Abstract
Citrus melanose is a fungal disease caused by Diaporthe citri F.A. Wolf. It is found in various citrus-growing locations across the world. The host range of D. citri is limited to plants of the Citrus genus. The most economically important hosts are Citrus reticulata (mandarin), C. sinensis (sweet orange), C. grandis or C. maxima (pumelo), and C. paradisi (grapefruit). In the life cycle of D. citri throughout the citrus growing season, pycnidia can be seen in abundance on dead branches, especially after rain, with conidia appearing as slimy masses discharged from the dead twigs. Raindrops can transmit conidia to leaves, twigs, and fruits, resulting in disease dispersion throughout small distances. Persistent rains and warm climatic conditions generally favor disease onset and development. The melanose disease causes a decline in fruit quality, which lowers the value of fruits during marketing and exportation. High rainfall areas should avoid planting susceptible varieties. In this article, information about the disease symptoms, history, geographic distribution, epidemiology, impact, and integrated management practices, as well as the pathogen morphology and identification, was reviewed and discussed.
1. Introduction
1.1. Major Fungal Diseases on Citrus
Several citrus diseases are currently documented in China and around the world. The generally occurring fungal diseases include melanose, gummosis, and stem-end rot caused by Diaporthe spp.; branch cankers caused by Botryosphaeriaceae [1,2]; scab caused by Elsinoë spp. [3,4,5,6,7,8,9]; black rot caused by Alternaria spp. [10,11,12,13,14]; greasy leaf spot caused by Cercosporoid genus [15,16]; anthracnose caused by Colletotrichum spp. [17,18,19,20,21,22,23,24,25]; and blue and green mold caused by Penicillium spp. [26,27,28]. Among these fungal diseases, melanose, gummosis, and stem-end rot caused by Diaporthe spp. have a significant impact on citrus production [29,30]. At the same time, some Diaporthe spp. have also been reported as endophytes and/or saprobes on citrus [29,30,31,32,33,34,35,36,37].
Melanose disease was not a major problem in citrus crops prior to the 1990s. However, the accumulation of a large number of dead branches or trees results in an increase in fungal inocula in old citrus orchards worldwide. Currently, melanose has become the major fungal disease of citrus in China, dramatically reducing the commercial value of citrus fruits (Figure 1). Diaporthe spp., have been isolated from citrus hosts in many citrus-growing regions of China, e.g., Jiangxi, Zhejiang, Guangxi, Guangdong, Shaanxi, Fujian, Hunan, Chongqing, Yunnan, etc.

Figure 1.
The typical symptoms of melanose disease in the field with different citrus tissue causal agents by Diaporthe species: (A) pumelo fruit (C. maxima) from Chongqing; (B,C) orange fruits (C. sinensis) from Chongqing; (D) young orange leaf (C. sinensis var. Brasliliensis) from Guizhou; (E) mandarin leaf (Citrus sp.) from Zhejiang; (F) orange fruits (C. sinensis) from Chongqing; (G) citrus fruit (C. changshan-huyou) from Zhejiang; and (H) mandarin fruit (C. reticulata) from Zhejiang.
1.2. Diaporthe Species Associated with Citrus
Previous studies about Diaporthe spp. have largely concentrated on species identification, especially the species associated with specific hosts. The molecular taxonomy of the genus Diaporthe related to citrus and allied taxa has made great advances in recent years. The phylogenies based on multiple loci provide a more robust and comprehensible taxonomy and nomenclature for D. citri and will serve as a starting point for field study by plant pathologists, breeders, and mycologists. Such information may be used to improve disease management and the deployment of citrus cultivars with species-specific and/or broad-spectrum resistance.
All citrus species, including grapefruit, clementine, lemon, lime, mandarin, orange, satsuma, and tangerine, are susceptible to melanose. Phomopsis citri was first recorded as a citrus parasitic fungus causing stem-end rot symptoms in Florida, USA [38]. Its teleomorph (sexual stage) is D. citri [39]. In addition to D. citri, many other Diaporthe species were also detected in citrus hosts. They could be pathogens, endophytes, or saprobes on citrus [29,31,40,41,42,43,44,45]. The summary of the global distribution of Diaporthe species associated with citrus hosts and their allied genera confirmed with DNA sequences is shown in Table 1.

Table 1.
Summary of the global distribution of Diaporthe species associated with Citrus hosts and their allied genera confirmed with DNA sequences.
1.3. Identification and Molecular Diagnostics
Citrus melanose is caused by D. citri, which belongs to Kingdom Fungi; Ascomycota; Sordariomycetes; Diaporthales; Diaporthaceae; Diaporthe [57,58,59,60,61,62,63,64]. The genus of Diaporthe was established by Nitschke [65]. Phomopsis is the anamorphic (asexual stage) name of Diaporthe [38,63,66,67,68,69,70]. The genus Diaporthe shows high species diversity; more than 1200 species named “Diaporthe” and about 1050 species named “Phomopsis” have been recorded in MycoBank lists (http://www.mycobank.org; accessed on 9 June 2021).
1.3.1. Morphological Characteristics
For taxonomy of Diaporthe species, morphological characterization based on conidia morphology, fruiting body structure, and culture characteristics has been the basis of this study [71,72,73,74]. On PDA culture medium, mycelium is typically fan-shaped and white in color [75]. Teleomorphic ascomata, which are usually immersed in the substrate erumpent through pseudostromata mostly surrounding the ascomata, have more or less elongated perithecial necks. The pseudostromata are distinct and often delimited by dark lines [76]. The perithecia are circular, flattened at the base, with long black beaks [39,77]. The perithecia generally remain within the plant’s bark but protrude out of the stem surface, which makes them easily visible under a dissecting microscope. Asci are unitunicate and clavate to cylindrical, loosening from the ascogenous cells at an early stage and lying free in the ascocarp. Ascospores are biseriate to uniseriate, and there are two oil droplets or guttulae within each cell, which are fusoid, ellipsoid to cylindrical, septate, straightly constricted at the septum, inequilateral or curved, hyaline, and sometimes with appendages [76,78]. Because ascospores are forcibly ejected from the asci, they become windborne and are responsible for the long-distance spread of the pathogen [39]. Upon finding a suitable substrate, spores may germinate, producing hyphae that quickly become septate mycelium [77].
The anamorphic state of this fungus is the most important stage for the disease cycle. The pycnidia (asexual fruiting bodies) of D. citri are scattered on the substratum and are dark in color, ovoid, thick-walled, and erumpent. Conidiophores are hyaline and branched, and occasionally, they are short and 1–2 septate. Conidiogenous cells were phialidic, hyaline, and slightly tapering toward the apex [30,37]. Generally, they are multiseptate and filiform with enteroblastic and monophiladic conidiogenesis [79,80]. It may produce three types of hyaline, non-septate conidia, namely, alpha, beta conidia [81], as well as an intermediate between these two conidial types, namely, gamma conidia [82,83,84]. The alpha conidia are functional, aseptate, single-celled, hyaline, fusiform, and usually biguttulate but sometimes lack guttula (lipid drop) or have more guttulae. The beta conidia tend to be produced in older pycnidia and are also aseptate, hyaline, long, slender, rod-shaped structures. They may be filiform and straight, but more often they are hooked at one end, lack guttula, and do not germinate [85]. The gamma conidia are hyaline, multiguttulate, fusiform to subcylindrical, with an acute or rounded apex, while the base is sometimes truncate [73,82,83,84,86,87]. The asexual morphology and cultural characteristics of D. citri are shown in Figure 2.
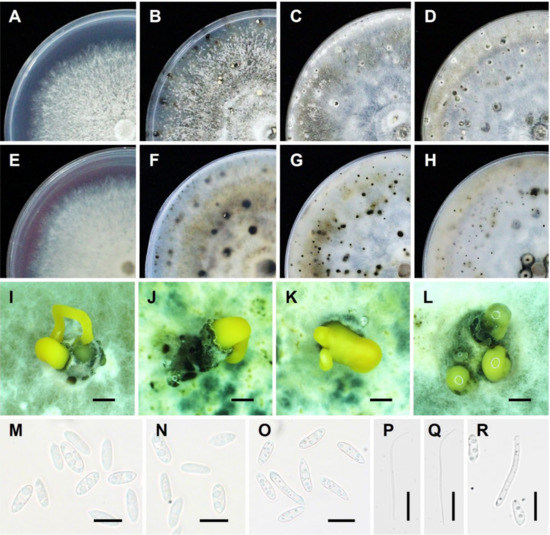
Figure 2.
