Detection of Genomic Regions Controlling the Antioxidant Enzymes, Phenolic Content, and Antioxidant Activities in Rice Grain through Association Mapping
Abstract
:1. Introduction
2. Results
2.1. Phenotyping of the Population for Antioxidant Traits in Rice
2.2. Genotype-by-Trait Biplot Analysis for Relatedness among the Germplasm Lines for the Antioxidant Traits
2.3. Nature of Association among Antioxidant Traits
2.4. Genetic Diversity Parameters Analysis
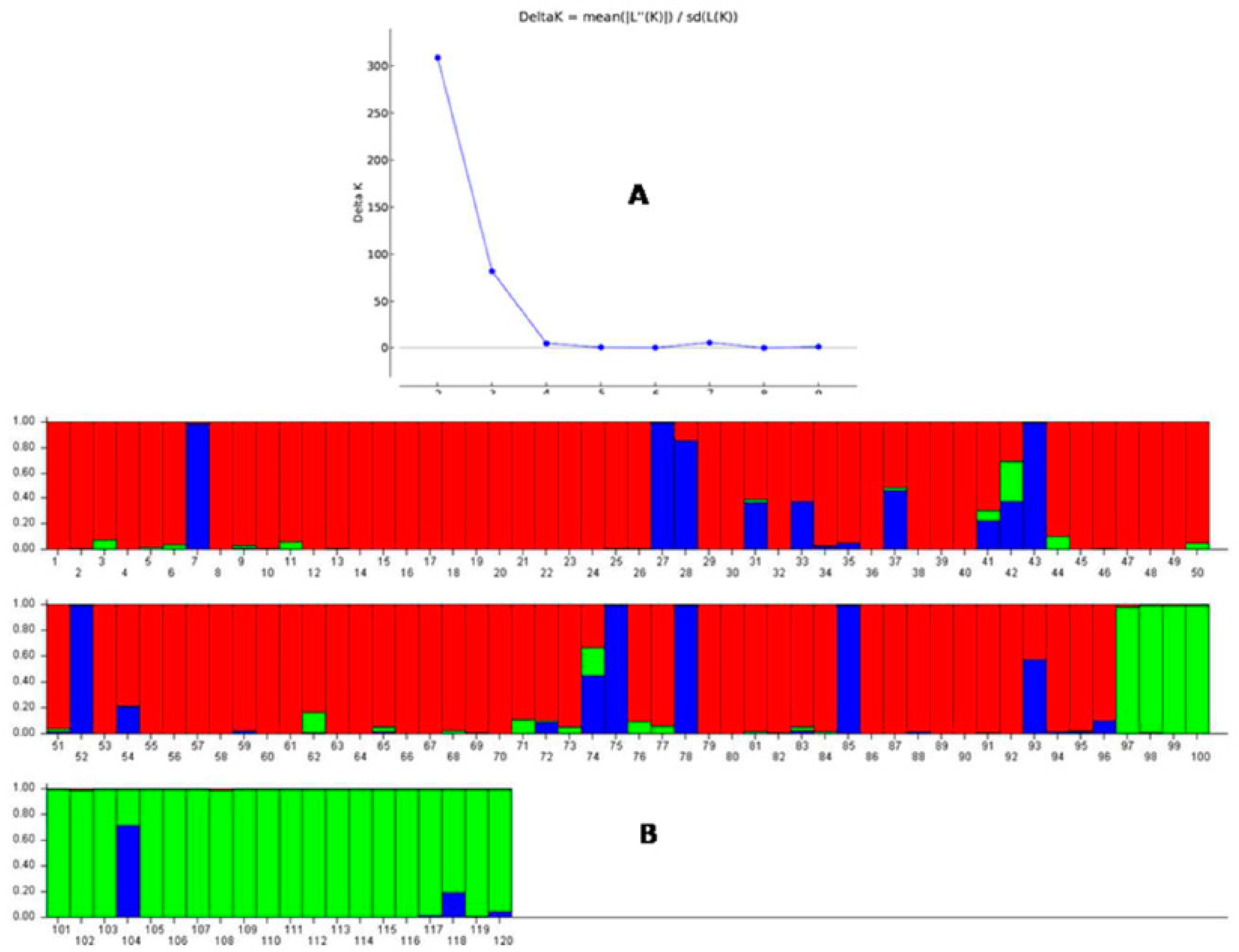
2.5. Population Genetic Structure Analysis
2.6. Molecular Variance (AMOVA) and LD Decay Plot Analysis
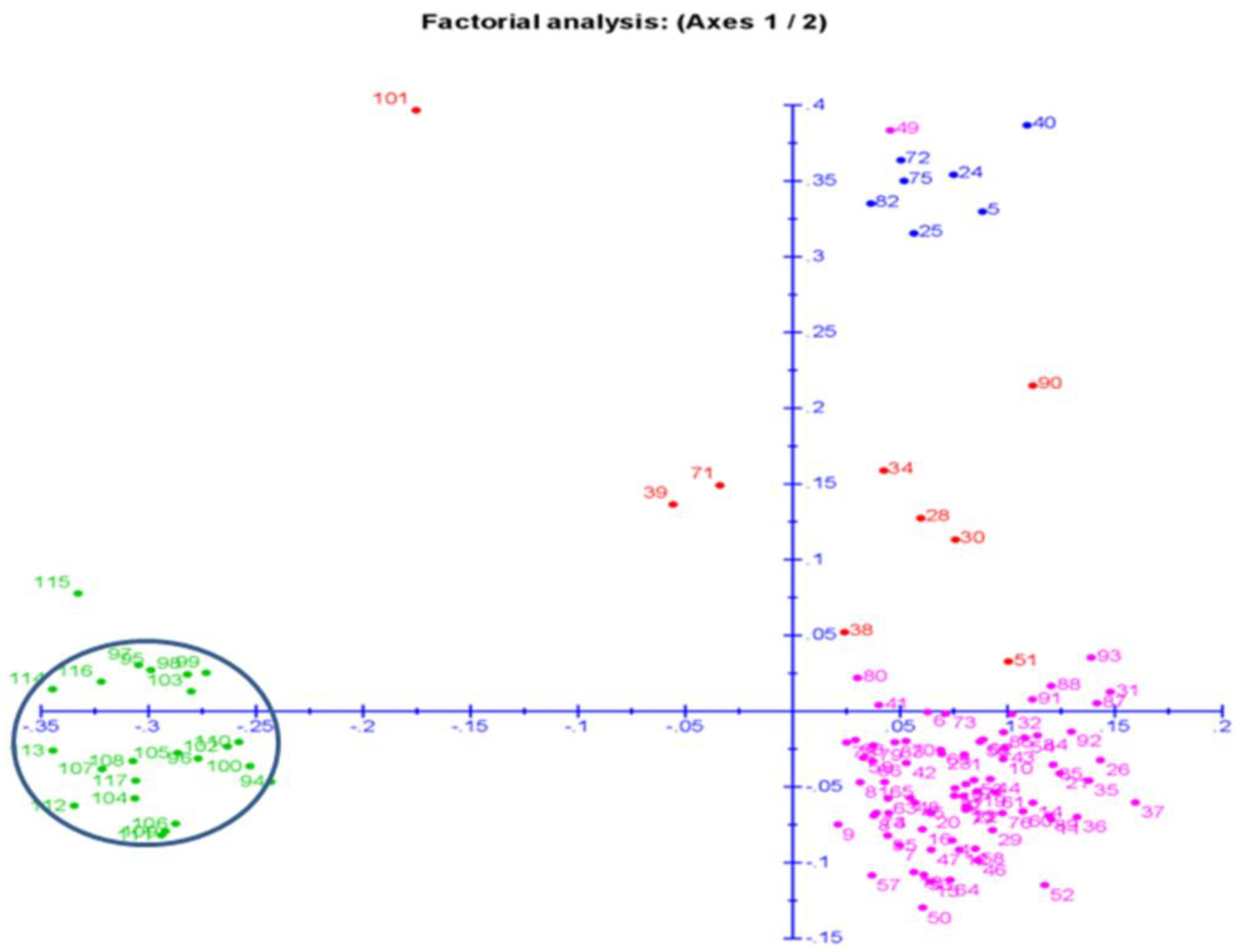
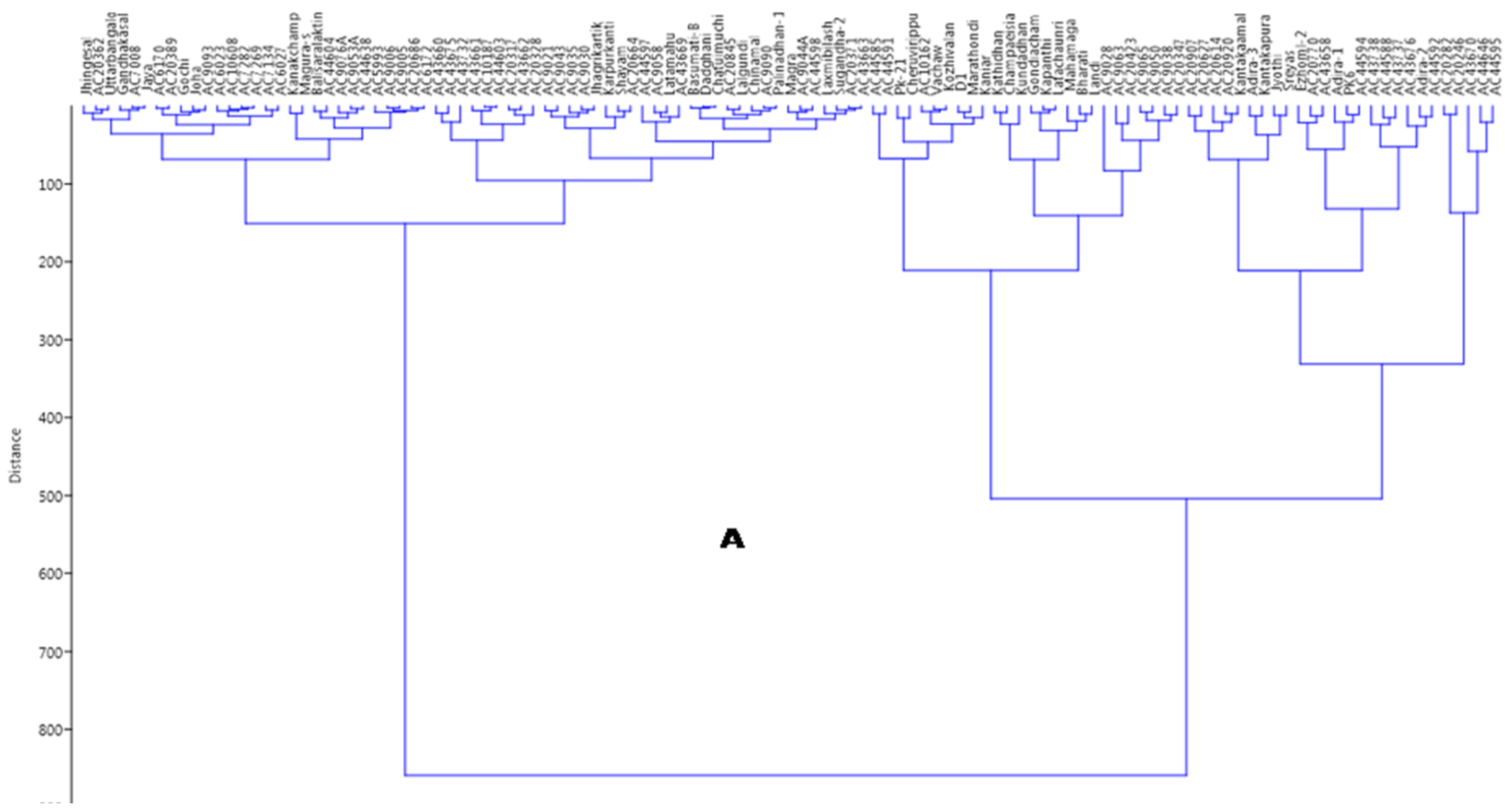
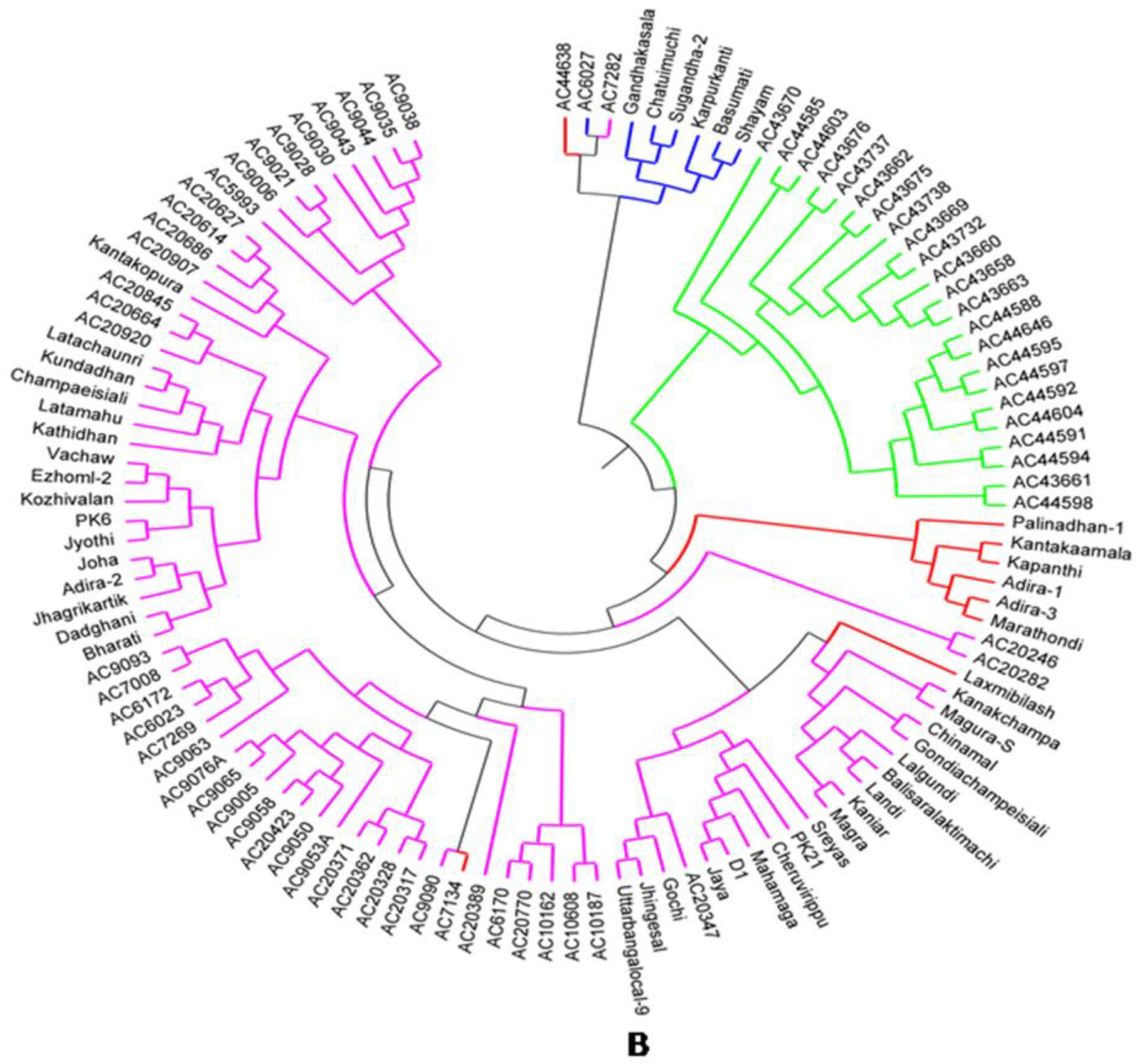
2.7. Principal Coordinates and Cluster Analyses for Genetic Relatedness among the Germplasm Lines
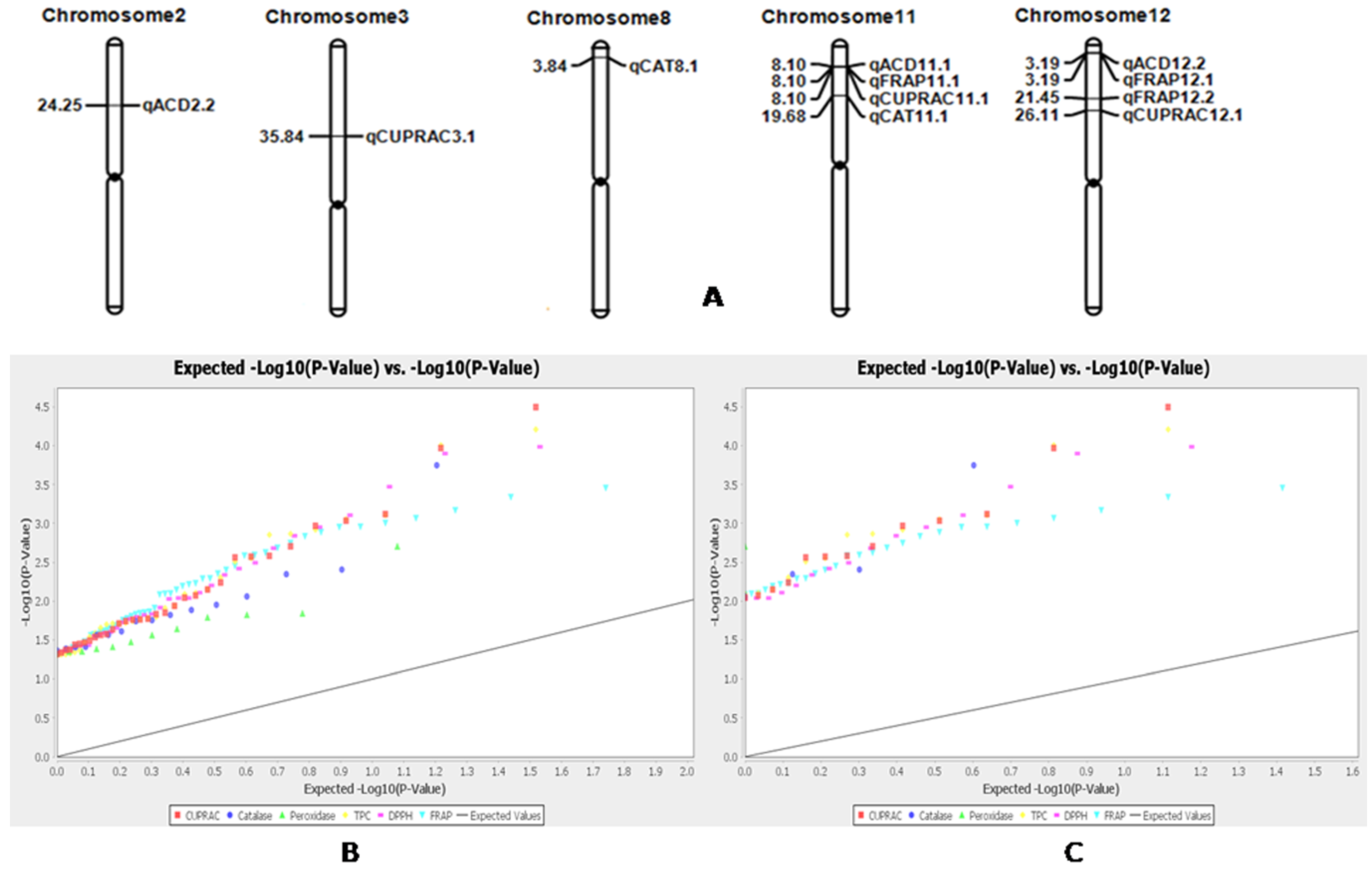
2.8. Marker–Trait Association for Antioxidant Traits in Rice
3. Discussion
4. Materials and Methods
4.1. Seed Material
4.2. Phenotyping for the Antioxidant Traits
4.2.1. Statistical Analysis
4.2.2. Genomic DNA Isolation, PCR Analysis and Selection of SSR Markers
4.3. Molecular Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of variance |
| CAT | Catalase |
| CUPRAC | Cupric ion reducing antioxidant capacity |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| FDR | False discovery rate |
| FRAP | Ferric reducing antioxidant power |
| PEROX | Guaicol peroxidase |
| PIC | Polymorphic information content |
| RBD | Randomized block design |
| TPC | Total phenolics content |
References
- Ferreira, I.C.F.R.; Abreu, R.M.V. Stress oxidativo, antioxidantes efitoquı´micos. Bioanalise 2007, 2, 32–39. [Google Scholar]
- Govindaraj, M.; Masilamani, P.V.; Albert, A.; Bhaskaran, M. Role of antioxidant in seed quality—A review. Agric. Rev. 2017, 38, 180–190. [Google Scholar] [CrossRef]
- Zhu, C.; Chen, J. Changes in soluble sugar and antioxidant enzymes in peanut seeds during ultra dry storage and after accelerated aging. Seed Sci. Technol. 2007, 35, 387–401. [Google Scholar] [CrossRef]
- Demirkaya, M.; Dietz, K.J.; Sivritepe, H.O. Changes in antioxidant enzymes during ageing of onion seeds. Not. Bot. Horti. Agrobo. 2010, 38, 49–52. [Google Scholar] [CrossRef]
- Islam, M.; Begum, H.; Ali, M.; Kamruzzaman, M.; Hoque, S.; Hoque, M. DNA fingerprinting and genetic diversities in some Bangladeshi aus rice (Oryza sativa L.) genotypes. SAARC J. Agric. 2017, 15, 123–137. [Google Scholar] [CrossRef] [Green Version]
