Haploid Induction in Tomato (Solanum lycopersicum L.) via Gynogenesis
Abstract
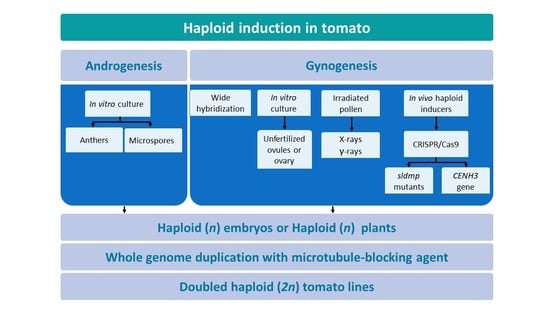
1. Introduction
2. Importance of Doubled Haploids
3. Doubled Haploids in Tomato
4. Gynogenesis in Agricultural Crops
4.1. In Vitro Culture of Ovaries or Ovules
4.2. Irradiated Pollen
4.3. Wide Hybridization
4.4. In Vivo Haploid Induction
5. Gynogenesis in Tomato
5.1. Wide Hybridization
5.2. Unfertilized Ovule Culture
5.3. Irradiated Pollen
5.4. In Vivo Haploid Inducers
6. Future Perspectives
7. Materials and Methods
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Data. 2021. Available online: http://www.fao.org/faostat/en/#data (accessed on 13 October 2021).
- Seguí-Simarro, J.M. Androgenesis in Solanaceae. In In Vitro Embryogenesis in Higher Plants. Methods in Molecular Biology; Germanà, M.A., Lambardi, M., Eds.; Springer Science + Business Media: New York, NY, USA, 2016; pp. 209–244. ISBN 978-1-4939-3061-6. [Google Scholar]
- Yan, G.; Liu, H.; Wang, H.; Lu, Z.; Wang, Y.; Mullan, D.; Hamblin, J.; Liu, C. Accelerated generation of selfed pure line plants for gene identification and crop breeding. Front. Plant Sci. 2017, 8, 1786. [Google Scholar] [CrossRef] [PubMed]
- Bal, U.; Abak, K. Haploidy in tomato (Lycopersicum esculentum Mill.): A critical review. Euphytica 2007, 158, 1–9. [Google Scholar] [CrossRef]
- Hooghvorst, I.; Ramos-Fuentes, E.; López-Cristofannini, C.; Ortega, M.; Vidal, R.; Serrat, X.; Nogués, S. Antimitotic and hormone effects on green double haploid plant production through anther culture of Mediterranean japonica rice. Plant Cell Tiss. Organ Cult. 2018, 134, 205–215. [Google Scholar] [CrossRef]
- Duyen, D.V.; Kwon, Y.; Kabange, N.R.; Lee, J.-Y.; Lee, S.-M.; Kang, J.-W.; Park, H.; Cha, J.-K.; Cho, J.-H.; Shin, D.; et al. Novel QTL associated with aerenchyma-mediated radial oxygen loss (ROL) in rice (Oryza sativa L.) under iron (II) sulfide. Plants 2022, 11, 788. [Google Scholar] [CrossRef]
- Qu, Y.; Liu, Z.; Zhang, Y.; Yang, J.; Li, H. Improving the sorting efficiency of maize haploid kernels using an NMR-based method with oil content double thresholds. Plant Methods 2021, 17, 2. [Google Scholar] [CrossRef]
- Trampe, B.; Batîru, G.; Pereira da Silva, A.; Frei, U.K.; Lübberstedt, T. QTL mapping for haploid inducibility using genotyping by sequencing in maize. Plants 2022, 11, 878. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, L.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef]
- Wiśniewska, H.; Majka, M.; Kwiatek, M.; Gawłowska, M.; Surma, M.; Adamski, T.; Kaczmarek, Z.; Drzazga, T.; Lugowska, B.; Korbas, M.; et al. Production of wheat-doubled haploids resistant to eyespot supported by marker-assisted selection. Electron. J. Biotechnol. 2019, 37, 11–17. [Google Scholar] [CrossRef]
- Al-Ashkar, I.; Alderfasi, A.; El-Hendawy, S.; Al-Suhaibani, N.; El-Kafafi, S.; Seleiman, M.F. Detecting salt tolerance in doubled haploid wheat lines. Agronomy 2019, 9, 211. [Google Scholar] [CrossRef]
- Palacios, M.A.; Seguí-Simarro, J.M. Anther culture in sweet pepper (Capsicum annuum L.). In Doubled Haploid Technology; Seguí-Simarro, J.M., Ed.; Methods in Molecular Biology; Humana: New York, NY, USA, 2021; pp. 279–292. ISBN 9781071613345. [Google Scholar]
- Mir, R.; Calabuig-Serna, A.; Seguí-Simarro, J.M. Doubled haploids in eggplant. Biology 2021, 10, 685. [Google Scholar] [CrossRef]