Asexual morphology and cultural characteristics of D. citri: (A,E) culture on PDA medium after 7 days; (B,F) culture on potato dextrose agar (PDA) medium after 30 days; (C,G) culture on corn meal agar (CMA) medium after 30 days; (D,H) culture on oatmeal agar (OMA) medium after 30 days; (I–L) conidiomata sporulating on PDA medium after 30 days; (M–O) Alpha conidia; (P,Q) Beta conidia; and (R) Alpha and Gamma conidia. Note: (A–D) surface and (E–H) reversed sides of colony culture. Scale bar: (I–L) = 200 μm; (M–R) = 10 μm.
1.3.2. Molecular Identification
Currently, four nuclear genome sequences of D. citri have been deposited in GenBank with the accession numbers JACTAD000000000, JADAZQ000000000, JADAZP000000000, and JADAZO000000000 for strains NFHF-8-4, ZJUD2, ZJUD14, and Q7, respectively [88,89]. The genome assembly sizes of ZJUD2 (59.5 Mp) and ZJUD14 (52.0 Mp) were relatively shorter, while NFHF-8-4 and Q7 contained longer assembly size (more than 63 Mp) [88,89].
Taylor et al. [90] proposed genealogical concordance phylogenetic species recognition (GCPSR), which compares individual gene sequences to find inconsistencies, and it has been shown to be very useful in defining species boundaries in morphologically conserved fungi [91]. Although each cluster in combined trees is usually considered a separate lineage, the common approach of concatenating sequenced data to delimit species without using the GCPSR principle overestimates the real diversity of species placement [91,92,93,94,95,96]. Since the widespread use of DNA sequences [35], genus Diaporthe species identification has progressed beyond host association and morphological characterization [73,81]. The Diaporthe genus is commonly represented by using traditional molecular barcoding for fungal species identification based on nuclear ribosomal internal transcribed spacer regions (ITS) [70,97,98,99]. As a result, some Diaporthe species have been reported to be perplexing, with contradictory findings when only the ITS sequence is used to produce a phylogenetic tree [35,67,99,100,101,102].
According to previous studies, multi-locus phylogenetic analysis has been proved more efficient to identify isolates at the species level [29,35,102,103,104,105]. Several loci, including large subunit of the ribosomal DNA (LSU), intergenic spacers of the ribosomal DNA (IGS), ITS, translation elongation factor 1-α gene (TEF1-α), ß-tubulin gene (TUB2), histone 3 gene (HIS3), calmodulin gene (CAL), actin gene (ACT), DNA-lyase gene (APN2), 60S ribosomal protein L37 gene (FG1093), and mating type genes (MAT-1-1-1 and MAT-1-2-1), are demonstrated as efficient tools to determine Diaporthe species. Even molecular sequences are already being used to identify species and rebuild phylogenies, complete genome sequences for Diaporthe species are still in the future. Currently, the most frequently used molecular loci in this genus are the ITS, TEF1-α, TUB2, HIS3, and CAL [35,58,98,104]. Among them, TEF1-α is the most efficient tool in resolving the phylogenetic signal of the D. eres species complex [101,106]. Similarly, the highly variable TEF1-α was also shown to be the most efficient locus in distinguishing Diaporthe species [99,101,104,106,107]. Although the ITS region showed the relatively limited delimitation of Diaporthe species in phylogenetic analyses, it is still informative and should not be excluded from concatenation analysis of multi-locus DNA sequences [58,94,98,104]. A summary of universal and species-specific primers used for species determination within the Diaporthe genus is shown in Table 2.

Table 2.
Summary of published universal primers and species-specific primers used for species determination within Diaporthe spp.
1.3.3. Molecular Diagnosis
A conventional species-specific PCR method has been developed to distinguish D. citri from other Diaporthe species [32,46,117]. The PCR-based technique showed outstanding specificity and sensitivity, indicating that it may be used to effectively detect D. citri in practice. Effective PCR with citrus tissues infected by D. citri, as well as modern PCR, isothermal amplification, or any technique that is fast, low-cost, and accurate for alternative detection of certain diseases, should be developed, because such methods may also be applied for phytosanitary detection in plant quarantine.
1.3.4. Genetic Populations
Although D. citri and other Diaporthe infections are well-known, information about their diversity, population genetics, reproductive methods, and pathogenicity is limited [125,126]. Our understanding of the infection process, host range, and fungicide resistance of D. citri would improve if we understood its population genetics in nature. Such information is also useful for making long-term management strategies for this disease [127,128,129]. In China, the population genetics of D. citri was analyzed by using polymorphic simple sequence repeat (SSR) markers and the mating type idiomorphs. The majority of the analyzed samples came from southern China, including Fujian, Zhejiang, Jiangxi, Hunan, and Guizhou provinces. It was shown that alleles at the 14 SSR loci were not substantially different from linkage equilibrium, and most subpopulations exhibited equal frequencies of the two mating types. The findings suggest that teleomorphic reproduction is important in D. citri populations in southern China, and the ascospores seem to be a major contributor to citrus disease [46]. The presence of significant genetic differences among different geographical populations, however, does not eliminate the possibility of migration. Closely related strains were detected from many geographically diverse regions. They also found signs of genetic mixing between two extremely distinct genetic populations. These findings imply that D. citri populations are evolving, which might be accelerated by either increasing human impacts through frequent citrus seedling exchange or by global climate change [46].
2. Epidemiology, Life Cycle, and Symptomatology
Citrus melanose is caused by D. citri, which attacks foliage, fruits, and twigs when they are immature. Since mature tissues are more immune to pathogen attack, the first 8 to 9 weeks of the citrus growing season are the most vulnerable to pathogen attack. Melanose signs can differ depending on the severity of the infection. At the end of the susceptibility cycle, the flyspeck melanose symptoms appear [130,131].
The fungal inocula can be scattered over a wide range since ascospores are released forcefully and can be spread over a long distance. D. citri is primarily a saprophyte that feeds on and receives its nutrition from dead wood [44,132]. Perithecia and pycnidia are only found on dead and dying twigs and fruits showing stem-end rot. The conidia provided by pycnidia are the primary source of inoculum [133]. Ascospores are ejected forcibly and play a significant role in long-distance dispersal [132]. As a result of the widespread dissemination of a vast number of ascospores, the number of cases of infection is rising [134]. When ascospores or conidia of Diaporthe land on the surface of a plant, the disease will be triggered. Pathogens thrive in dry environments with temperatures ranging from 17 to 35 °C [44].
The germination of spores requires approximately 10 to 24 h of moisture, depending on the temperature [44,134], and the germination and formation of a germ tube takes 36 to 48 h [133]. After that, the citrus melanose pathogen directly penetrates the cuticle layer tissue and infects the plant.
D. citri could overwinter on debris, e.g., mummy fruits, dead stems, branches, and dry leaves. Perithecia could form on debris next year. Ascospores are produced in proportion to the amount of dead wood present in a canopy. These spores contribute slightly to the disease severity of an orchard, but they are carried by the wind and spread across long distances. Conidia, developed in mature pycnidia, can continuously infect citrus during the growing season. Conidia can be dispersed to nearby citrus trees with raindrops, which most probably cause the majority of fruit infections (Figure 3). Nevertheless, conidia can also be transmitted through the air over long distances when rainfall is scarce.