- Sanghamitra, P.; Sah, R.P.; Bagchi, T.B.; Sharma, S.G.; Kumar, A.; Munda, S.; Sahu, R.K. Evaluation of variability and environmental stability of grain quality and agronomic parameters of pigmented rice (O. sativa L.). J. Food Sci. Technol. 2018, 55, 879–890. [Google Scholar] [CrossRef]
- Mbanjo, E.G.N.; Kretzschmar, T.; Jones, H.; Ereful, N.; Blanchard, C.; Boyd, L.A.; Sreenivasulu, N. The Genetic Basis and Nutritional Benefits of Pigmented Rice Grain. Front Genet. 2020, 11, 229. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Jin, L.; Zhang, G.; Lu, Y.; Shen, Y.; Bao, J. Association mapping of grain color, phenolic content, flavonoid content and antioxidant capacity in dehulled rice. Theor. Appl. Genet. 2011, 122, 1005–1016. [Google Scholar] [CrossRef]
- Shao, Y.; Bao, J. Polyphenols in whole rice grain: Genetic diversity and health benefits. Food Chem. 2015, 180, 86–97. [Google Scholar] [CrossRef]
- Sahoo, S.; Sanghamitra, P.; Nanda, N.; Pawar, S.; Pandit, E.; Bastia, R.; Muduli, K.C.; Pradhan, S.K. Association of molecular markers with physio-biochemical traits related to seed vigour in rice. Physiol. Mol. Biol. Plants 2020, 26, 1989–2003. [Google Scholar] [CrossRef]
- Yamane, K.; Mitsuya, S.; Taniguchi, M.; Hiroshi, M. Transcription Profiles of Genes Encoding Catalase and Ascorbate Peroxidase in the Rice Leaf Tissues under Salinity. Plant Prod. Sci. 2010, 13, 164–168. [Google Scholar] [CrossRef]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate Peroxidase and Catalase Activities and Their Genetic Regulation in Plants Subjected to Drought and Salinity Stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef] [Green Version]
- Rossatto, T.; de-Amaral, M.N.; Benitez, L.C.; Vighi, I.L.; Braga, E.; de Magalhães, J.; Maia, A.M.M.; da Silva, P.L. Gene expression and activity of antioxidant enzymes in rice plants, cv. BRS AG, under saline stress. Physiol. Mol. Biol. Plants 2017, 23, 865–875. [Google Scholar] [CrossRef] [Green Version]
- Alam, N.B.; Ghosh, A. Comprehensive analysis and transcript profiling of Arabidopsis thaliana and Oryza sativa catalase gene family suggests their specific roles in development and stress responses. Plant Physiol. Biochem. 2018, 123, 54–64. [Google Scholar] [CrossRef]
- Xu, T.Y.; Sun, J.; Chang, H.L.; Zheng, H.L.; Wang, J.G.; Liu, H.L.; Yang, L.M.; Zhao, H.W.; Zou, D.T. QTL mapping for anthocyanin and proanthocyanidin content in red rice. Euphytica 2017, 213, 243. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Menezes-Benavente, L.; Margis, R.; Margis-Pinheiro, M. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: Inferences from the rice genome. J. Mol. Evol. 2004, 59, 761–770. [Google Scholar] [CrossRef]