- Dong, Y.Q.; Zhao, W.X.; Li, X.H.; Liu, X.C.; Gao, N.N.; Huang, J.H.; Wang, W.Y.; Xu, X.L.; Tang, Z.H. Androgenesis, gynogenesis, and parthenogenesis haploids in cucurbit species. Plant Cell Rep. 2016, 35, 1991–2019. [Google Scholar] [CrossRef] [PubMed]
- Niazian, M.; Shariatpanahi, M.E.; Abdipour, M.; Oroojloo, M. Modeling callus induction and regeneration in an anther culture of tomato (Lycopersicon esculentum L.) using image processing and artificial neural network method. Protoplasma 2019, 256, 1317–1332. [Google Scholar] [CrossRef] [PubMed]
- Forster, B.P.; Heberle-Bors, E.; Kasha, K.J.; Touraev, A. The resurgence of haploids in higher plants. Trends Plant Sci. 2007, 12, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.; Badenes, M.L.; Del Mar Naval, M. Induced parthenogenesis by gamma-irradiated pollen in loquat for haploid production. Breed. Sci. 2016, 66, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Kundu, M.; Dubey, A. Effect of gamma ray irradiated pollen technique on seed development pattern in Citrus. Indian J. Genet. Plant Breed. 2020, 80, 450–458. [Google Scholar] [CrossRef]
- Keleş, D.; Özcan, C.; Pınar, H.; Ata, A.; Denli, N.; Yücel, N.; Taşkın, H.; Büyükalaca, S. First report of obtaining haploid plants using tissue culture techniques in spinach. HortScience 2016, 51, 742–749. [Google Scholar] [CrossRef]
- Hooghvorst, I.; Nogués, S. Opportunities and challenges in doubled haploids and haploid inducer-mediated genome-editing systems in cucurbits. Agronomy 2020, 10, 1441. [Google Scholar] [CrossRef]
- Zayachkovskaya, T.; Domblides, E.; Zayachkovsky, V.; Kan, L.; Domblides, A.; Soldatenko, A. Production of gynogenic plants of red beet (Beta vulgaris L.) in unpollinated ovule culture in vitro. Plants 2021, 10, 2703. [Google Scholar] [CrossRef]
- Takamura, Y.; Takahashi, R.; Hikage, T.; Hatakeyama, K.; Takahata, Y. Production of haploids and doubled haploids from unfertilized ovule culture of various wild species of gentians (Gentiana spp.). Plant Cell Tiss. Organ Cult. 2021, 146, 505–514. [Google Scholar] [CrossRef]
- Rajcan, I.; Boersma, J.G.; Shaw, E.J. Plant genetic techniques: Plant breeder’s toolbox: In Comprehensive Biotechnology; Moo-Young, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 133–147. ISBN 978-0-08-088504-9. [Google Scholar]
- Asif, M. Progress and Opportunities of Doubled Haploid Production; Springer International Publishing: New York, NY, USA, 2013; p. 75. ISBN 978-3-319-00732-8. [Google Scholar]
- Kelliher, T.; Starr, D.; Wang, W.; McCuiston, J.; Zhong, H.; Nuccio, M.L.; Martin, B. Maternal haploids are preferentially induced by CENH3-tailswap transgenic complementation in maize. Front. Plant Sci. 2016, 7, 414. [Google Scholar] [CrossRef]
- Maqbool, M.A.; Beshir, A.; Khokhar, E.S. Doubled haploids in maize: Development, deployment, and challenges. Crop Sci. 2020, 60, 2815–2840. [Google Scholar] [CrossRef]
- Chaikam, V.; Molenaar, W.; Melchinger, A.E.; Boddupalli, P.M. Doubled haploid technology for line development in maize: Technical advances and prospects. Theor. Appl. Genet. 2019, 132, 3227–3243. [Google Scholar] [CrossRef] [PubMed]