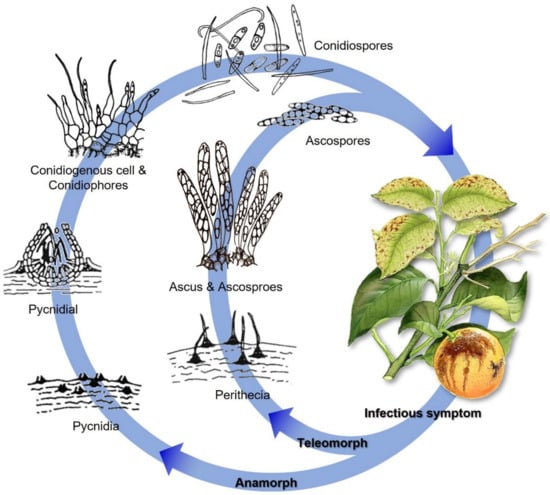
Figure 3.
Representative Diaporthe disease cycle: melanose disease cycle on citrus caused by D. citri. Revised and redrawn from Burnett [135], Timmer et al. [75], and Udayanga et al. [63].
Symptoms appear as discrete small, sunken, brown spots about one week after infection, which later become raised and filled with reddish-brown gum. The leaf pustules are initially surrounded by a yellow halo. Diseased areas regreen later and create corky pustules. On fruits, pustules can grow relatively large and can crack, creating a pattern of mudcake. The severity of the disease is determined mainly by the amount of inoculum-bearing dead wood in the canopy of the tree and the duration of the wetting period following rainfall or sprinkler irrigation. Wet, rainy conditions, especially when rain showers occur late in the day, and fruits staying continuously wet on warm nights are conducive to infection.
3. Geographic Distribution and Host Associations
The USDA’s Agricultural Research Service’s Systematic Mycology and Microbiology Laboratory (SMML database: https://nt.ars-grin.gov/fungaldatabases/, accessed on 10 December 2021) and the Centre for Agriculture and Bioscience International (CABI database: https://www.cabi.org/isc/, accessed on 15 January 2022) obtained some information about D. citri, including the geographic distribution and host associations. According to the SMML and CABI databases, D. citri has been discovered on citrus hosts and related species all over the world. The D. citri is the most predominant species in the Diaporthe genus, which occurs widely in citrus-growing countries, e.g., China, Philippines, Japan, Korea, Thailand, Myanmar, Cambodia, Fiji, Mauritius, United States, Mexico, Haiti, Cuba, Dominican Republic, Panama, Puerto Rico, Venezuela, Trinidad and Tobago, Brazil, Cyprus, Portugal (Azores Islands), New Zealand, Niue, Samoa, Tonga, Cook Islands, Cote d’Ivoire, and Zimbabwe, which has also been summarized previously [32]. A global geographic distribution map of D. citri associated with the citrus hosts is available on the CABI database (accessed and last modified on 16 November 2021) and is shown in Figure 4. The green disease-free areas may mean that no data is in the CABI database, but this does not necessarily mean that the disease is absent.
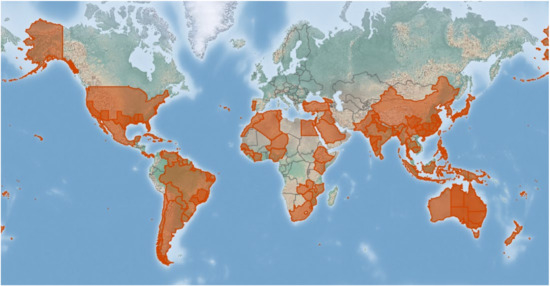
Figure 4.
A global geographic distribution map of D. citri associated with the citrus-host plant is available on the CABI database.
4. Main Management Approaches of Melanose Disease
The yield is almost unaffected by melanose disease, and the juice processing is unaffected as well. However, the quality of the fruit for marketing and exportation suffers the consequences. In order to avoid poor quality and fruit deterioration caused by citrus melanose, integrated management practices should be implemented. Integrated pest management (IPM) is now largely recognized as the most effective way to protect plants. Its ultimate objective will be to maintain pest populations below economically injurious levels without or just with minimal pesticides. Although IPM must rely on pesticides currently, minimizing chemical inputs while maintaining crop quality at an economically viable level is a basic requirement for plant protection. To achieve this goal, it is critical to understand the disease epidemiology at various points in time while performing pest control [132,136].
Currently, no resistance cultivars are available for melanose control in practice. The removal of dead wood to reduce the pressure of melanose fungus is both time-consuming and labor-intensive. Nevertheless, pruning dead branches should be performed on a regular basis. Proper pruning enhances air circulation within the canopy of the plant, keeping it dry and reducing opportunities for pathogens to survive and cause infections. It will also improve the effectiveness of fungicide infiltration into the foliage [43,137]. Furthermore, avoid planting sensitive citrus cultivars or species in high-rainfall zones, such as sweet orange, grapefruit, and pumelo [137,138]. Other management practices, such as citrus plantations in low-rainfall and sunny zones, should be implemented. Interplanting citrus with non-susceptible hosts is also a feasible measure [137,139].
4.1. Chemical Control
Application of fungicides is still the most commonly used method to control melanose disease on citrus. Many fungicides have been tested for melanose control. Copper is a protective compound, which forms a layer on the surface of plant tissue, e.g., fruit, protecting it from infection. The gap in the protective copper layer, however, grows larger as the fruit grows and expands. If conditions are favorable for the pathogen infection, the copper layer needs to be renewed through another spray. The melanose fungus stored in dead wood is slightly affected by copper spraying. The use of copper fungicides before flowering will not reduce infection. A copper fungicide must be applied on the fruit surface to provide efficient melanose control. In the case of serious infection in late summer, additional protectant spray should be applied [140]. Applications of pyraclostrobin to the spring flush growth of citrus trees are much more efficient for controlling melanose, scab, and Alternaria brown spot than those of famoxadone or copper hydroxide [44,141,142,143,144]. Bushong and Timmer [145] demonstrated that azoxystrobin was a highly effective preventative spray for melanose, whereas benomyl and fenbuconazole were not. As post-infection treatments for melanose, none of the fungicides are successful. In Japan, dithianon and mancozeb were used to spray alternately from June to August to control this disease [146]. In Pakistan, five chemicals were tested at recommended doses, including penflufen, copper hydroxide, tebuconazole plus trifloxystrobin, and difenoconazole, for controlling melanose disease. When used as a protectant, copper hydroxide was found to be the most effective for the management of citrus melanose [147]. Whereas Anwar et al. [148] evaluated six different fungicides for citrus melanose control, the use of mancozeb led to a significant inhibition of fungal growth. Similarly, several chemicals, including mancozeb and fenbuconazole, were found to be effective in controlling citrus melanose in China and other countries [40,43,148,149,150].
4.2. Biological Control
Although chemical control plays an important role in managing plant diseases, overuse of chemical pesticides has raised severe issues about food contamination, environmental pollution, and phytotoxicity. Biocontrol is a viable option as it is friendly to the environment. Biological control of plant diseases with antagonistic bacteria is a viable alternative to chemical control. Many antagonistic bacteria are known to play important roles in the sustainability of natural ecosystems, and some of them can be employed as inoculants to stimulate plant growth and resistance.
For melanose control, more and more biocontrol candidates have been developed, e.g., Burkholderia gladioli: TRH423-3, MRL408-3, Pseudomonas pudia: THJ609-3, and P. fluorescens: TRH415-2, and selected for their antifungal effectiveness against D. citri using dual-culture testing. Disease suppression was observed after pretreatment with the rhizobacterial strains, with varying degrees of protection rates for each rhizobacterial strain. Following the pathogen inoculation, subsequent treatment with the rhizobacterial strains also enhanced protection rates. The rhizobacterial strains might be especially useful in organic citrus production where chemicals are strictly forbidden [151]. Similarly, pre-treatment with P. putida strain THJ609-3 resulted in a decreased disease incidence. When the infection behaviors of D. citri and necrosis deposits on plant tissues were examined using a fluorescent microscope, it was shown that the process of disease development was reduced after being treated with the bacterial strain, especially the conidia germination rates, which were significantly lower after being pretreated with the strain THJ609-3. Furthermore, morphological abnormalities of the germ tubes were also observed. These results pointed to the bacterial-direct antifungal action on the leaf surfaces as a potential cause of disease reduction [152]. Thiobacillus species were used to generate bio-sulfur, which was investigated as an alternative to managing citrus melanose. It was found that melanose disease severity was lower on bio-sulfur pretreated citrus leaves than on untreated leaves, suggesting that bio-sulfur might be applied as an environmentally friendly alternative to control citrus melanose [153]. Bacillus velezensis CE 100, an effective biocontrol agent, has been used to control D. citri. In dual culture plates, D. citri mycelial growth was significantly suppressed by strain CE 100, suggesting that some volatile substances inhibited the growth of D. citri. It was also observed that the bacterial culture filtrate (BCF) of strain CE 100 inhibited D. citri growth. Microscopic examination indicated that BCF had a substantial impact on the pathogen hyphal shape, most probably the result of numerous cell-wall disintegrating enzymes and metabolites generated by strain CE 100. Interestingly, D. citri conidial germination was decreased by approximately 80% when 50% BCF of strain CE 100 was used [154].
