mori, I.C.; Hirayama, T.; Kaminaka, H.; et al. Allantoin, a stress-related purine metabolite, can activate jasmonate signaling in a MYC2-regulated and abscisic acid-dependent manner. J. Exp. Bot. 2016, 67, 2519–2532. [Google Scholar] [CrossRef] [PubMed]
- Bharti, N.; Pandey, S.S.; Barnawal, D.; Patel, V.K.; Kalra, A. Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 2016, 6, 34768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, J.; González, J.E. The novel genes emmABC, are associated with exopolysaccharide production, motility, stress adaptation, and symbiosis in Sinorhizobium meliloti. J. Bacteriol. 2009, 191, 5890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasotia, A.; Varma, A.; Tuteja, N.; Choudhary, D. Amelioration of soybean plant from saline-induced condition by exopolysaccharide producing Pseudomonas-mediated expression of high affinity K-transporter (HKT1) gene. Curr. Sci. 2016, 111, 1961–1967. [Google Scholar] [CrossRef]
- Kwon, Y.S.; Ryu, C.-M.; Lee, S.; Park, H.B.; Han, K.S.; Lee, J.H.; Lee, K.; Chung, W.S.; Jeong, M.-J.; Kim, H.K.; et al. Proteome analysis of Arabidopsis seedlings exposed to bacterial volatiles. Planta 2010, 232, 1355–1370. [Google Scholar] [CrossRef]
- Sunita, K.; Mishra, I.; Mishra, J.; Prakash, J.; Arora, N.K. Secondary metabolites from halotolerant plant growth promoting rhizobacteria for ameliorating salinity stress in plants. Front. Microbiol. 2020, 11, 567768. [Google Scholar] [CrossRef]
- Rajkumar, M.; Nagendran, R.; Lee, K.J.; Lee, W.H.; Kim, S.Z. Infuence of plant growth promoting bacteria and Cr on the growth of indian mustard. Chemosphere 2006, 62, 741–748. [Google Scholar] [CrossRef]
- Sheng, X.F.; Xia, J.J. Improvement of rape (Brassica napus) plant growth and cadmium uptake by cadmium-resistant bacteria. Chemosphere 2006, 64, 1036–1042. [Google Scholar] [CrossRef]
- Wani, P.A.; Khan, M.S.; Zaidi, A. Effect of metal tolerant plant growth promoting Bradyrhizobium sp. (vigna) on growth, symbiosis, seed yield and metal uptake by green gram plants. Chemosphere 2007, 70, 36–45. [Google Scholar] [CrossRef]
- Egamberdieva, D. Alleviation of salt stress by plant growth regulators and IAA producing bacteria in wheat. Acta Physiol. Plant. 2009, 31, 861–864. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Kucharova, Z. Selection for root colonising bacteria stimulating wheat growth in saline soils. Biol. Fertil. Soils 2009, 45, 563–571. [Google Scholar] [CrossRef]
- Bianco, C.; Defez, R. Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium melilotistrain. J. Exp. Bot. 2009, 60, 3097–3107. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Defez, R. Improvement of phosphate solubilization and Medicago plant yield by an indole-3-acetic acid-overproducing strain of Sinorhizobium meliloti. Appl. Environ. Microbiol. 2010, 76, 4626–4632. [Google Scholar] [CrossRef] [Green Version]
- De Bashan, L.E.; Hernandez, J.P.; Nelson, K.N.; Bashan, Y.; Maier, R.M. Growth of quailbush in acidic, metalliferous desert mine tailings: Effect of Azospirillum brasilense Sp6 on biomass production and rhizosphere community structure. Microb. Ecol. 2010, 60, 915–927. [Google Scholar] [CrossRef] [Green Version]
- Banaei-Asl, F.; Farajzadeh, D.; Bandehagh, A.; Komatsu, S. Comprehensive proteomic analysis of canola leaf inoculated with a plant growth-promoting bacterium, Pseudomonas fluorescens, under salt stress. Biochim. Biophys. Acta 2016, 1864, 1222–1236. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.; Li, J.; Dong, Y.; Liu, H.; Peng, J.; Hu, Y.; Sun, Y. Profiling of plant growth-promoting metabolites by phosphate-solubilizing bacteria in maize rhizosphere. Plants 2021, 10, 1071. [Google Scholar] [CrossRef]