- Cerrudo, D.; Cao, S.; Yuan, Y.; Martinez, C.; Suarez, E.A.; Babu, R.; Zhang, X.; Trachsel, S. Genomic selection outperforms marker assisted selection for grain yield and physiological traits in a maize doubled haploid population across water treatments. Front. Plant Sci. 2018, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.A.; Schilling, R.K.; Tilbrook, J.; Brien, C.; Dowling, K.; Rabie, H.; Short, L.; Trittermann, C.; Garcia, A.; Barrett-Lennard, E.G.; et al. Mapping of novel salt tolerance QTL in an Excalibur × Kukri doubled haploid wheat population. Theor. Appl. Genet. 2018, 131, 2179–2196. [Google Scholar] [CrossRef]
- Khumalo, T.P.; Barnard, A.; Dube, E.; Tsilo, T.J. Characterization of vegetative vigor of two doubled-haploid wheat populations. J. Crop Improv. 2021, 19, 1542–7528. [Google Scholar] [CrossRef]
- Anshori, M.F.; Purwoko, B.S.; Dewi, I.S.; Ardie, S.W.; Suwarno, W.B. A new approach to select doubled haploid rice lines under salinity stress using indirect selection index. Rice Sci. 2021, 28, 368–378. [Google Scholar] [CrossRef]
- Akbar, M.R.; Purwoko, B.P.; Dewi, I.S.; Suwarno, W.B.; Sugiyanta, S.; Anshori, M.F. Agronomic and yield selection of doubled haploid lines of rainfed lowland rice in advanced yield trials. Biodiversitas 2021, 22, 3006–3012. [Google Scholar] [CrossRef]
- Bonga, J.M.; Klimaszewska, K.K.; Aderkas, P.V. Recalcitrance in clonal propagation, in particular of conifers. Plant Cell Tiss. Organ Cult. 2010, 100, 241–254. [Google Scholar] [CrossRef]
- Corral-Martínez, P.; Nuez, F.; Seguí-Simarro, J.M. Genetic, quantitative and microscopic evidence for fusion of haploid nuclei and growth of somatic calli in cultured ms1035 tomato anthers. Euphytica 2011, 178, 215–228. [Google Scholar] [CrossRef]
- Julião, S.A.; Carvalho, C.R.; da Silva, T.C.R.; Koehler, A.D. Multiploidy occurrence in tomato calli from anther culture. Afr. J. Biotechnol. 2015, 14, 2846–2855. [Google Scholar] [CrossRef][Green Version]
- Sharp, W.R.; Dougall, D.K.; Paddock, E.F. Haploid plantlets and callus from immature pollen grains of Nicotiana and Lycopersicon. Bull. Torrey Bot. Club. 1971, 98, 219–222. [Google Scholar] [CrossRef]
- Gresshoff, P.M.; Doy, C.H. Development and differentiation of haploid Lycopersicon esculentum (Tomato). Planta 1972, 107, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Seguí-Simarro, J.M.; Nuez, F. Meiotic metaphase I to telophase II as the most responsive stage during microspore development for callus induction in tomato (Solanum lycopersicum) anther cultures. Acta Physiol. Plant. 2005, 27, 675–685. [Google Scholar] [CrossRef]
- Ahmadi, B.; Shariatpanahi, M.E.; Asghari-Zakaria, R.; Zare, N.; Azadi, P. Efficient microspore embryogenesis induction in tomato (Lycopersicon esculentum Mill.) using shed microspore culture. J. Pure Appl. Microbiol. 2015, 9, 21–29. [Google Scholar] [CrossRef]
- Kumar, S.; Jindal, S.K.; Sarao, N.K.; Dhaliwal, M.S. Callus induction and plant regeneration of tomato through anther culture. Veg. Sci. 2020, 47, 23–27. [Google Scholar]
- Seguí-Simarro, J.M.; Nuez, F. Androgenesis induction from tomato anther cultures: Callus characterization. Acta Hortic. 2006, 725, 855–861. [Google Scholar] [CrossRef]
- Seguí-Simarro, J.M.; Nuez, F. Embryogenesis induction, callogenesis, and plant regeneration by in vitro culture of tomato isolated microspores and whole anthers. J. Exp. Bot. 2007, 5, 1119–1132. [Google Scholar] [CrossRef]