5. Conclusions
In this paper, the history of citrus melanose, pathogen morphology, molecular identification, population studies, epidemiology of disease symptoms and life cycle, global distribution, and integrated disease management are documented. At present, there are no cases of plants bred or engineered specifically for resistance to diseases caused by D. citri. Does the teleomorph D. citri have higher opportunities for surviving on different hosts? Does genetic recombination play an important role in survival or in variability in this species? Are teleomorph ascospores spread differently from anamorphic conidia? Do ascospores and conidia infect citrus tissues in the same way? Which defense reactions occur in infected plants? At what level is the pathogen D. citri recognized by the plant? Are signal cascades of defensive reactions known? These are some of the questions about pathogen epidemiology that still need to be answered as they directly impact disease management. However, more understanding of the molecular mechanisms that confer virulence on D. citri is helpful in the development of alternative disease management strategies, especially when it is urgent to develop environmentally friendly approaches or tools to maintain the plant health in the future.
Author Contributions
Conceptualization, C.C.; writing—original draft preparation, C.C. and X.L.; writing—review and editing, C.C., X.L., Y.L. and C.L.; supervision, C.L.; project administration and funding acquisition, Y.L. and C.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Fundamental Research Funds for the Central Universities (No. 2662020ZKPY018) and the National Key Research and Development Program of China (No. 2017YFD020200103).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank Margaret Senior (https://www.dpi.nsw.gov.au/about-us/services/collections/scientific-illustrations/senior/citrus/melanose-citrus (accessed on 1 June 2021); Department of Primary Industries, Government of New South Wales) for providing an illustration of melanose infected by D. citri to re-draw the general life cycle of citrus melanose disease. The authors also sincerely thank the reviewers for their contributions during the revision process.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
References
- BeZerra, J.D.P.; Crous, P.W.; Aiello, D.; Gullino, M.L.; Polizzi, G.; Guarnaccia, V. Genetic diversity and pathogenicity of Botryosphaeriaceae species associated with symptomatic citrus plants in Europe. Plants 2021, 10, 492. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.E.; Wang, W.; Crous, P.W.; Wang, H.K.; Jiao, C.; Huang, F.; Pu, Z.X.; Zhu, Z.R.; Li, H.Y. Species of Botryosphaeriaceae associated with citrus branch diseases in China. Persoonia 2021, 47, 106–135. [Google Scholar] [CrossRef]
- Chung, K.R. Elsinoë fawcettii and Elsinoë australis: The fungal pathogens causing citrus scab. Mol. Plant Pathol. 2011, 12, 123–135. [Google Scholar] [CrossRef]
- Fan, X.L.; Barreto, R.W.; Groenewald, J.Z.; Bezerra, J.D.; Pereira, O.L.; Cheewangkoon, R.; Mostert, L.; Tian, C.M.; Crous, P.W. Phylogeny and taxonomy of the scab and spot anthracnose fungus Elsinoë (Myriangiales, Dothideomycetes). Stud. Mycol. 2017, 87, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Gopal, K.; Govindarajulu, B.; Ramana, K.T.V.; Kumar, C.S.K.; Gopi, V.; Sankar, T.G.; Lakshmi, L.M.; Lakshmi, T.N.; Sarada, G. Citrus scab (Elsinoë fawcettii): A review. J. Agric. Allied Sci. 2014, 3, 49–58. [Google Scholar]
- Hyun, J.W.; Yi, S.H.; MacKenzie, S.J.; Timmer, L.W.; Kim, K.S.; Kang, S.K.; Kwon, H.M.; Lim, H.C. Pathotypes and genetic relationship of worldwide collections of Elsinoë spp. causing scab disease of citrus. Phytopathology 2009, 99, 721–728. [Google Scholar] [CrossRef] [Green Version]
- Miles, A.K.; Tan, Y.P.; Shivas, R.G.; Drenth, A. Novel pathotypes of Elsinoë australis associated with Citrus australasica and Simmondsia chinensis in Australia. Trop. Plant Pathol. 2015, 40, 26–34. [Google Scholar] [CrossRef]
- Timmer, L.W.; Priest, M.; Broadbent, P.; Tan, M.K. Morphological and pathological characterization of species of Elsinoë causing scab diseases of citrus. Phytopathology 1996, 86, 1032–1038. [Google Scholar] [CrossRef]
- Whiteside, J.O. Biological characteristics of Elsinoë fawcetti pertaining to the epidemiology of sour orange scab. Phytopathology 1975, 65, 1170–1177. [Google Scholar] [CrossRef]
- Peever, T.L.; Carpenter-Boggs, L.; Timmer, L.W.; Carris, L.M.; Bhatia, A. Citrus black rot is caused by phylogenetically distinct lineages of Alternaria alternata. Phytopathology 2005, 95, 512–518. [Google Scholar] [CrossRef] [Green Version]
- Timmer, L.W.; Peever, T.L.; Solel, Z.; Akimitsu, K. Alternaria diseases of citrus—Novel pathosystems. Phytopathol. Mediterr. 2003, 42, 99–112. [Google Scholar]
- Timmer, L.W.; Solel, Z.; Orozco-Santos, M. Alternaria Brown Spot of Mandarins. In Compendium of Citrus Diseases; Timmer, L.W., Garnsey, S.M., Graham, J.H., Eds.; The American Phytopathological Society Press: Saint Paul, MI, USA, 2000. [Google Scholar]
- Woudenberg, J.H.C.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woudenberg, J.H.C.; Seidl, M.F.; Groenewald, J.Z.; de Vries, M.; Stielow, J.B.; Thomma, B.P.H.J.; Crous, P.W. Alternaria section alternaria: Species, formae speciales or pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pretorius, M.C.; Crous, P.W.; Groenewald, J.Z.; Braun, U. Phylogeny of some cercosporoid fungi from Citrus. Sydowia 2003, 55, 286–305. [Google Scholar]
- Huang, F.; Groenewald, J.Z.; Zhu, L.; Crous, P.W.; Li, H.Y. Cercosporoid diseases of Citrus. Mycologia 2015, 107, 1151–1171. [Google Scholar] [CrossRef] [Green Version]
- Bragança, C.A.D.; Damm, U.; Baroncelli, R.; Júnior, N.S.M.; Crous, P.W. Species of the Colletotrichum acutatum complex associated with anthracnose diseases of fruit in Brazil. Fungal Biol. 2016, 120, 547–561. [Google Scholar] [CrossRef]
- Damm, U.; Cannon, P.F.; Woudenberg, J.H.C.; Johnston, P.R.; Weir, B.S.; Tan, Y.P.; Shivas, R.G.; Crous, P.W. The Colletotrichum boninense species complex. Stud. Mycol. 2012, 73, 75–78. [Google Scholar] [CrossRef] [Green Version]
- Damm, U.; Woudenberg, J.H.C.; Cannon, P.F.; Crous, P.W. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Divers. 2009, 39, 45–87. [Google Scholar]
- Guarnaccia, V.; Groenewald, J.Z.; Polizzi, G.; Crous, P.W. High species diversity in Colletotrichum associated with citrus diseases in Europe. Persoonia 2017, 39, 32–50. [Google Scholar] [CrossRef]
- Honger, J.O.; Offei, S.K.; Oduro, K.A.; Odamtten, G.T.; Nyaku, S.T. Identification and molecular characterisation of Colletotrichum species from avocado, citrus and pawpaw in Ghana. S. Afr. J. Plant Soil 2016, 33, 177–185. [Google Scholar] [CrossRef]