- Bowler, C.; Montagu, M.V.; Inze, D. Superoxide dismutase and stress tolerance. Ann. Rev. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Jin, L.; Xiao, P.; Lu, Y.; Shao, Y.F.; Shen, Y.; Bao, J.S. Quantitative trait loci for brown rice color, total phenolics and, flavonoid contents and antioxidant capacity in rice grain. Cereal Chem. 2009, 86, 609–615. [Google Scholar] [CrossRef]
- Shao, Y.; Tang, F.; Huang, Y.; Xu, F.; Chen, Y.; Tong, C.; Chen, H.; Bao, J. Analysis of genotype × environment interactions for polyphenols and antioxidant capacity of rice by association mapping. J. Agric. Food Chem. 2014, 62, 5361–5368. [Google Scholar] [CrossRef]
- Xu, F.; Jinsong, B.; Tae-Sung, K.; Yong-Jin, P. Genome-wide association mapping of polyphenol contents and antioxidant capacity in whole-grain rice. J. Agric. Food Chem. 2016, 64, 4695–4703. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Naveenkumar, R.; Barik, S.R.; Mohanty, S.P.; Nayak, D.K.; Ghritlahre, S.K.; Rao, D.S.; Reddy, J.N.; et al. Linkage disequilibrium mapping for grain Fe and Zn enhancing QTLs useful for nutrient dense rice breeding. BMC Plant Biol. 2020, 20, 57. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh, B.I.; Yamasaki, M.; Doebley, J.F.; Mc Mullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed model method for association mapping that accounts for multiple levels of relatedness. Nature Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Pandit, E.; Tasleem, S.; Barik, S.R.; Mohanty, D.P.; Nayak, D.K.; Mohanty, S.P.; Das, S.; Pradhan, S.K. Genome-wide association mapping reveals multiple QTLs governing tolerance response for seedling stage chilling stress in indica rice. Front. Plant Sci. 2017, 8, 552. [Google Scholar] [CrossRef] [Green Version]
- Pawar, S.; Pandit, E.; Arjun, A.; Wagh, M.; Bal, D.; Panda, S.; Bastia, D.N.; Pradhan, S.K.; Mohanty, I.C. Genetic variation and association of molecular markers for iron toxicity tolerance in rice. Oryza 2017, 54, 356–366. [Google Scholar] [CrossRef]
- Oselebe, H.O.; Ogah, O.; Odo, M.I.; Ogbu, K.I. Determination of Phytochemical and Antioxidant Properties of Some Rice Varieties and Hybrids Grown in Ebonyi State, Nigeria. Nig. J. Biotech. 2013, 26, 60–67. [Google Scholar]
- Goufo, P.; Trindade, H. Rice antioxidants: Phenolic acids, flavonoids, anthocyanins, proanthocyanidins, tocopherols, tocotrienols, γ-oryzanol, and phytic acid. Food Sci. Nutr. 2014, 2, 75–104. [Google Scholar] [CrossRef]