- Moreno, A.; Claveria, E.; Pujol, M.; Dolcet-Sanjuan, R. Development of a methodology for the production of doubled haploid lines in Solanum lycopersicum L. Acta Hortic. 2012, 935, 95–100. [Google Scholar] [CrossRef]
- Khan, P.S.S.V.; Vijayalakshmi, G.; Raja, M.M.; Naik, M.L.; Germanà, M.A.; Terry, R.G. Doubled haploid production in onion (Allium cepa L.): From gynogenesis to chromosome doubling. Plant Cell Tiss. Organ Cult. 2020, 142, 1–22. [Google Scholar] [CrossRef]
- Panahandeh, J.; Farhadi, N. Haploid induction via in vitro gynogenesis in Persian shallot (Allium hirtifolium). J. Hortic. Res. 2019, 27, 91–98. [Google Scholar] [CrossRef]
- Doi, H.; Takahata, Y. Haploid and doubled haploid plant production in gentian (Gentiana spp.). In The Gentianaceae: Biotechnology and Applications; Rybczyński, J., Davey, M., Mikuła, A., Eds.; Springer: Berlin, Heidelberg, 2015; Volume 2, pp. 187–197. ISBN 9783642541018. [Google Scholar]
- Doi, H.; Yokoi, S.; Hikage, T.; Nishihara, M.; Tsutsumi, K.-I.; Takahata, Y. Gynogenesis in gentians (Gentiana triflora, G. scabra): Production of haploids and doubled haploids. Plant Cell Rep. 2011, 30, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Bal, U.; Abak, K. Attempts of haploidy induction in tomato (Lycopersicon esculentum Mill.) via gynogenesis II: In vitro non-fertilized ovary culture. Pak. J. Biol. Sci. 2003, 6, 750–755. [Google Scholar] [CrossRef][Green Version]
- Wang, S.M.; Lan, H.; Jia, H.H.; Xie, K.D.; Wu, X.M.; Chen, C.L.; Guo, W.W. Induction of parthenogenetic haploid plants using gamma irradiated pollens in ″Hirado Buntan″ pummelo (Citrus grandis [L.] Osbeck). Sci. Hortic. 2016, 207, 233–239. [Google Scholar] [CrossRef]
- Lotfi, M.; Alan, A.R.; Henning, M.J.; Jahn, M.M.; Earle, E.D. Production of haploid and doubled haploid plants of melon (Cucumis melo L.) for use in breeding for multiple virus resistance. Plant Cell Rep. 2003, 21, 1121–1128. [Google Scholar] [CrossRef] [PubMed]
- Nasertorabi, M.; Madadkhah, E.; Moghbeli, E.; Grouh, M.S.H.; Soleimani, A. Production of haploid lines from parthenogenetic Iranian melon plants obtained of irradiated pollen (Cucumis melo L.). Int. Res. J. Appl. Basic Sci. 2012, 3, 1585–1589. [Google Scholar]
- Jedidi, E.; Kamiri, M.; Poullet, T.; Ollitrault, P.; Froelicher, Y. Efficient haploid production on ′Wilking′ mandarin by induced gynogenesis. Acta Hortic. 2015, 1065, 495–500. [Google Scholar] [CrossRef]
- Kurtar, E.S.; Seymen, M.; Kal, Ü. An overview of doubled haploid plant production in Cucurbita species. Yüzüncü Yıl Üniversitesi Tarım Bilimleri Dergisi 2020, 30, 510–520. [Google Scholar] [CrossRef]
- Niu, Z.; Jiang, A.; Abu Hammad, W.; Oladzadabbasabadi, A.; Xu, S.S.; Mergoum, M.; Elias, E.M. Review of doubled haploid production in durum and common wheat through wheat × maize hybridization. Plant Breed. 2014, 133, 313–320. [Google Scholar] [CrossRef]
- Santra, M.; Wang, H.; Seifert, S.; Haley, S. Doubled haploid laboratory protocol for wheat using wheat–maize wide hybridization. In Wheat Biotechnology; Methods in Molecular Biology; Bhalla, P., Singh, M., Eds.; Humana Press: New York, NY, USA, 2017; pp. 234–249. ISBN 978-1-4939-7337-8. [Google Scholar]
- Piosik, Ł.; Zenkteler, E.; Zenkteler, M. Development of haploid embryos and plants of Lactuca sativa induced by distant pollination with Helianthus annuus and H. tuberosus. Euphytica 2016, 208, 439–451. [Google Scholar] [CrossRef]