- Huang, F.; Chen, G.Q.; Hou, X.; Fu, Y.S.; Cai, L.; Hyde, K.D.; Li, H.Y. Colletotrichum species associated with cultivated citrus in China. Fungal Divers. 2013, 61, 61–74. [Google Scholar] [CrossRef]
- Peng, L.J.; Yang, Y.L.; Hyde, K.D.; Bahkali, A.H.; Liu, Z.Y. Colletotrichum species on citrus leaves in Guizhou and Yunnan provinces, China. Cryptogam. Mycol. 2012, 33, 267–283. [Google Scholar]
- Wang, W.; de Silva, D.D.; Moslemi, A.; Edwards, J.; Ades, P.K.; Crous, P.W.; Taylor, P.W.J. Colletotrichum species causing anthracnose of citrus in Australia. J. Fungi 2021, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, G.J.; Eckert, J.W.; Pitt, J.I. Revised description of Penicillium ulaiense and its role as a pathogen of citrus fruits. Phytopathology 1994, 84, 719–727. [Google Scholar] [CrossRef]
- Tashiro, N.; Manabe, K.; Ide, Y. First report of whisker mold, a postharvest disease on citrus caused by Penicillium ulaiense (in Japan). J. Gen. Plant Pathol. 2012, 78, 140–144. [Google Scholar] [CrossRef]
- Louw, J.P.; Korsten, L. Pathogenicity and host susceptibility of Penicillium spp. on citrus. Plant Dis. 2015, 99, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Guarnaccia, V.; Crous, P.W. Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 2017, 8, 317–334. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Hou, X.; Dewdney, M.M.; Fu, Y.S.; Chen, G.Q.; Hyde, K.D.; Li, H.Y. Diaporthe species occurring on citrus in China. Fungal Divers. 2013, 61, 237–250. [Google Scholar] [CrossRef]
- Huang, F.; Udayanga, D.; Wang, X.H.; Hou, X.; Mei, X.F.; Fu, Y.S.; Hyde, K.D.; Li, H.Y. Endophytic Diaporthe associated with citrus: A phylogenetic reassessment with seven new species from China. Fungal Biol. 2015, 119, 331–347. [Google Scholar] [CrossRef]
- Chaisiri, C.; Liu, X.Y.; Lin, Y.; Li, J.B.; Xiong, B.; Luo, C.X. Phylogenetic analysis and development of molecular tool for detection of Diaporthe citri causing melanose disease of citrus. Plants 2020, 9, 329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danggomen, A.; Visarathanonth, N.; Manoch, L.; Piasai, O. Morphological studies of endophytic and plant pathogenic Phomopsis liquidambaris and Diaporthe phaseolorum (P. phaseoli anamorph) from healthy plants and diseased fruits. Thai J. Agric. Sci. 2013, 46, 157–164. [Google Scholar]
- Dong, Z.Y.; Manawasinghe, I.S.; Huang, Y.H.; Shu, Y.X.; Phillips, A.J.; Dissanayake, A.J.; Hyde, K.D.; Xiang, M.M.; Luo, M. Endophytic Diaporthe associated with Citrus grandis cv. Tomentosa in China. Front. Microbiol. 2021, 11, 609387. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.R.; Glienke, C.; Videira, S.I.R.; Lombard, L.; Groenewald, J.Z.; Crous, P.W. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 2013, 31, 1–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.P.; Edwards, J.; Grice, K.R.E.; Shivas, R.G. Molecular phylogenetic analysis reveals six new species of Diaporthe from Australia. Fungal Divers. 2013, 61, 251–260. [Google Scholar] [CrossRef]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Hyde, K.D. Species limits in Diaporthe: Molecular re-assessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia 2014, 32, 83–101. [Google Scholar] [CrossRef] [Green Version]
- Fawcett, H.S. The cause of stem-end rot of citrus fruits (Phomopsis citri n. sp.). Phytopathology 1912, 2, 109–113. [Google Scholar]
- Wolf, F.A. The perfect stage of the fungus which causes melanose of citrus. J. Agric. Res. 1926, 33, 621–625. [Google Scholar]
- Chen, G.Q.; Jiang, L.Y.; Xu, F.S.; Li, H.Y. In vitro and in vivo screening of fungicides for controlling citrus melanose caused by Diaporthe citri. J. Zhejiang Univ. (Agric. Life Sci.) 2010, 36, 440–444. (In Chinese) [Google Scholar]
- Douanla-Meli, C.; Langer, E.; Mouafo, F.T. Fungal endophyte diversity and community patterns in healthy and yellowing leaves of Citrus limon. Fungal Ecol. 2013, 6, 212–222. [Google Scholar] [CrossRef]
- Habib, W.; Gerdes, E.; Antelmi, I.; Baroudy, F.; Choueiri, E.; Nigro, F. Diaporthe foeniculina associated with severe shoot blight of lemon in Lebanon. Phytopathol. Mediterr. 2015, 54, 149–150. [Google Scholar]
- Jiang, L.Y.; Xu, F.S.; Huang, Z.D.; Huang, F.; Chen, G.Q.; Li, H.Y. Occurrence and control of citrus melanose caused by Diaporthe citri. Acta Agric. Zhejiangensis 2012, 24, 647–653. (In Chinese) [Google Scholar]
- Mondal, S.N.; Vicent, A.; Reis, R.F.; Timmer, L.W. Saprophytic colonization of citrus twigs by Diaporthe citri and factors affecting pycnidial production and conidial survival. Plant Dis. 2007, 91, 387–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murali, T.S.; Suryanarayanan, T.S.; Geeta, R. Endophytic Phomopsis species: Host range and implications for diversity estimates. Can. J. Microbiol. 2006, 52, 673–680. [Google Scholar] [CrossRef]
- Xiong, T.; Zeng, Y.T.; Wang, W.; Li, P.D.; Gai, Y.P.; Jiao, C.; Zhu, Z.R.; Xu, J.P.; Li, H.Y. Abundant genetic diversity and extensive differentiation among geographic populations of the citrus pathogen Diaporthe citri in Southern China. J. Fungi 2021, 7, 749. [Google Scholar] [CrossRef] [PubMed]
- Guarnaccia, V.; Crous, P.W. Species of Diaporthe on Camellia and Citrus in the Azores Islands. Phytopathol. Mediterr. 2018, 57, 307–319. [Google Scholar]
- Yang, Q.; Jiang, N.; Tian, C.M. New species and records of Diaporthe from Jiangxi province, China. MycoKeys 2021, 77, 41–64. [Google Scholar] [CrossRef]
- Vakalounakis, D.J.; Ntougias, S.; Kavroulakis, N.; Protopapadakis, E. Neofusicoccum parvum and Diaporthe foeniculina associated with twig and shoot blight and branch canker of citrus in Greece. J. Phytopathol. 2019, 167, 527–537. [Google Scholar] [CrossRef]
- Tekiner, N.; Tozlu, E.; Guarnaccia, V. First report of Diaporthe foeniculina causing fruit rot of lemon in Turkey. J. Gen. Plant Pathol. 2020, 102, 277. [Google Scholar] [CrossRef] [Green Version]
- Juybari, H.Z.; Tajick Ghanbary, M.A.; Karimi, H.; Arzanlou, K.; Seasonal, M. Seasonal, tissue and age influences on frequency and biodiversity of endophytic fungi of Citrus sinensis in Iran. For. Pathol. 2019, 6, e12559. [Google Scholar] [CrossRef]
- Chaisiri, C.; Liu, X.Y.; Yin, W.X.; Luo, C.X.; Lin, Y. Morphology characterization, molecular phylogeny, and pathogenicity of Diaporthe passifloricola on Citrus reticulata cv. Nanfengmiju in Jiangxi province, China. Plants 2021, 10, 218. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.J.; Wei, X.; Xia, P.L.; Yi, J.P.; Yu, Z.H.; Deng, J.X.; Li, Q.L. Diaporthe taoicola and D. siamensis, two new records on Citrus sinensis in China. Mycobiology 2021, 49, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.Y.; Chung, W.C.; Huang, H.C.; Chung, W.H.; Chung, W.H. Identification of endophytic fungi of medicinal herbs of Lauraceae and Rutaceae with antimicrobial property. Taiwania 2012, 57, 229–241. [Google Scholar]