- Naik, K.; Mishra, S.; Srichandan, H.; Singh, P.K.; Sarangi, P.K. Plant growth promoting microbes: Potential link to sustainable agriculture and environment. Biocatal. Agric. Biotechnol. 2019, 21, 101326. [Google Scholar] [CrossRef]
- Nacoon, S.; Jogloy, S.; Riddech, N.; Mongkolthanaruk, W.; Kuyper, T.W.; Boonlue, S. Interaction between phosphate solubilizing bacteria and arbuscular mycorrhizal fungi on growth promotion and tuber inulin content of Helianthus tuberosus L. Sci. Rep. 2020, 10, 4916. [Google Scholar] [CrossRef] [Green Version]
- Stephen, J.; Shabanamol, S.; Rishad, K.S.; Jisha, M.S. Growth enhancement of rice (Oryza sativa) by phosphate solubilizing Gluconacetobacter sp. (MTCC 8368) and Burkholderia sp. (MTCC 8369) under greenhouse conditions. 3 Biotech 2015, 5, 831–837. [Google Scholar] [CrossRef] [Green Version]
- Jayaprakashvel, M.; Mathivanan, N. Management of plant diseases by microbial metabolites. In Bacteria in Agrobiology: Plant Nutrient Management; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 237–265. [Google Scholar] [CrossRef]
- Nakkeeran, S.; Marimuthu, T.; Raguchander, T.E. Exploring, DAPG. and phenazine producing PGPR strains and fungal antagonists for the management of Noni diseases. WNRF Tech. Bull. 2013, 11. [Google Scholar]
- Reimer, D.; Bode, H.B. A natural prodrug activation mechanism in the biosynthesis of nonribosomal peptides. Nat. Prod. Rep. 2014, 31, 154–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majed, R.; Faille, C.; Kallassy, M.; Gohar, M. Bacillus cereus biofilms-same, only different. Front. Microbiol. 2016, 7, 16. [Google Scholar] [CrossRef]
- Gururani, M.A.; Upadhyaya, C.P.; Baskar, V.; Venkatesh, J.; Nookaraju, A.; Park, S.W. Plant growth-promoting rhizobacteria enhance abiotic stress tolerance in Solanum tuberosum through inducing changes in the expression of ROS-scavenging enzymes and improved photosynthetic performance. J. Plant Growth Regul. 2013, 32, 245–258. [Google Scholar] [CrossRef]
- Babu, A.G.; Kim, J.D.; Oh, B.T. Enhancement of heavy metal phytoremediation by Alnus firma with endophytic Bacillus thuringiensis GDB-1. J. Hazard. Mater. 2013, 250–251, 477–483. [Google Scholar] [CrossRef]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Crowley, D.E. Microbial siderophores in the plant rhizosphere. In Iron Nutrition in Plants and Rhizospheric Microorganisms; Barton, L.L., Abadía, J., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 169–198. [Google Scholar] [CrossRef]
- Rajkumar, M.; Ae, N.; Prasad, M.N.V.; Freitas, H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010, 28, 142–149. [Google Scholar] [CrossRef]
- Shaikh, S.S.; Sayyed, R.Z. Role of plant growth-promoting rhizobacteria and their formulation in biocontrol of plant diseases. In Plant Microbes Symbiosis: Applied Facets; Maheshwari, D.K., Ed.; Springer: New Delhi, India, 2015; pp. 337–351. [Google Scholar] [CrossRef]
- Sah, S.; Singh, N.; Singh, R. Iron acquisition in maize (Zea mays L.) using Pseudomonas siderophore. 3 Biotech 2017, 7, 121. [Google Scholar] [CrossRef] [Green Version]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant-microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Terrer, C.; Phillips, R.P.; Hungate, B.A.; Rosende, J.; Pett-Ridge, J.; Craig, M.E.; van Groenigen, K.J.; Keenan, T.F.; Sulman, B.N.; Stocker, B.D.; et al. A trade-off between plant and soil carbon storage under elevated CO2. Nature 2021, 591, 599–603. [Google Scholar] [CrossRef]
- Song, L.; Lu, H.-Z.; Xu, X.-L.; Li, S.; Shi, X.-M.; Chen, X.; Wu, Y.; Huang, J.-B.; Chen, Q.; Liu, S.; et al. Organic nitrogen uptake is a significant contributor to nitrogen economy of subtropical epiphytic bryophytes. Sci. Rep. 2016, 6, 30408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ninnemann, H.; Maier, J. Indications for the occurrence of nitric oxide synthases in fungi and plants and the involvement in photoconidiation of Neurospora crassa. Photochem. Photobiol. 1996, 64, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14 (Suppl. S1), 165–183. [Google Scholar] [CrossRef] [Green Version]