- Wei, Y.; Zhu, M.; Qiao, H.; Li, F.; Zhang, S.; Zhang, S.; Zhang, H.; Sun, R. Characterization of interspecific hybrids between flowering Chinese cabbage and broccoli. Sci. Hortic. 2018, 240, 552–557. [Google Scholar] [CrossRef]
- Wei, Y.; Li, F.; Zhang, S.; Zhang, S.; Zhang, H.; Qiao, H.; Sun, R. Characterization of interspecific hybrids between Chinese cabbage (Brassica rapa) and red cabbage (Brassica oleracea). Sci. Hortic. 2019, 250, 33–37. [Google Scholar] [CrossRef]
- Bal, U.; Abak, K. Attempts of haploidy induction in tomato (Lycopersicon esculentum Mill.) via gynogenesis I: Pollination with Solanum sisymbriifolium Lam. Pollen. Pak. J. Biol. Sci. 2003, 6, 745–749. [Google Scholar] [CrossRef][Green Version]
- Chambonnet, D. In situ induction of haploid gynogenesis in tomato. Rep. Tom. Gen. Coop. 2012, 62, 5–22. [Google Scholar]
- Uliana Trentin, H.; Frei, U.K.; Lübberstedt, T. Breeding maize maternal haploid inducers. Plants 2020, 9, 614. [Google Scholar] [CrossRef] [PubMed]
- Gilles, L.M.; Khaled, A.; Laffaire, J.-B.; Chaignon, S.; Gendrot, G.; Laplaige, J.; Bergès, H.; Beydon, G.; Bayle, V.; Barret, P.; et al. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. Eur. Mol. Biol. Organ. 2017, 36, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Sun, J.; Li, R.; Yan, S.; Chen, W.; Guo, L.; Qin, G.; Wang, P.; Luo, C.; Huang, W.; et al. A reactive oxygen species burst causes haploid induction in maize. Mol. Plant. 2022, 15, 943–955. [Google Scholar] [CrossRef]
- Tang, H.; Zhang, S.; Yu, M.; Wang, K.; Yu, Y.; Qiu, Y.; Chang, Y.; Lin, Z.; Du, L.; Fu, D.; et al. Effects of TaMTL-Edited mutations on grain phenotype and storage component composition in wheat. Agriculture 2022, 12, 587. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, B.; Li, M.; Wang, D.; Jiao, Y.; Qi, X.; Wang, M.; Liu, Z.; Chen, C.; Wang, Y.; et al. A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis. Nat. Plants 2020, 6, 466–472. [Google Scholar] [CrossRef]
- Li, Y.; Li, D.; Xiao, Q.; Wang, H.; Wen, J.; Tu, J.; Shen, J.; Fu, T.; Yi, B. An in planta haploid induction system in Brassica napus. J. Integr. Plant Biol. 2022, 64, 1140–1144. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Y.; Chen, B.; Liu, J.; Wang, D.; Li, M.; Qi, X.; Liu, C.; Boutilier, K.; Chen, S. Establishment of a dmp based maternal haploid induction system for polyploid Brassica napus and Nicotiana tabacum. J. Integr. Plant Biol. 2022, 64, 1281–1294. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Zhang, J.; Jia, M.; Cao, L.; Yu, J.; Zhao, D. Haploid induction in allotetraploid tobacco using DMPs mutation. Planta 2022, 255, 98. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Xia, X.; Jiang, T.; Li, L.; Zhang, P.; Niu, L.; Cheng, H.; Wang, K.; Lin, H. In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula. Plant Biotechnol. J. 2022, 20, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Chen, B.; Wang, D.; Zhu, X.; Li, M.; Zhang, J.; Chen, M.; Wang, M.; Riksen, T.; Liu, J.; et al. In vivo maternal haploid induction in tomato. Plant Biotechnol. J. 2022, 20, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Op Den Camp, R.H.M.; Van Dijk, P.L.; Gallard, A. Method for the Production of Haploid and Subsequent Doubled Haploid plants. Patent WO 2017/200386 A1 2017. Available online: https://patents.google.com/patent/WO2017058022A1/en (accessed on 29 May 2022).