- Mahadevakumar, S.; Yadav, V.; Tejaswini, G.S.; Sandeep, S.N.; Janardhana, G.R. First report of Phomopsis citri associated with dieback of Citrus lemon in India. Plant Dis. 2014, 98, 1281. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, F.; Samsampour, D.; Seyahooei, M.A.; Bagheri, A.; Soltani, J. Diversity and spatiotemporal distribution of fungal endophytes associated with Citrus reticulata cv. Siyahoo. Curr. Microbiol. 2019, 76, 279–289. [Google Scholar] [CrossRef]
- Hongsanan, S.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Samarakoon, M.C.; Jeewon, R.; Zhao, Q.; Al-Sadi, A.M.; Bahkali, A.H. An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Divers. 2017, 84, 25–41. [Google Scholar] [CrossRef]
- Hyde, K.D.; Nilsson, R.H.; Alias, S.A.; Ariyawansa, H.A.; Blair, J.E.; Cai, L.; de Cock, A.W.A.M.; Dissanayake, A.J.; Glockling, S.L.; Goonasekara, I.D.; et al. One stop shop: Backbones trees for important phytopathogenic genera: I (2014). Fungal Divers. 2014, 67, 21–125. [Google Scholar] [CrossRef] [Green Version]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The faces of fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Bhat, J.D.; Dayarathne, M.C.; Huang, S.K.; Norphanphoun, C.; Senanayake, I.C.; Perera, R.H.; et al. Families of Sordariomycetes. Fungal Divers. 2016, 79, 1–317. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Huang, S.K.; Abdel-Wahab, M.A.; Daranagama, D.A.; Dayarathne, M.; D’souza, M.J.; Goonasekara, I.D.; et al. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 2015, 72, 199–301. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Crous, P.W.; Groenewald, J.Z.; Maharachchikumbura, S.S.N.; Jeewon, R.; Phillips, A.J.L.; Bhat, J.D.; Perera, R.H.; Li, Q.R.; Li, W.J.; et al. Families of Diaporthales based on morphological and phylogenetic evidence. Stud. Mycol. 2017, 86, 217–296. [Google Scholar] [CrossRef] [PubMed]
- Udayanga, D.; Liu, X.Z.; McKenzie, E.H.C.; Chukeatirote, E.; Bahkali, A.H.A.; Hyde, K.D. The genus Phomopsis: Biology, applications, species concepts and names of common phytopathogens. Fungal Divers. 2011, 50, 189–225. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Rajeshkumar, K.C.; Hawksworth, D.L.; Madrid, H.; Kirk, P.M.; Braun, U.; Singh, R.V.; Crous, P.W.; Kukwa, M.; et al. Notes for genera: Ascomycota. Fungal Divers. 2017, 86, 1–594. [Google Scholar]
- Nitschke, T.R.J. Pyrenomycetes germanici. In Die Kernpilze Deutschlands Bearbeitet von dr. Th. Nitschke; Eduard Trewendt: Breslau, Germany, 1870; Volume 2, pp. 161–320. [Google Scholar]
- Diogo, E.L.F.; Santos, J.M.; Phillips, A.J.L. Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Divers. 2010, 44, 107–115. [Google Scholar] [CrossRef]
- Farr, D.F.; Castlebury, L.A.; Rossman, A.Y.; Putnam, M.L. A new species of Phomopsis causing twig dieback of Vaccinium vitis-idaea (lingonberry). Mycol. Res. 2002, 106, 745–752. [Google Scholar] [CrossRef]
- Gao, Y.H.; Liu, F.; Duan, W.J.; Crous, P.W.; Cai, L. Diaporthe is paraphyletic. IMA Fungus 2017, 8, 153–187. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.; Sun, S.K. Phomopsis fruit rot of subtropical peach in Taiwan. Plant Pathol. Bull. 2003, 12, 212–214. [Google Scholar]
- Santos, J.M.; Phillips, A.J.L. Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Divers. 2009, 34, 111–125. [Google Scholar]
- Brayford, D. Variation in Phomopsis isolates from Ulmus species in the British Isles and Italy. Mycol. Res. 1990, 94, 691–697. [Google Scholar] [CrossRef]
- Chi, P.K.; Jiang, Z.D.; Xiang, M.M. Flora Fungorum Sinicorum; Science Press: Beijing, China, 2007; Volume 34. (In Chinese) [Google Scholar]
- Mostert, L.; Crous, P.W.; Kang, J.C.; Phillips, A.J.L. Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: Morphological, cultural, molecular and pathological characterization. Mycologia 2001, 93, 146–167. [Google Scholar] [CrossRef]
- Uecker, F.A. A world list of Phomopsis names with notes on nomenclature, morphology and biology. Mycol. Mem. 1988, 13, 1–231. [Google Scholar]
- Timmer, L.W.; Mondal, S.N.; Peres, N.A.R.; Bhatia, A. Fungal Diseases of Fruit and Foliage of Citrus Trees. In Diseases of Fruits and Vegetables; Naqvi, S.A.M.H., Ed.; Springer: Dordrecht, The Netherlands, 2004; Volume 1, pp. 247–290. [Google Scholar]
- Wehmeyer, L.E. The Genus Diaporthe Nitschke and Its Segregates; Science Series; University of Michigan Studies: Ann Arbor, MI, USA, 1933; Volume 9, pp. 1–349. [Google Scholar]
- Whiteside, J.O.; Garnsey, S.M.; Timmer, L.W. Melanose. In Compendium of Citrus Diseases; Whiteside, J.O., Ed.; APS Press: St. Paul, MN, USA, 1988; pp. 20–21. [Google Scholar]
- Muntanola-Cvetković, M.; Mihaljčević, M.; Petrov, M. On the identity of the causative agent of a serious Phomopsis-Diaporthe disease in sunflower plants. Nova Hedwig. 1981, 34, 417–435. [Google Scholar]
- Punithalingam, E. Phomopsis anacardii. CMI Descr. Pathog. Fungi Bact. 1985, 826, 1–2. [Google Scholar]
- Cristescu, C. A new species of Phomopsis Sacc. (Mitosporic fungi) in Romania. Rom. J. Biol.-Plant Biol. 2003, 48, 45–49. [Google Scholar]
- Rehner, S.A.; Uecker, F.A. Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete phomopsis. Can. J. Bot. 1994, 72, 1666–1674. [Google Scholar] [CrossRef]
- Cristescu, C. The morphology and anatomy of structure somatic and reproductive of species of Phomopsis (Sacc.) Bubák. Bul. Grădinii Bot. Iaşi Tomul 2007, 14, 19–27. [Google Scholar]
- Rosskopf, E.N.; Charudattan, R.; Shabana, Y.M.; Benny, G.L. Phomopsis amaranthicola, a new species from Amaranthus sp. Mycologia 2000, 92, 114–122. [Google Scholar] [CrossRef]
- Rosskopf, E.N.; Charudattan, R.; Valero, J.T.; Stall, W.M. Field evaluation of Phomopsis amaranthicola, a biological control agent of Amaranthus spp. Plant Dis. 2000, 84, 1225–1230. [Google Scholar] [CrossRef] [Green Version]
- Sutton, B.C. The Coelomycetes. Fungi Imperfecti with Pycnidia, Acervuli and Stromata; Commonwealth Mycological Institute: Kew, Surrey, UK, 1980. [Google Scholar]
- Punithalingam, E. Studies on Sphaeropsidales in culture, II. Mycol. Pap. 1974, 136, 1–63. [Google Scholar]
- Rodeva, R.; Stoyanova, Z.; Pandeva, R. A new fruit disease of pepper in Bulgaria caused by Phomopsis capsici. Acta Hortic. 2009, 830, 551–556. [Google Scholar] [CrossRef]