- Bartosz, G. Oxidative stress in plants. Acta Physiol. Plant. 1997, 19, 47–64. [Google Scholar] [CrossRef]
- Hancock, J.T. Harnessing evolutionary toxins for signaling: Reactive oxygen species, nitric oxide and hydrogen sulfide in plant cell regulation. Front. Plant Sci. 2017, 8, 189. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, S.; Lata, C.; Chauhan, P.S.; Nautiyal, C.S. Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol. Biochem. 2016, 99, 108–117. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, H.; Ruan, W.; Deng, M.; Wang, F.; Peng, J.; Luo, J.; Chen, Z.; Yi, K. Abnormal inflorescence meristem1 functions in salicylic acid biosynthesis to maintain proper reactive oxygen species levels for root meristem activity in rice. Plant Cell 2017, 29, 560–574. [Google Scholar] [CrossRef] [Green Version]
- Grover, M.; Madhubala, R.; Ali, S.Z.; Yadav, S.K.; Venkateswarlu, B. Influence of Bacillus spp. strains on seedling growth and physiological parameters of sorghum under moisture stress conditions. J. Basic Microbiol. 2014, 54, 951–961. [Google Scholar] [CrossRef]
- Zhou, C.; Ma, Z.; Zhu, L.; Xiao, X.; Xie, Y.; Zhu, J.; Wang, J. Rhizobacterial strain Bacillus megaterium BOFC15 induces cellular polyamine changes that improve plant growth and drought resistance. Int. J. Mol. Sci. 2016, 17, 976. [Google Scholar] [CrossRef]
- Forni, C.; Duca, D.; Glick, B.R. Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil 2017, 410, 335–356. [Google Scholar] [CrossRef]
- Goswami, M.; Deka, S. Plant growth-promoting rhizobacteria-alleviators of abiotic stresses in soil: A review. Pedosphere 2020, 30, 40–61. [Google Scholar] [CrossRef]
- Rosa, P.A.L.; Shintate Galindo, F.; da Silva Oliveira, C.E.; Jalal, A.; Mortinho, E.S.; Fernandes, G.C.; Rocha Marega, E.M.; Buzetti, S.; Teixeira Filho, M.C.M. Inoculation with plant growth-promoting bacteria to reduce phosphate fertilization requirement and enhance technological quality and yield of sugarcane. Microorganisms 2022, 10, 192. [Google Scholar] [CrossRef] [PubMed]
- Shaterian, J.; Waterer, D.; De Jong, H.; Tanino, K.K. Differential stress responses to NaCl salt application in early- and late-maturing diploid potato (Solanum sp.) clones. Environ. Exp. Bot. 2005, 54, 202–212. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Kaur, H.; Kaur, J.; Gera, R. Plant growth promoting rhizobacteria: A boon to agriculture. Int. J. Cell Sci. Biotechnol. 2016, 5, 17–22. [Google Scholar] [CrossRef]
- Reid, M.S. The role of ethylene in flower senescence. In Proceedings of the IV International Symposium on Postharvest Physiology of Ornamental Plants, Herzliya, Israel, 20–25 March 1988; p. 261. [Google Scholar]
- Li, Q.; Saleh-Lakha, S.; Glick, B.R. The effect of native and ACC deaminase-containing Azospirillum brasilense Cd1843 on the rooting of carnation cuttings. Can. J. Microbiol. 2005, 51, 511–514. [Google Scholar] [CrossRef]
- Liu, F.C.; Xing, S.J.; Ma, H.L.; Du, Z.Y.; Ma, B.Y. Cytokinin-producing, plant growth-promoting rhizobacteria that confer resistance to drought stress in Platycladus orientalis container seedlings. Appl. Microbiol. Biotechnol. 2013, 97, 9155–9164. [Google Scholar] [CrossRef]
- Kang, B.G.; Kim, W.T.; Yun, H.S.; Chang, S.C. Use of plant growth-promoting rhizobacteria to control stress responses of plant roots. Plant Biotechnol. Rep. 2010, 4, 179–183. [Google Scholar] [CrossRef]