- Di Giacomo, M.; Luciani, M.D.; Cambiaso, V.; Zorzoli, R.; Rubén, R.G.; Pereira da Costa, J.H. Tomato near isogenic lines to unravel the genetic diversity of S. pimpinellifolium LA0722 for fruit quality and shelf life breeding. Euphytica 2020, 216, 126. [Google Scholar] [CrossRef]
- Awais Ghani, M.; Mehran Abbas, M.; Amjad, M.; Ziaf, K.; Ali, B.; Shaheen, T.; Saeed Awan, F.; Nawaz Khan, A. Production and characterization of tomato derived from interspecific hybridisation between cultivated tomato and its wild relatives. J. Hortic. Sci. Biotechnol. 2020, 95, 506–520. [Google Scholar] [CrossRef]
- Chetelat, R.T.; Qin, X.; Tan, M.; Burkart-Waco, D.; Moritama, Y.; Huo, X.; Wills, T.; Pertuzé, R. Introgression lines of Solanum sitiens, a wild nightshade of the Atacama Desert, in the genome of cultivated tomato. Plant J. 2019, 100, 836–850. [Google Scholar] [CrossRef]
- Ali, A.A.M.; Romdhane, W.B.; Tarroum, M.; Al-Dakhil, M.; Al-Doss, A.; Alsadon, A.A.; Hassairi, A. Analysis of salinity tolerance in tomato introgression lines based on morpho-physiological and molecular traits. Plants 2021, 10, 2594. [Google Scholar] [CrossRef]
- Prasanna, H.C.; Sinha, D.P.; Rai, G.K.; Krishna, R.; Kashyap, S.P.; Singh, N.K.; Singh, M.; Malathi, V.G. Pyramiding Ty-2 and Ty-3 genes for resistance to monopartite and bipartite tomato leaf curl viruses of India. Plant Pathol. 2015, 64, 256–264. [Google Scholar] [CrossRef]
- Baek, Y.S.; Covey, P.A.; Petersen, J.J.; Chetelat, R.T.; McClure, B.; Bedinger, P.A. Testing the SI × SC rule: Pollen–pistil interactions in interspecific crosses between members of the tomato clade (Solanum section Lycopersicon, Solanaceae). Am. J. Bot. 2015, 102, 302–311. [Google Scholar] [CrossRef]
- Marin-Montes, I.M.; Lobato-Ortiz, R.; Carrillo-Castañeda, G.; Rodríguez-Pérez, J.E.; García-Zavala, J.J.; Hernández-Rodríguez, M.; Velasco-García, A.M. Genetic parameters of an interspecific cross between S. lycopersicum L. and S. habrochaites Knapp & Spooner. Rev. Chap. Ser. Hortic. 2020, 26, 111–123. [Google Scholar] [CrossRef]
- Piosik, Ł.; Ruta-Piosik, M.; Zenkteler, M.; Zenkteler, E. Development of interspecific hybrids between Solanum lycopersicum L. and S. sisymbriifolium Lam. via embryo calli. Euphytica 2019, 215, 31. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, X.; Du, Y.; Zhu, D.; Guo, Y.; Gao, J.F.; Snyder, J. Haploid induction via in vitro gynogenesis in tomato (Solanum lycopersicum L.). J. Int. Agric. 2014, 13, 2122–2131. [Google Scholar] [CrossRef]
- Nishiyama, I.; Tsukuda, S. Radiobiological studies in plant, I. Effects of X-rays upon pollen germination and fertility. Jpn. J. Genet. 1959, 34, 363–370. [Google Scholar] [CrossRef]
- Nishiyama, I.; Tsukuda, S. Effects of X- and gamma-irradiations on pollen fertility of Lycopersicum pimpinellifolium. Jpn. J. Genet. 1961, 36, 423–427. [Google Scholar] [CrossRef]
- Akbudak, N.; Seniz, V. In vitro and in vivo behavior of gamma irradiated tomato (Lycopersicon esculentum) pollen. N. Z. J. Crop Hortic. Sci. 2009, 37, 361–367. [Google Scholar] [CrossRef]
- Choun-Sea, L.; Chen-Tran, H.; Yu-Hsuan, Y.; Po-Xing, Z.; Fu-Hui, W.; Qiao-Wei, C.; Yu-Lin, W.; Ting-Li, W.; Steven, L.; Jin-Jun, Y.; et al. DNA-free CRISPR-Cas9 gene editing of wild tetraploid tomato Solanum peruvianum using protoplast regeneration. Plant Physiol. 2022, 188, 1917–1930. [Google Scholar] [CrossRef]
- Xuhu, G.; Jianguo, Z.; Zhiwen, C.; Jun, Q.; Yongfang, Z.; Hong, S.; Zhongli, H. CRISPR/Cas9-targeted mutagenesis of SlCMT4 causes changes in plant architecture and reproductive organ in tomato. Hortic. Res. 2022, 9, uhac081. [Google Scholar] [CrossRef]