- Sanghamitra, P.; Bagchi, T.B.; Sharma, S.G.; Sarkar, S.; Basak, N. Characterization of red and purple-pericarp rice (Oryza sativa L.) based on physico-chemical and antioxidative properties of grains. Oryza 2017, 54, 57–64. [Google Scholar]
- Mahender, A.; Anandan, A.; Pradhan, S.K.; Pandit, E. Rice grain nutritional traits and their enhancement using relevant genes and QTLs through advanced approaches. SpringerPlus 2016, 5, 2086. [Google Scholar] [CrossRef] [Green Version]
- Sanghamitra, P.; Nanda, N.; Barik, S.R.; Sahoo, S.; Pandit, E.; Bastia, R.; Bagchi, T.B.; Pradhan, S.K. Genetic structure and molecular markers-trait association for physiological traits related to seed vigour. Plant Gene 2021, 28. [Google Scholar] [CrossRef]
- Pawar, S.; Pandit, E.; Mohanty, I.C.; Saha, D.; Pradhan, S.K. Population genetic structure and association mapping for iron toxicity tolerance in rice. PLoS ONE 2021, 16, e0246232. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Pandit, E.; Guru, M.; Nayak, D.K.; Tasleem, S.; Barik, S.R.; Mohanty, D.P.; Mohanty, S.P.; Patra, B.C.; Pradhan, S.K. Genetic diversity, population structure, marker validation and kinship analysis for seedling stage cold tolerance in indica rice. Oryza 2018, 55, 396–405. [Google Scholar] [CrossRef]
- Mohapatra, S.; Pandit, E.; Barik, S.R.; Patra, B.C.; Pradhan, S.K. Genetic diversity and population structure in early duration rice genotypes. Oryza 2017, 54, 158–168. [Google Scholar] [CrossRef]
- Barik, S.R.; Pandit, E.; Sanghamitra, P.; Mohanty, S.P.; Behera, A.; Mishra, J.; Nayak, D.K.; Bastia, R.; Moharana, A.; Sahoo, A.; et al. Unraveling the genomic regions controlling the seed vigour index, root growth parameters and germination per cent in rice. PLoS ONE 2022. [Google Scholar] [CrossRef]
- Pandit, E.; Panda, R.K.; Sahoo, A.; Pani, D.R.; Pradhan, S.K. Genetic relationship and structure analyses of root growth angle for improvement of drought avoidance in early and mid-early maturing rice genotypes. Rice Sci. 2020, 27, 124–132. [Google Scholar] [CrossRef]
- Anandan, A.; Anumalla, M.; Pradhan, S.K.; Ali, J. Population Structure, Diversity and Trait Association Analysis in Rice (Oryza sativa L.) Germplasm for early seedling vigor (ESV) Using Trait Linked SSR Markers. PLoS ONE 2016, 11, e0152406. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.K.; Barik, S.R.; Sahoo, A.; Mohapatra, S.; Nayak, D.K.; Mahender, A.; Meher, J.; Anandan, A.; Pandit, E. Population structure, genetic diversity and molecular marker-trait association analysis for high temperature stress tolerance in rice. PLoS ONE 2016, 11, 123. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Bharati, B.; Chatopadhyay, K.; Singh, S.; Dash, P.; Reddy, J.N. Association mapping reveals multiple QTLs for grain protein content in rice useful for biofortification. Mol. Genet. Genom. 2019, 294, 963–983. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro methods. In Methods in Enzymology; Colowick, S.P., Kaplan, N.O., Eds.; Academic Press: Cambridge, UK, 1984; Volume 105, pp. 114–121. [Google Scholar]
- Putter, J. Peroxidase. In Methods of Enzymatic Analysis; Bergmeyer, H.U., Ed.; Academic Press: Cambridge, UK, 1974; pp. 685–690. [Google Scholar]
- Zilic, S.; Hadzi-Taskovic, S.V.; Dodig, D.; Maksimovic, V.; Maksimovic, M.; Basic, Z. Antioxidant activity of small grain cereals caused by phenolics and lipid soluble antioxidants. J. Cereal Sci. 2011, 54, 417–424. [Google Scholar] [CrossRef]
- Zhou, H.M.; Zhu, K.X.; Qian, H.F. Antioxidant and free radical-scavenging activities of wheat germ protein hydrolysates (WGPH) prepared with alcalase. Process Biochem. 2006, 41, 1296–1302. [Google Scholar] [CrossRef]
- Mau, J.L.; Lin, H.C.; Chen, C.C. Antioxidant properties of several medicinal mushrooms. J. Agric. Food Chem. 2002, 50, 6072–6077. [Google Scholar] [CrossRef] [PubMed]
- Apak, R.; Guçlu, K.; Ozyurek, M.; Çelik, S.E. Mechanism of antioxidant capacity assays and the CUPRAC (cupric ion reducing antioxidant capacity) assay. Microchimica Acta 2008, 160, 413–419. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [PubMed] [Green Version]
- Barik, S.R.; Pandit, E.; Pradhan, S.K.; Singh, S.; Mohapatra, T. QTL mapping for relative water content trait at reproductive stage drought tolerance in rice. Ind. J. Genet. Plant Breed. 2018, 78, 401–408. [Google Scholar]
- Barik, S.R.; Pandit, E.; Pradhan, S.K.; Mohanty, S.P.; Mohapatra, T. Genetic mapping of morpho-physiological traits involved during reproductive stage drought tolerance in rice. PLoS ONE 2019, 14, e0214979. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.K.; Pandit, E.; Nayak, D.K.; Behera, L.; Mohapatra, T. Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol. 2019, 19, 352. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Muse, S.V. Power Marker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; Vonholdt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evannomethod. Conserva. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: http://darwin.cirad.fr/Darwin.
- Pandit, E.; Pawar, S.; Barik, S.R.; Mohanty, S.P.; Meher, J.; Pradhan, S.K. Marker-Assisted Backcross Breeding for Improvement of Submergence Tolerance and Grain Yield in the Popular Rice Variety ‘Maudamani’. Agronomy 2021, 11, 1263. [Google Scholar] [CrossRef]