- Gamalero, E.; Glick, B.R. Bacterial modulation of plant ethylene levels. Plant Physiol. 2015, 169, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 2004, 42, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Sandhya, V.; Ali, S.Z.; Grover, M.; Reddy, G.; Venkateswarlu, B. Effect of plant growth promoting Pseudomonas spp. on compatible solutes, antioxidant status and plant growth of maize under drought stress. Plant Growth Regul. 2010, 62, 21–30. [Google Scholar] [CrossRef]
- Belimov, A.A.; Safronova, V.I.; Sergeyeva, T.A.; Egorova, T.N.; Matveyeva, V.A.; Tsyganov, V.E.; Borisov, A.Y.; Tikhonovich, I.; Kluge, C.; Preisfeld, A.; et al. Characterization of plant growth promoting rhizobacteria isolated from polluted soils and containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol. 2001, 47, 642–652. [Google Scholar] [CrossRef]
- Dos Santos, R.M.; Diaz, P.A.E.; Lobo, L.L.B.; Rigobelo, E.C. Use of plant growth-promoting rhizobacteria in maize and sugarcane: Characteristics and applications. Front. Sustain. Food Syst. 2020, 4, 136. [Google Scholar] [CrossRef]
- Khan, A.L.; Haloa, B.A.; Alia, A.E.S.; Al-Hosnia, K.; Al-Harrasia, J.H.A.; Lee, I.-J. Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanum lycopersicum. Electron. J. Biotechnol. 2016, 21, 58–64. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Sun, Y.; Sheng, H.; Li, H.; Liu, X. Effects of the inoculations using bacteria producing ACC deaminase on ethylene metabolism and growth of wheat grown under different soil water contents. Plant Physiol. Biochem. 2018, 125, 178–184. [Google Scholar] [CrossRef]
- Ali, S.; Kim, W.C. Plant growth promotion under water: Decrease of waterlogging-induced ACC and ethylene levels by ACC deaminase-producing bacteria. Front. Microbiol. 2018, 9, 1096. [Google Scholar] [CrossRef]
- Glick, B.R. Bacterial ACC deaminase and the alleviation of plant stress. Adv. Appl. Microbiol. 2004, 56, 291–312. [Google Scholar]
- Glick, B.R. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 2005, 251, 1–7. [Google Scholar] [CrossRef]
- Rashid, S.; Charles, T.C.; Glick, B.R. Isolation and characterization of new plant growth-promoting bacterial endophytes. Appl. Soil Ecol. 2012, 61, 217–224. [Google Scholar] [CrossRef]
- Nagaraju, Y.; Mahadevaswamy; Naik, N.M.; Gowdar, S.B.; Narayanarao, K.; Satyanarayanarao, K. ACC deaminase-positive halophilic bacterial isolates with multiple plant growth-promoting traits improve the growth and yield of chickpea (Cicer arietinum L.) under salinity stress. Front. Agron. 2021, 3, 681007. [Google Scholar] [CrossRef]
- Win, K.T.; Tanaka, F.; Okazaki, K.; Ohwaki, Y. The ACC deaminase expressing endophyte Pseudomonas spp. Enhances NaCl stress tolerance by reducing stress-related ethylene production, resulting in improved growth, photosynthetic performance, and ionic balance in tomato plants. Plant Physiol. Biochem. 2018, 127, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Vanderstraeten, L.; Depaepe, T.; Bertrand, S.; Van Der Straeten, D. The ethylene precursor acc affects early vegetative development independently of ethylene signaling. Front. Plant Sci. 2019, 10, 1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Su, C.; Dong, C.-H. Genome-wide transcriptomic and proteomic exploration of molecular regulations in quinoa responses to ethylene and salt stress. Plants 2021, 10, 2281. [Google Scholar] [CrossRef] [PubMed]
- Bomle, D.V.; Kiran, A.; Kumar, J.K.; Nagaraj, L.S.; Pradeep, C.K.; Ansari, M.A.; Alghamdi, S.; Kabrah, A.; Assaggaf, H.; Dablool, A.S.; et al. Plants saline environment in perception with rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase. Int. J. Mol. Sci. 2021, 22, 11461. [Google Scholar] [CrossRef] [PubMed]
- Pospisilova, J. Participation of phytohormones in the stomatal regulation of gas exchange during water stress. Biol. Plant. 2003, 46, 491–506. [Google Scholar] [CrossRef]