- Yang, S.H.; Kim, E.; Park, H.; Koo, Y. Selection of the high efficient sgRNA for CRISPR-Cas9 to edit herbicide related genes, PDS, ALS, and EPSPS in tomato. Appl. Biol. Chem. 2022, 65, 13. [Google Scholar] [CrossRef]
- Tang, Y.; Li, H.; Liu, C.; He, Y.; Wang, H.; Zhao, T.; Xu, X.; Li, J.; Yang, H.; Jiang, J. CRISPR-Cas9-mediated mutagenesis of the SlSRM1-like gene leads to abnormal leaf development in tomatoes. BMC Plant Biol. 2022, 22, 13. [Google Scholar] [CrossRef]
- Masayuki, I.; Tetsuya, Y.; Momoko, M.; Yusuke, K.; Akihito, K.; Kazuhiro, I. Tomato brown rugose fruit virus resistance generated by quadruple knockout of homologs of TOBAMOVIRUS MULTIPLICATION1 in tomato. Plant Physiol. 2022, 189, 679–689. [Google Scholar] [CrossRef]
- Yonggang, Q.; Xiuying, S.; Yun, S.; Xiaoming, Z. Tetraploidy inducement of small-fruit-type tomato with colchicine. Chin. Agric. Sci. Bull. 2007, 23, 19. [Google Scholar]
- Praça, M.M.; Carvalho, C.R.; Clarindo, W.R. A practical and reliable procedure for in vitro induction of tetraploid tomato. Sci. Hortic. 2009, 122, 501–505. [Google Scholar] [CrossRef]
- Cola, G.P.A.; Marques, A.M.; Damasceno, S.; Carvalho, C.R.; Clarindo, W.R. In vitro polyploidization in Solanum lycopersicum Mill. “Santa Cruz Kada Gigante”. Cytologia 2014, 79, 351–358. [Google Scholar] [CrossRef]
| Species | Common Name | Pathway | Ploidy Level Determination | Haploid Plants | Reference |
|---|---|---|---|---|---|
| S. lycopersicum L. | Tomato | Anther culture | Flow cytometry | 3 | Corral-Martínez et al. [34] |
| Anther culture | Flow cytometry | 0 | Julião et al. [35] | ||
| Anther culture | Burn’s technique | 0 | Sharp et al. [36] | ||
| Anther culture | Flow cytometry | 0 | Seguí–Simarro et al. [38] | ||
| Anther culture | Flow cytometry | 0 | Seguí–Simarro et al. [41] | ||
| Anther culture | Flow cytometry | 0 | Seguí–Simarro et al. [42] | ||
| Anther culture | Flow cytometry | 0 | Moreno et al. [43] |
| Species | Common Name | Pathway | Ploidy Level Determination | Haploid Induction Rate | Reference |
|---|---|---|---|---|---|
| Beta vulgaris L. | Red beet | Unfertilized ovule culture | Flow cytometry and chromosome counting | 25% | Zayachkovskaya et al. [21] |
| Gentiana spp. | Gentians | Unfertilized ovule culture | Flow cytometry and molecular marker analysis | 32.5% | Takamura et al. [22] |
| Allium hirtifolium Boiss | Persian shallot | Unfertilized ovary | Squash root | 0–77% | Panahandeh et al. [45] |
| Gentiana triflora | Gentians | Unfertilized ovules | Flow cytometry and Feulgen staining | 23.5–56% | Doi et al. [47] |
| Solanum lycopersicum L. | Tomato | Non-fertilized ovary culture | - | 0% | Bal et al. [48] |
| Species | Common Name | Pathway | Ploidy Level Determination | Haploid Induction Rate | Reference |
|---|---|---|---|---|---|
| Eriobotrya japonica (Thunb.) Lindl. | Loquat | γ–irradiated pollen | Flow cytometry | 0.007–0.008% | Blasco et al. [17] |
| Citrus grandis (L.) Osbeck | Pummelo | γ–irradiated pollen | Flow cytometry | 1%s | Wang et al. [49] |
| Spinacia oleracea L. | Spinach | γ-irradiated pollen | Flow cytometry | - | Keleş et al. [19] |
| Cucumis melo L. | Melon | γ-irradiated pollen | Flow cytometry | 14–33% | Lotfi et al. [50] |
| Cucumis melo L. | Melon | γ-irradiated pollen | Chromosome counting | 23.65% | Nasertorabi et al. [51] |
| Citrus reticulata | Mandarin | γ-irradiated pollen | Flow cytometry | 2.58–8.33% | Jedidi et al. [52] |
| Species | Common Name | Pathway | Ploidy Level Determination | Haploid Induction Rate | Reference |
|---|---|---|---|---|---|