- Liu, X.Y.; Chaisiri, C.; Lin, Y.; Yin, W.X.; Luo, C.X. Whole-genome sequence of Diaporthe citri isolate NFHF-8-4 the causal agent of citrus melanose. Mol. Plant Microbe. Interact. 2021, 34, 845–847. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.P.; Xiong, T.; Xiao, X.; Li, P.D.; Zeng, Y.T.; Li, L.; Riely, B.K.; Li, H.Y. The genome sequence of the citrus melanose pathogen Diaporthe citri and two citrus-related Diaporthe species. Phytopathology 2021, 111, 779–783. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.W.; Jacobson, D.J.; Kroken, S.; Kasuga, T.; Geiser, D.M.; Hibbett, D.S.; Fisher, M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Ecol. 2000, 31, 21–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Wang, M.; Damm, U.; Crous, P.W.; Cai, L. Species boundaries in plant pathogenic fungi: A Colletotrichum case study. BMC Evol. Biol. 2016, 16, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achari, S.R.; Kaur, J.; Dinh, Q.; Mann, R.; Sawbridge, T.; Summerell, B.A.; Edwards, J. Phylogenetic relationship between australian Fusarium oxysporum isolates and resolving the species complex using the multispecies coalescent model. BMC Genom. 2020, 21, 248. [Google Scholar] [CrossRef] [Green Version]
- Hilário, S.; Gonçalves, M.F.M.; Alves, A. Using genealogical concordance and coalescent-based species delimitation to assess species boundaries in the Diaporthe eres complex. J. Fungi 2021, 7, 507. [Google Scholar] [CrossRef]
- Hilário, S.; Santos, L.; Alves, A. Diaporthe amygdali, a species complex or a complex species? Fungal Biol. 2021, 125, 505–518. [Google Scholar] [CrossRef]
- Kubatko, L.S.; Degnan, J.H. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol. 2007, 56, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Stewart, J.E.; Timmer, L.W.; Lawrence, C.B.; Pryor, B.M.; Peever, T.L. Discord between morphological and phylogenetic species boundaries: Incomplete lineage sorting and recombination results in fuzzy species boundaries in an asexual fungal pathogen. BMC Evol. Biol. 2014, 14, 38. [Google Scholar] [CrossRef]
- Castlebury, L. The Diaporthe vaccinii complex of fruit pathogens. Inoculum 2005, 56, 12. [Google Scholar]
- Marin-Felix, Y.; Hernández-Restrepo, M.; Wingfield, M.J.; Akulov, A.; Carnegie, A.J.; Cheewangkoon, R.; Gramaje, D.; Groenewald, J.Z.; Guarnaccia, V.; Halleen, F.; et al. Genera of phytopathogenic fungi: GOPHY 2. Stud. Mycol. 2019, 92, 47–133. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.M.; Correia, V.G.; Phillips, A.J.L.; Spatafora, J.W. Primers for mating-type diagnosis in Diaporthe and Phomopsis: Their use in teleomorph induction in vitro and biological species definition. Fungal Biol. 2010, 114, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Farr, D.F.; Castlebury, L.A.; Rossman, A.Y. Morphological and molecular characterization of Phomopsis vaccinii and additional isolates of phomopsis from blueberry and cranberry in the Eastern United States. Mycologia 2002, 94, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Chukeatirote, E.; Hyde, K.D. Insights into the genus Diaporthe: Phylogenetic species delimitation in the D. eres species complex. Fungal Divers. 2014, 67, 203–229. [Google Scholar] [CrossRef] [Green Version]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Chukeatirote, E.; Hyde, K.D. The Diaporthe sojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biol. 2015, 119, 383–407. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Liu, M.; Zhang, W.; Chen, Z.; Udayanga, D.; Chukeatirote, E.; Li, X.H.; Yan, J.Y.; Hyde, K.D. Morphological and molecular characterisation of Diaporthe species associated with grapevine trunk disease in China. Fungal Biol. 2015, 119, 283–294. [Google Scholar] [CrossRef]
- Santos, L.; Alves, A.; Alves, R. Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ 2017, 5, e3120. [Google Scholar] [CrossRef] [Green Version]
- Van Niekerk, J.M.; Groenewald, J.Z.; Farr, D.F.; Fourie, P.H.; Halleen, F.; Crous, P.W. Reassessment of Phomopsis species on grapevine. Australas. Plant Pathol. 2005, 34, 27–39. [Google Scholar] [CrossRef]
- Chaisiri, C.; Liu, X.Y.; Lin, Y.; Fu, Y.P.; Zhu, F.X.; Luo, C.X. Phylogenetic and haplotype network analyses of Diaporthe eres species in China based on sequences of multiple loci. Biology 2021, 10, 179. [Google Scholar] [CrossRef]
- Castlebury, L.A.; Farr, D.F.; Rossman, A.Y. Phylogenetic distinction of Phomopsis isolates from cucurbits. Inoculum 2001, 52, 25. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for desianing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Tacke, B.K.; Casper, H.C. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA 2000, 97, 7905–7910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, D.M.; Castlebury, L.A.; Rossman, A.Y.; White, J.F. New molecular markers for fungal phylogenetics: Two genes for species-level systematic in the Sordariomycetes (Ascomycota). Mol. Phylogenet. Evol. 2012, 64, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Myllys, L.; Stenroos, S.; Thell, A. New genes for phylogenetic studies of lichenized fungi: Glyceraldehyde-3-phosphate dehydrogenase and betatubulin genes. Lichenologist 2002, 34, 237–246. [Google Scholar] [CrossRef]
- Crous, P.W.; Groenewald, J.Z.; Risède, J.M.; Simoneau, P.; Hywel-Jones, N.L. Calonectria species and their Cylindrocladium anamorphs: Species with clavate vesicles. Stud. Mycol. 2004, 50, 415–430. [Google Scholar] [CrossRef] [Green Version]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Appl. Environ. Microb. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Pecchia, S.; Mercatelli, E.; Vannacci, G. Intraspecific diversity within Diaporthe helianthi: Evidence from rDNA intergenic spacer (IGS) sequence analysis. Mycopathologia 2004, 157, 317–326. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Choi, C.W.; Jung, K.E.; Kim, M.J.; Yoon, S.H.; Park, S.M.; Jin, S.B.; Hyun, J.W. Development of molecular marker to detect citrus melanose caused by Diaporthe citri from citrus melanose-like symptoms. Plant Pathol. J. 2021, 37, 681–686. [Google Scholar] [CrossRef]
- Crous, P.W.; Schoch, C.L.; Hyde, K.D.; Wood, A.R.; Gueidan, C.; de Hoog, G.S.; Groenewald, J.Z. Phylogenetic lineages in the Capnodiales. Stud. Mycol. 2009, 64, 17–47. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Crytococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilário, S.; Amaral, I.A.; Gonçalves, M.F.M.; Lopes, A.; Santos, L.; Alves, A. Diaporthe species associated with twig blight and dieback of Vaccinium corymbosum in Portugal, with description of four new species. Mycologia 2020, 112, 293–308. [Google Scholar] [CrossRef]
- Li, K.N.; Rouse, D.I.; German, T.L. PCR primers that allow intergenetic differentiation of ascomycetes and their application to Verticillium spp. Appl. Environ. Microb. 1994, 60, 4324–4331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donnell, K.; Cigelnik, E.; Nirenberg, H.I. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 1998, 90, 465–493. [Google Scholar] [CrossRef]