- Dodd, I.C. Soil moisture heterogeneity during deficit irrigation alters root-to-shoot signalling of abscisic acid. Funct. Plant Biol. 2007, 34, 439–448. [Google Scholar] [CrossRef]
- Munemasa, S.; Hauser, F.; Park, J.; Waadt, R.; Brandt, B.; Schroeder, J.I. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 2015, 28, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Sah, S.K.; Reddy, K.R.; Li, J.X. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [Green Version]
- Dodd, I.C.; Zinovkina, N.Y.; Safronova, V.I.; Belimov, A.A. Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 2010, 157, 361–379. [Google Scholar] [CrossRef]
- Forchetti, G.; Masciarelli, O.; Alemano, S.; Alvarez, D.; Abdala, G. Endophytic bacteria in sunflower (Helianthus annuus L.): Isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 2007, 76, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Sgroy, V.; Cassán, F.; Masciarelli, O.; Del Papa, M.F.; Lagares, A.; Luna, V. Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl. Microbiol. Biotechnol. 2009, 85, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.C.; Travaglia, C.N.; Bottini, R.; Piccoli, P.N. Participation of abscisic acid and gibberellins produced by endophytic Azospirillum in the alleviation of drought effects in maize. Botany 2009, 87, 455–462. [Google Scholar] [CrossRef]
- Yao, L.; Wu, Z.; Zhen, Y.; Kaleem, I.; Li, C. Growth promotion and protection against salt stress by Pseudomonas putida Rs-198 on cotton. Eur. J. Soil Biol. 2010, 46, 49–54. [Google Scholar] [CrossRef]
- Bresson, J.; Vasseur, F.; Dauzat, M.; Labadie, M.; Varoquaux, F.; Touraine, B.; Vile, D. Interact to survive: Phyllobacterium brassicacearum improves Arabidopsis tolerance to severe water deficit and growth recovery. PLoS ONE 2014, 9, e107607. [Google Scholar] [CrossRef]
- Belimov, A.A.; Dodd, I.C.; Safronova, V.I.; Dumova, V.A.; Shaposhnikov, A.I.; Ladatko, A.G.; Davies, W.J. Abscisic acid metabolizing rhizobacteria decrease ABA concentrations in planta and alter plant growth. Plant Physiol. Biochem. 2014, 74, 84–91. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Harshavardhan, V.T.; Govind, G.; Seiler, C.; Kohli, A. Contrapuntal role of ABA: Does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 2012, 506, 265–273. [Google Scholar] [CrossRef]
- Pandey, A.; Sharma, M.; Pandey, G.K. Emerging roles of strigolactones in plant responses to stress and development. Front. Plant Sci. 2016, 7, 434. [Google Scholar] [CrossRef] [Green Version]
- Brivanlou, A.H.; Darnell, J.E., Jr. Signal transduction and the control of gene expression. Science 2002, 295, 813–818. [Google Scholar] [CrossRef] [Green Version]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Guillaumot, D.; Guillon, S.; Morsomme, P.; Batoko, H. ABA, porphyrins and plant TSPO-related protein. Plant Signal. Behav. 2000, 4, 1087–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhee, C.; Zapotoczny, G.; Masquelier, D.; Ghislain, M.; Batoko, H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 2011, 23, 785–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hachez, C.; Veljanovski, V.; Reinhardt, H.; Guillaumot, D.; Vanhee, C.; Chaumont, F.; Batoko, H. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interaction and autophagic degradation. Plant Cell 2014, 26, 4974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akpinar, B.A.; Avsar, B.; Lucas, S.J.; Budak, H. Plant abiotic stress signaling. Plant Signal. Behav. 2012, 7, 1450–1455. [Google Scholar] [CrossRef]
- Roychoudhury, A.; Paul, S.; Basu, S. Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 2013, 32, 985–1006. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Skalak, J.; Nicolas, K.L.; Vankova, R.; Hejatko, J. Signal integration in plant abiotic stress responses via multistep phosphorelay signaling. Front. Plant Sci. 2021, 12, 644823. [Google Scholar] [CrossRef]