| Triticum aestivum L. | Wheat | Wheat × maize crossing | - | - | Wiśniewska et al. [10] |
| Lactuca sativa L. | Lettuce | Cross-pollination with Helianthus annus L. | Flow cytometry and chromosome counting | 15% | Piosik et al. [56] |
| Lactuca sativa L. | Lettuce | Cross-pollination with Helianthus tuberosus L. | Flow cytometry and chromosome counting | 16% | Piosik et al. [56] |
| Solanum lycopersicum L. | Tomato | Cross-pollination with S. sisymbriifolium Lam. | Chromosome counting | 0% | Bal et al. [59] |
| Solanum lycopersicum L. | Tomato | Cross-pollination with S. sisymbriifolium Lam. | Flow cytometry and chromosome counting | ~10% cells haploids | Chambonnet [60] |
| Species | Common Name | Pathway | Ploidy Level Determination | Haploid Induction Rate | Reference |
|---|---|---|---|---|---|
| Zea mays L. | Maize | Inducer inbred lines | Morphological markers | 2.5–15.7% | Qu et al. [7] |
| Zea mays L. | Maize | BHI Bulk | Embryo coloration (R1-nj) | 11.2–16.8% | Trampe et al. [8] |
| Zea mays L. | Maize | Frame-shift mutation in MATRILINEAL (MTL) | Flow cytometry | 6.7% | Kelliher et al. [9] |
| Zea mays L. | Maize | Eliminate native CENH3- gene | Flow cytometry | 0.05–0.31% | Kelliher et al. [25] |
| Zea mays L. | Maize | Inducer lines (NOT LIKE DAD) | Morphological markers | 0–3.59% | Gilles et al. [62] |
| Triticum aestivum L. | Wheat | Edited the MTL alleles using CRISPR/Cas9 | Chromosome counting | 0–15.6% | Tang et al. [64] |
| Arabidopsis thaliana | Arabidopsis | Edited the DMP genes using CRISPR/Cas9 | Flow cytometry | 0–4.41% | Zhong et al. [65] |
| Brassica napus L. | Oilseed rape | Knocked out of BnaDMP using CRISPR/Cas9 | Flow cytometry | 1.5 +-0.63% | Li et al. [66] |
| Brassica napus L. | Oilseed rape | DMP CRISPR/Cas9 mutagenesis | Flow cytometry | 0–4.44% | Zhong et al. [67] |
| Nicotiana tabacum | Tobacco | DMP CRISPR/Cas9 mutagenesis | Flow cytometry | 0–1.63% | Zhong et al. [67] |
| Nicotiana tabacum | Tobacco | DMP CRISPR/Cas9 mutagenesis | Flow cytometry and cytological observation | 1.52–1.75% | Zhang et al. [68] |
| Medicago truncatula Gaertn | Barrel medic | DMP CRISPR/Cas9 mutagenesis | Flow cytometry | 0.29–0.82% | Wang et al. [69] |
| Solanum lycopersicum L. | Tomato | DMP CRISPR/Cas9 mutagenesis | Flow cytometry | 0.5–3.7% | Zhong et al. [70] |
| Solanum lycopersicum L. | Tomato | Edition of the CENH3 gen with GFP-tailswap disruption | Flow cytometry | 0.2–2.3% | Op Den Camp et al. [71] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marin-Montes, I.M.; Rodríguez-Pérez, J.E.; Robledo-Paz, A.; de la Cruz-Torres, E.; Peña-Lomelí, A.; Sahagún-Castellanos, J. Haploid Induction in Tomato (Solanum lycopersicum L.) via Gynogenesis. Plants 2022, 11, 1595. https://doi.org/10.3390/plants11121595
Marin-Montes IM, Rodríguez-Pérez JE, Robledo-Paz A, de la Cruz-Torres E, Peña-Lomelí A, Sahagún-Castellanos J. Haploid Induction in Tomato (Solanum lycopersicum L.) via Gynogenesis. Plants. 2022; 11(12):1595. https://doi.org/10.3390/plants11121595
Chicago/Turabian StyleMarin-Montes, Ivan Maryn, Juan Enrique Rodríguez-Pérez, Alejandrina Robledo-Paz, Eulogio de la Cruz-Torres, Aureliano Peña-Lomelí, and Jaime Sahagún-Castellanos. 2022. "Haploid Induction in Tomato (Solanum lycopersicum L.) via Gynogenesis" Plants 11, no. 12: 1595. https://doi.org/10.3390/plants11121595
APA StyleMarin-Montes, I. M., Rodríguez-Pérez, J. E., Robledo-Paz, A., de la Cruz-Torres, E., Peña-Lomelí, A., & Sahagún-Castellanos, J. (2022). Haploid Induction in Tomato (Solanum lycopersicum L.) via Gynogenesis. Plants, 11(12), 1595. https://doi.org/10.3390/plants11121595