- Mohapatra, S.; Bastia, A.K.; Meher, J.; Sanghamitra, P.; Pradhan, S.K. Development of submergence tolerant, bacterial blight resistant and high yielding near isogenic lines of popular variety, ‘Swarna’ through marker-assisted breeding approach. Front. Plant Sci. 2021. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]

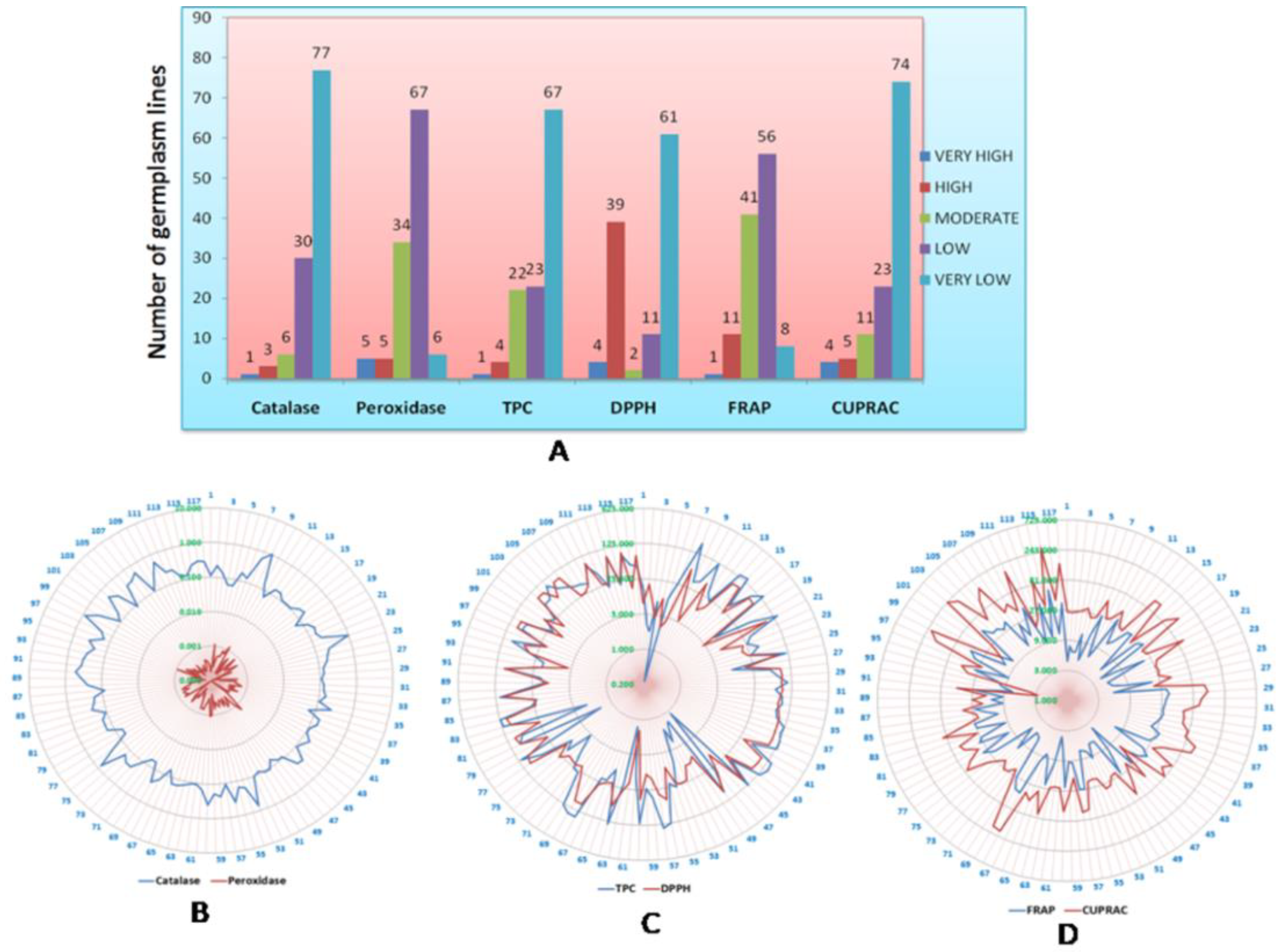



| Sl. No. | Kernel Color | Genotype/Vernacular Name/Accession No. | Catalase | Peroxidase | TPC | DPPH | FRAP | CUPRAC |
|---|---|---|---|---|---|---|---|---|
| 1 | White | AC.5993 | 0.108 | 0.0002 | 14.432 | 8.149 | 4.142 | 25.167 |
| 2 | White | AC.6221 | 0.216 | 0.0003 | 3.136 | 19.613 | 7.313 | 24.958 |
| 3 | White | AC.6023 | 0.130 | 0.0011 | 2.295 | 8.149 | 6.007 | 25.833 |
| 4 | White | AC.6172 | 0.065 | 0.0003 | 9.295 | 4.420 | 6.045 | 27.458 |
| 5 | White | AC.6027 | 0.065 | 0.0005 | 0.232 | 11.050 | 11.978 | 31.500 |
| 6 | White | AC.6007 | 0.173 | 0.0005 | 11.250 | 3.039 | 4.590 | 22.875 |
| 7 | White | AC.9021 | 0.130 | 0.0005 | 34.091 | 4.144 | 12.360 | 24.958 |
| 8 | Red | AC.9028 | 0.628 | 0.0008 | 205.568 | 58.011 | 27.575 | 49.958 |
| 9 | White | AC.9030 | 1.082 | 0.0007 | 35.000 | 11.326 | 5.821 | 26.417 |
| 10 | White | AC.9035 | 0.152 | 0.0013 | 41.364 | 5.249 | 8.955 | 29.958 |
| 11 | Red | AC.9038 | 0.065 | 0.0002 | 118.523 | 42.064 | 31.306 | 44.542 |
| 12 | White | AC.9043 | 0.152 | 0.0005 | 31.477 | 12.155 | 12.052 | 25.792 |
| 13 | White | AC.9044A | 0.152 | 0.0006 | 23.295 | 16.713 | 14.813 | 23.917 |
| 14 | Red | AC.20920 | 0.108 | 0.0002 | 134.886 | 81.107 | 24.963 | 117.875 |
| 15 | Red | AC.20907 | 0.108 | 0.0003 | 148.182 | 86.645 | 27.164 | 118.083 |
| 16 | White | AC.20845 | 0.108 | 0.0008 | 34.773 | 22.801 | 5.075 | 21.208 |
| 17 | Red | AC.20770 | 0.152 | 0.0005 | 94.545 | 84.202 | 23.694 | 89.125 |
| 18 | Red | AC.20627 | 0.173 | 0.0003 | 144.545 | 82.736 | 28.470 | 134.958 |
| 19 | White | AC.20686 | 0.065 | 0.0004 | 8.636 | 4.560 | 4.179 | 24.125 |
| 20 | White | AC.20664 | 0.130 | 0.0007 | 45.455 | 13.192 | 7.276 | 25.792 |
| 21 | Red | AC.20614 | 0.173 | 0.0003 | 148.523 | 85.342 | 20.858 | 101.208 |
| 22 | White | Jhagrikartik | 0.152 | 0.0002 | 34.545 | 8.143 | 4.739 | 32.042 |
| 23 | White | Dadghani | 0.195 | 0.0004 | 26.136 | 21.010 | 5.000 | 28.083 |
| 24 | White | Shayam | 0.974 | 0.0001 | 39.318 | 18.567 | 5.299 | 29.542 |
| 25 | White | Basumati-B | 0.238 | 0.0001 | 26.364 | 22.801 | 7.500 | 27.875 |
| 26 | Red | Bharati | 0.195 | 0.0002 | 114.545 | 80.130 | 20.709 | 41.292 |