- Wang, K.; Wang, T.; Ren, C.; Dou, P.; Miao, Z.; Liu, X.; Huang, D.; Wang, K. Aqueous Extracts of Three Herbs Allelopathically Inhibit Lettuce Germination but Promote Seedling Growth at Low Concentrations. Plants 2022, 11, 486. [Google Scholar] [CrossRef]
- Batoko, H.; Dagdas, Y.; Baluska, F.; Sirko, A. Understanding and exploiting autophagy signaling in plants. Essays Biochem. 2017, 61, 675–685. [Google Scholar] [CrossRef]
- Van Rensburg, H.C.J.; Van den Ende, W.; Signorelli, S. Autophagy in Plants: Both a Puppet and a Puppet Master of Sugars. Front. Plant Sci. 2019, 10, 14. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.-Y.; Bassham, D.C. Combating stress: The interplay between hormone signaling and autophagy in plants. J. Exp. Bot. 2020, 71, 1723–1733. [Google Scholar] [CrossRef]
- Liu, F.; Marshall, R.S.; Li, F. Understanding and exploiting the roles of autophagy in plants through multi-omics approaches. Plant Sci. 2018, 274, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An intracellular degradation pathway regulating plant survival and stress response. Front Plant Sci. 2020, 11, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Wang, T.; Han, J.; Li, M.; Zhao, Y.; Su, T.; Ma, C. Plant Autophagy: An intricate process controlled by various signaling pathways. Front. Plant Sci. 2021, 12, 754982. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Bassham, D.C. Chapter one—New insight into the mechanism and function of autophagy in plant cells. In International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 320, pp. 1–40. [Google Scholar]
- Bao, Y.; Song, W.-M.; Wang, P.; Yu, X.; Li, B.; Jiang, C.; Shiu, S.-H.; Zhang, H.; Bassham, D.C. COST1 regulates autophagy to control plant drought tolerance. Proc. Natl. Acad. Sci. USA 2020, 117, 7482–7493. [Google Scholar] [CrossRef]
- Qi, H.; Xia, F.-N.; Xiao, S. Autophagy in plants: Physiological roles and post-translational regulation. J. Integr. Plant Biol. 2021, 63, 161–179. [Google Scholar] [CrossRef]
- Liu, Y.; Bassham, D.C. TOR Is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 2010, 5, e11883. [Google Scholar] [CrossRef] [Green Version]
- Signorelli, S.; Tarkowski, L.P.; Van den Ende, W.; Bassham, D.C. Linking autophagy to abiotic stress responses. Trends Plant Sci. 2019, 24, 413–430. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.-J.; Liu, C.-X.; Shao, S.-J.; Zhou, J. Molecular mechanisms of autophagy regulation in plants and their application in agriculture. Front. Plant Sci. 2021, 11, 618944. [Google Scholar] [CrossRef]
- Boycheva Woltering, S.; Isomo, E. Knowing when to self-eat. Fine-tuning autophagy through ATG8 iso-forms in plants. Front. Plant Sci. 2020, 11, 579875. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Bassham, D.C. Autophagy during drought: Function, regulation, and application. Plant J. 2022, 109, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Thanthridge, N.; Bhowmik, S.D.; Ferguson, B.J.; Kabbage, M.; Mundree, S.G.; Williams, B. Potential biotechnological application of autophagy for agriculture. Front. Plant Sci. 2021, 12, 760407. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.; Wang, D.; Gong, Q. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1459. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, S.; Hagihara, S.; Otomo, K.; Ishida, H.; Hidema, J.; Nemoto, T.; Izumi, M. Autophagy contributes to the quality control of leaf mitochondria. Plant Cell Physiol. 2021, 62, 229–247. [Google Scholar] [CrossRef]
- Kroemer, G.; Marino, G.; Levine, B. Autophagy and the integrated stress response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef] [Green Version]