- Aveskamp, M.M.; Verkley, G.J.M.; Gruyter, J.; Perelló, A.; Woudenberg, J.H.C.; Groenewald, J.Z.; Crous, P.W. DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia 2009, 101, 363–382. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Ruocco, M.; Baroncelli, R.; Cacciola, S.O.; Pane, C.; Monti, M.M.; Firrao, G.; Vergara, M.; Lio, G.M.S.; Vannacci, G.; Scala, F. Polyketide synthases of Diaporthe helianthi and involvement of DHPKS1 in virulence on sunflower. BMC Genom. 2018, 19, 27. [Google Scholar] [CrossRef]
- Says-Lesage, V.; Roeckel-Drevet, P.; Viguié, A.; Tourvieille, J.; Nicolas, P.; Labrouhe, D.T. Molecular variability within Diaporthe/Phomopsis helianthi from France. Phytopathology 2002, 92, 308–313. [Google Scholar] [CrossRef] [Green Version]
- Croll, D.; McDonald, B.A. The genetic basis of local adaptation for pathogenic fungi in agricultural ecosystems. Mol. Ecol. 2017, 26, 2027–2040. [Google Scholar] [CrossRef]
- Stukenbrock, E.H.; McDonald, B.A. The origins of plant pathogens in agro-ecosystems. Annu. Rev. Phytopathol. 2008, 46, 75–100. [Google Scholar] [CrossRef] [Green Version]
- Xu, J. Evolutionary Genetics of Fungi; Horizon Scientific Press: Norfolk, UK, 2005. [Google Scholar]
- Timmer, L.W.; Garnsey, S.M.; Graham, J.H. Scab Diseases, revised edition: 31–32 ed.; American Phytopathological Society Press: St. Paul, MN, USA, 2000; p. 92. [Google Scholar]
- Kuhara, S. The Application of the Epidemiologic Simulation Model “Melan” to Control Citrus Melanose Caused by Diaporthe Citri (Faw.) Wolf; ASPAC Food and Fertilizer Technology Center: Taipei, China, 1999; p. 481. [Google Scholar]
- Kucharek, T.; Whiteside, J.; Brown, E. Melanose and Phomopsis Stem-End Rot of Citrus. In Plant Pathology Fact Sheet, Florida Cooperative Extension Service; Institute of Food and Agricultural Sciences, University of Florida: Gainesville, FL, USA, 2000; pp. 26–30. [Google Scholar]
- Bach, W.J.; Wolf, F.A. The isolation of the fungus that causes citrus melanose and the pathological anatomy of the host. J. Agric. Res. 1928, 37, 243–252. [Google Scholar]
- Agostini, J.P.; Bushong, P.M.; Bhatia, A.; Timmer, L.W. Influence of environmental factors on severity of citrus scab and melanose. Plant Dis. 2003, 87, 1102–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burnett, H.C. Melanose Diaporthe Citri (Faw.) Wolf; Florida Department of Agriculture and Consumer Services, Divition of Plant Industry: Gainesville, FL, USA, 1962; p. 1. [Google Scholar]
- Hardy, S.; Donovan, N. Managing Melanose in Citrus; Profitable & sustainable primary industries, New South Wales Department of Primary Industries: Orange, NSW, Australia, 2007; Volume 751, pp. 1–3. [Google Scholar]
- Nelson, S. Citrus Melanose. In Plant Disease; Cooperative Extension Service, College of tropical Agriculture and Human Resources, University of Hawaii: Honolulu, HI, USA, 2008; pp. 1–5. [Google Scholar]
- Barkley, P.B.; Schubert, T.; Schutte, G.C.; Godfrey, K.; Hattingh, V.; Telford, G.; Beattie, G.A.C.; Hoffman, K.M. Invasive Pathogens in Plant Biosecurity. Case Study: Citrus Biosecurity; Springer: Dordrecht, The Netherlands, 2014; pp. 547–592. [Google Scholar]
- Rehman, F.U.; Kalsoom, M.; Sultan, A.; Adnan, M.; Junaid, S.; Akram, H.; Tariq, M.A.; Shafique, T.; Zafar, M.I. Citrus melanose and quality degradation of fruit by this disease: A review. J. Biogeneric Sci. Res. 2020, 3, 1–4. [Google Scholar] [CrossRef]
- Dewdney, M.M.; Timmer, L.W. Florida Citrus Pest Management Guide. Available online: http://edis.ifas.ufl.edu (accessed on 19 August 2021).
- Driscoll, P.J. Copper toxicity on florida citrus—Why did it happen? Proc. Fla. State Hort. Soc. 2004, 117, 124–127. [Google Scholar]
- Narciso, J.; Widmer, W.; Ference, C.; Ritenour, M.; Diaz, R. Use of carnauba based carrier for copper sprays reduces infection by Xanthomonas citri subsp. citri and Diaporthe citri in Florida commercial grapefruit groves. Agric. Sci. 2012, 3, 962–970. [Google Scholar]
- Stover, E.D.; Ciliento, S.; Albrigo, G. Copper fungicide spray timings for melanose control in grapefruit: Comparison of computer modelling of copper residues vs. calendar sprays. Hortscience 2004, 39, 886A-886. [Google Scholar] [CrossRef]
- Timmer, L.W.; Zitko, S.E. Evaluation of copper fungicides and rates of metallic copper for control of melanose on grapefruit in florida. Plant Dis. 1996, 80, 166–169. [Google Scholar] [CrossRef]
- Bushong, P.M.; Timmer, L.W. Evaluation of postinfection control of citrus scab and melanose with benomyl, fenbuconazole, and azoxystrobin. Plant Dis. 2007, 84, 1246–1249. [Google Scholar] [CrossRef] [Green Version]
- Inuma, T. Decreasing the frequency of control of citrus melanose by using the fungicides dithianon for satsuma mandarin cultivation (in Japanese). Ann. Rept. Kansai Pl. Prot. 2014, 56, 85–87. [Google Scholar] [CrossRef] [Green Version]
- Idrees, M.; Naz, S.; Ehetisham-Ul-Haq, M.; Mehboob, S.; Kamran, M.; Ali, S.; Iqbal, M. Protectant and curative efficacy of different fungicides against citrus melanose caused by Phomopsis citri under in vivo conditions. Int. J. Biosci. 2019, 15, 194–199. [Google Scholar]
- Anwar, U.; Mubeen, M.; Iftikhar, Y.; Zeshan, M.A.; Shakeel, Q.; Sajid, A.; Umer, M.; Abbas, A. Efficacy of different fungicides against citrus melanose disease in Sargodha, Pakistan. Pak. L. Phytopathol. 2021, 33, 67–74. [Google Scholar] [CrossRef]
- Liu, X.; Wang, M.S.; Mei, X.F.; Jiang, L.Y.; Han, G.X.; Li, H.Y. Sensitivity evaluation of Diaporthe citri populations to mancozeb and screening of alternative fungicides for citrus melanose control. J. Plant Protec. 2018, 45, 373–381. (In Chinese) [Google Scholar]
- Qin, S.Y.; Zhou, X.Y.; Jiang, Y.L. Screening of fungicides for controlling citrus melanose in lab. Guangdong Agric. Sci. 2012, 39, 77–79. (In Chinese) [Google Scholar]
- Ko, Y.J.; Kang, S.Y.; Jeun, Y.C. Suppression of citrus melanose on the leaves treated with rhizobacterial strains after inoculation with Diaporthe citri. Res. Plant Dis. 2012, 18, 331–337. (In Korean) [Google Scholar] [CrossRef]
- Ko, Y.J.; Kim, J.S.; Kim, K.D.; Jeun, Y.C. Microscopical observation of inhibition-behaviors against Diaporthe citri by pre-treated with Pseudomonas putida strain THJ609-3 on the leaves of citrus plants. J. Microbiol. 2014, 52, 879–883. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.H.; Ko, E.J.; Kim, S.J.; Hyun, H.N.; Jeun, Y.C. Suppression of melanose caused by Diaporthe citri on citrus leaves pretreated with bio-sulfur. Plant Pathol. J. 2019, 35, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.R.; Maung, C.E.H.; Choi, T.G.; Kim, K.Y. Large scale cultivation of Bacillus velezensis CE 100 and effect of its culture on control of citrus melanose caused by Diaporthe citri. Korean J. Soil Sci. Fert. 2021, 54, 297–310. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).