| 27 | White | Joha | 0.173 | 0.0001 | 10.000 | 17.590 | 5.187 | 21.625 |
| 28 | Red | Adira-1 | 0.130 | 0.0002 | 51.250 | 78.372 | 32.127 | 123.292 |
| 29 | Red | Adira-2 | 0.130 | 0.0005 | 70.000 | 83.206 | 35.336 | 132.667 |
| 30 | Red | Adira-3 | 0.108 | 0.0002 | 103.864 | 80.407 | 31.381 | 107.875 |
| 31 | Red | PK6 | 0.130 | 0.0005 | 62.386 | 84.478 | 30.448 | 99.125 |
| 32 | Red | VAC.haw | 0.130 | 0.0005 | 84.889 | 77.075 | 28.784 | 58.917 |
| 33 | Red | Kozhivalan | 0.152 | 0.0006 | 87.893 | 72.392 | 29.463 | 61.833 |
| 34 | Red | Marathondi | 0.108 | 0.0007 | 71.023 | 78.880 | 27.440 | 60.583 |
| 35 | Red | Ezhoml-2 | 0.260 | 0.0003 | 78.182 | 88.295 | 24.142 | 76.208 |
| 36 | Red | Jyothi | 0.065 | 0.0005 | 121.818 | 87.786 | 21.716 | 75.792 |
| 37 | Red | Kantakapura | 0.152 | 0.0003 | 113.636 | 90.228 | 31.082 | 118.917 |
| 38 | Red | Kantakaamala | 0.152 | 0.0006 | 128.750 | 90.554 | 28.619 | 109.958 |
| 39 | Red | Kapanthi | 0.108 | 0.0004 | 131.705 | 89.902 | 24.515 | 43.074 |
| 40 | White | Karpurkanti | 0.108 | 0.0004 | 45.909 | 29.967 | 6.866 | 31.333 |
| 41 | Red | Kathidhan | 0.173 | 0.0004 | 148.295 | 89.577 | 22.836 | 76.208 |
| 42 | Red | Kundadhan | 0.130 | 0.0001 | 160.909 | 89.902 | 31.642 | 59.542 |
| 43 | Red | Champaeisiali | 0.108 | 0.0003 | 136.818 | 87.296 | 23.821 | 77.875 |
| 44 | White | Latamahu | 0.260 | 0.0007 | 33.295 | 24.202 | 14.104 | 11.875 |
| 45 | Red | Latachaunri | 0.238 | 0.0015 | 128.068 | 91.531 | 24.634 | 48.292 |
| 46 | White | AC.10608 | 0.087 | 0.0008 | 1.182 | 14.503 | 3.769 | 21.208 |
| 47 | White | AC.10187 | 0.087 | 0.0004 | 31.477 | 34.586 | 10.261 | 35.792 |
| 48 | Red | AC.10162 | 0.065 | 0.0007 | 83.409 | 76.796 | 24.888 | 64.333 |
| 49 | White | AC.7282 | 0.108 | 0.0008 | 1.477 | 9.807 | 5.784 | 22.250 |
| 50 | White | AC.7269 | 0.108 | 0.0004 | 1.722 | 12.569 | 5.709 | 23.500 |
| 51 | White | AC.7134 | 0.087 | 0.0007 | 3.636 | 12.431 | 5.746 | 31.208 |
| 52 | White | AC.7008 | 0.108 | 0.0005 | 1.864 | 23.260 | 9.701 | 32.458 |
| 53 | White | AC.9093 | 0.714 | 0.0004 | 8.750 | 24.309 | 7.201 | 20.792 |
| 54 | White | AC.9090 | 0.173 | 0.0006 | 26.591 | 22.238 | 6.866 | 19.958 |
| 55 | White | AC.9076A | 0.152 | 0.0005 | 21.250 | 9.945 | 6.418 | 20.792 |
| 56 | Red | AC.9065 | 0.390 | 0.0004 | 126.591 | 42.486 | 26.604 | 56.417 |
| 57 | Red | AC.9063 | 0.108 | 0.0007 | 152.500 | 31.878 | 26.007 | 61.208 |
| 58 | White | AC.9058 | 0.238 | 0.0003 | 36.591 | 15.193 | 9.739 | 16.833 |
| 59 | White | AC.9053A | 0.195 | 0.0010 | 23.523 | 12.017 | 9.515 | 28.708 |