- Avin-Wittenberg, T. Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 2019, 42, 1045–1053. [Google Scholar] [CrossRef]
- Cheng, L.; Zeng, Y.; Hu, S.; Zhang, N.; Cheung, K.C.P.; Li, B.; Leung, K.-S.; Jiamg, L. Systematic prediction of autophagy-related proteins using Arabidopsis thaliana interactome data. Plant J. 2021, 105, 708–720. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Ichimura, Y.; Ohsum, A. Atg8, a Ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Bu, F.; Yang, M.; Guo, X.; Huang, W.; Chen, L. Multiple functions of ATG8 family proteins in plant autophagy. Front. Cell Dev. Biol. 2020, 8, 466. [Google Scholar] [CrossRef]
- Gamalero, E.; Glick, B.R. Bacterial ACC deaminase and IAA: Interactions and consequences for plant growth in polluted environments. In Handbook of Phytoremediation; Golubev, I.A., Ed.; Nova Science: New York, NY, USA, 2010; pp. 763–774. [Google Scholar]
- Sun, L.; Zhang, M.; Ren, J.; Qi, J.; Zhang, G.; Leng, P. Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant Biol. 2010, 10, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, B.; Yin, C.-C.; He, S.-J.; Lu, X.; Zhang, W.-K.; Lu, T.-G.; Chen, S.-Y.; Zhang, J.-S. Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) Seedling. PLoS Genet. 2014, 10, e1004701. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Nang, M.P.S.H.; Oshima, K.; Ishibashi, Y. The ethylene signal mediates induction of gmatg8i in soybean plants under starvation stress. Biosci. Biotechnol. Biochem. 2011, 75, 1408–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depaepe, T.; Hendrix, S.; van Rensburg, H.C.J.; Van den Ende, W.; Cuypers, A.; Van der Straeten, D. At the crossroads of survival and death: The reactive oxygen species–ethylene–sugar triad and the unfolded protein response. Trends Plant Sci. 2021, 26, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Minina, E.A.; Moschou, P.N.; Vetukuri, R.R.; Sanchez-Vera, V.; Cardoso, C.; Liu, Q.; Elander, P.H.; Dalman, K.; Beganovic, M.; Lindberg Yilmaz, J.; et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 2018, 69, 1415–1432. [Google Scholar] [CrossRef]
- Lee, H.Y.; Park, H.L.; Park, C.; Chen, Y.-C.; Yoon, G.M. Reciprocal antagonistic regulation of E3 ligases controls ACC synthase stability and responses to stress. Proc. Natl. Acad. Sci. USA 2021, 118, e2011900118. [Google Scholar] [CrossRef]
- Mergemann, H.; Sauter, M. Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol. 2000, 124, 609–614. [Google Scholar] [CrossRef] [Green Version]
- Jia, W.; Ma, M.; Chen, J.; Wu, S. Plant morphological, physiological and anatomical adaption to flooding stress and the underlying molecular mechanisms. Int. J. Mol. Sci. 2021, 22, 1088. [Google Scholar] [CrossRef]
- Zhu, T.; Zou, L.; Li, Y.; Yao, X.; Xu, F.; Deng, X.; Zhang, D.; Lin, H. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol. J. 2018, 16, 2063–2076. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pérez, M.E.; Lemaire, S.D.; Crespo, J.L. Reactive oxygen species and autophagy in plants and Algae. Plant Physiol. 2012, 160, 156–164. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Mugume, Y.; Bassham, D.C. New advances in autophagy in plants: Regulation, selectivity and function. Semin. Cell Dev. Biol. 2018, 80, 113–122. [Google Scholar] [CrossRef]
- Verma, I.; Roopendra, K.; Sharma, A.; Chandra, A.; Kamal, A. Expression analysis of genes associated with sucrose accumulation and its effect on source–sink relationship in high sucrose accumulating early maturing sugarcane variety. Physiol. Mol. Biol. Plants 2019, 25, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, D.; Betsuyaku, E.; Terashima, I. Interspecific differences in how sink—Source imbalance causes photosynthetic downregulation among three legume species. Ann. Bot. 2019, 123, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Zhaowei, L.; Qian, Z.; Fangmin, C. Sugar starvation enhances leaf senescence and genes involved in sugar signaling pathways regulate early leaf senescence in mutant rice. Rice Sci. 2020, 27, 201–214. [Google Scholar] [CrossRef]
- Ashraf, M.; Ahmad, M.S.A.; Öztürk, M.; Aksoy, A. Crop improvement through different means: Challenges and prospects. In Crop Production for Agricultural Improvement; Ashraf, M., Öztürk, M., Ahmad, M., Aksoy, A., Eds.; Springer Science + Business Media: Dordrecht, The Netherlands, 2012. [Google Scholar] [CrossRef]
- Arora, N.K. Impact of climate change on agriculture production and its sustainable solutions. Environ. Sustain. 2019, 2, 95–96. [Google Scholar] [CrossRef] [Green Version]
- Arora, N.K.; Fatima, T.; Mishra, I.; Verma, M.; Mishra, J.; Mishra, V. Environmental sustainability: Challenges and viable solutions. Environ. Sustain. 2018, 1, 309–340. [Google Scholar] [CrossRef]
- Silva, L.C.R.; Lambers, H. Soil-plant-atmosphere interactions: Structure, function, and predictive scaling for climate change mitigation. Plant Soil 2021, 461, 5–27. [Google Scholar] [CrossRef]
- Zhang, Y.; Luan, Q.; Jiang, J.; Li, Y. Prediction and utilization of malondialdehyde in exotic pine under drought stress using near-infrared spectroscopy. Front. Plant Sci. 2021, 12, 735275. [Google Scholar] [CrossRef]
- Bashan, Y.; de-Bashan, L.E.; Prabhu, S.R. Advances in plant growth-promoting bacterial inoculant technology: Formulations and practical perspectives (1998–2013). Plant Soil 2014, 378, 1–33. [Google Scholar] [CrossRef] [Green Version]
- Shaaban, M.; Van Zwieten, L.; Bashir, S.; Younas, A.; Nunez-Delgado, A.; Afzal Chhajro, M.; Kubar, K.A.; Ali, U.; Shoaib Rana, M.; Mehmood, M.A.; et al. A concise review of biochar application to agriculturalsoils to improve soil conditions and fight pollution. J. Environ. Manag. 2018, 228, 429–440. [Google Scholar] [CrossRef]
- Bonfante, P.; Anca, I.A. Plants, mycorrhizal fungi, and bacteria: A network of interactions. Annu. Rev. Microbiol. 2009, 63, 363–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonfente, P.; Genre, A. Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonfante, P.; Requena, N. Dating in the dark: How roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 2011, 14, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Hamid, B.; Zaman, M.; Farooq, S.; Fatima, S.; Sayyed, R.Z.; Baba, Z.A.; Sheikh, T.A.; Reddy, M.S.; El Enshasy, H.; Gafur, A.; et al. Bacterial plant biostimulants: A sustainable way towards improving growth, productivity, and health of crops. Sustainability 2021, 13, 2856. [Google Scholar] [CrossRef]
- Chen, H.; Dong, J.; Wang, T. Autophagy in plant abiotic stress management. Int. J. Mol. Sci. 2021, 22, 4075. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koyro, H.-W.; Huchzermeyer, B. From Soil Amendments to Controlling Autophagy: Supporting Plant Metabolism under Conditions of Water Shortage and Salinity. Plants 2022, 11, 1654. https://doi.org/10.3390/plants11131654
Koyro H-W, Huchzermeyer B. From Soil Amendments to Controlling Autophagy: Supporting Plant Metabolism under Conditions of Water Shortage and Salinity. Plants. 2022; 11(13):1654. https://doi.org/10.3390/plants11131654
Chicago/Turabian StyleKoyro, Hans-Werner, and Bernhard Huchzermeyer. 2022. "From Soil Amendments to Controlling Autophagy: Supporting Plant Metabolism under Conditions of Water Shortage and Salinity" Plants 11, no. 13: 1654. https://doi.org/10.3390/plants11131654
APA StyleKoyro, H.-W., & Huchzermeyer, B. (2022). From Soil Amendments to Controlling Autophagy: Supporting Plant Metabolism under Conditions of Water Shortage and Salinity. Plants, 11(13), 1654. https://doi.org/10.3390/plants11131654