| 60 | Red | AC.9050 | 0.411 | 0.0011 | 116.477 | 38.370 | 23.657 | 56.833 |
| 61 | White | AC.9005 | 0.152 | 0.0003 | 13.409 | 1.657 | 5.784 | 24.542 |
| 62 | White | AC.20389 | 0.108 | 0.0004 | 1.409 | 18.730 | 3.769 | 23.917 |
| 63 | White | AC.20371 | 0.108 | 0.0005 | 19.773 | 28.664 | 10.187 | 24.958 |
| 64 | Red | AC.20423 | 0.130 | 0.0007 | 141.591 | 59.772 | 23.396 | 71.208 |
| 65 | White | AC.20362 | 0.152 | 0.0007 | 11.705 | 21.661 | 10.112 | 31.625 |
| 66 | White | AC.20328 | 0.152 | 0.0007 | 20.909 | 23.290 | 7.910 | 48.208 |
| 67 | White | AC.20317 | 0.065 | 0.0003 | 21.250 | 28.990 | 11.157 | 37.667 |
| 68 | Red | AC.20282 | 0.152 | 0.0003 | 169.091 | 87.948 | 42.127 | 202.250 |
| 69 | Red | AC.20246 | 0.238 | 0.0002 | 165.227 | 86.971 | 39.366 | 187.875 |
| 70 | Red | AC.20347 | 0.065 | 0.0003 | 132.727 | 36.156 | 28.060 | 47.250 |
| 71 | White | Palinadhan-1 | 0.065 | 0.0004 | 27.386 | 19.577 | 5.448 | 21.208 |
| 72 | White | Chatuimuchi | 0.108 | 0.0003 | 25.795 | 21.824 | 5.037 | 26.625 |
| 73 | White | Uttarbangalocal-9 | 0.216 | 0.0006 | 11.364 | 20.521 | 4.776 | 27.875 |
| 74 | White | Gochi | 0.216 | 0.0004 | 5.227 | 19.479 | 11.381 | 21.208 |
| 75 | White | Sugandha-2 | 0.152 | 0.0002 | 16.932 | 27.850 | 6.679 | 26.000 |
| 76 | White | Jhingesal | 0.801 | 0.0001 | 11.136 | 14.658 | 4.813 | 36.208 |
| 77 | Red | Cheruvirippu | 0.195 | 0.0002 | 46.932 | 88.804 | 22.425 | 73.917 |
| 78 | Red | Mahamaga | 0.065 | 0.0007 | 111.750 | 77.608 | 31.903 | 55.792 |
| 79 | White | Jaya | 0.065 | 0.0005 | 1.705 | 24.427 | 10.299 | 34.958 |
| 80 | Red | D1 | 0.152 | 0.0006 | 78.864 | 79.898 | 30.261 | 53.292 |
| 81 | Red | Pk-21 | 0.152 | 0.0007 | 62.841 | 88.041 | 25.522 | 59.542 |
| 82 | White | Gandhakasala | 0.152 | 0.0004 | 2.727 | 24.427 | 9.739 | 28.083 |
| 83 | Red | Sreyas | 0.108 | 0.0007 | 117.500 | 84.860 | 26.604 | 91.208 |
| 84 | Red | GondiAC.hampeisiali | 0.238 | 0.0005 | 122.273 | 91.694 | 27.612 | 42.250 |
| 85 | White | Chinamal | 0.195 | 0.0004 | 28.864 | 23.779 | 11.567 | 23.292 |
| 86 | White | Magra | 0.173 | 0.0004 | 21.477 | 23.453 | 16.604 | 24.750 |
| 87 | Red | Landi | 0.087 | 0.0004 | 104.659 | 89.577 | 25.075 | 38.292 |
| 88 | White | Lalgundi | 0.238 | 0.0003 | 33.182 | 26.384 | 9.291 | 18.708 |
| 89 | White | BalisaralaktimAC.hi | 0.346 | 0.0002 | 14.773 | 12.378 | 13.955 | 27.250 |
| 90 | White | Laxmibilash | 0.541 | 0.0002 | 17.727 | 23.127 | 9.216 | 16.625 |
| 91 | Red | Kaniar | 0.390 | 0.0001 | 82.955 | 89.251 | 26.418 | 48.083 |
| 92 | White | Kanakchampa | 0.368 | 0.0004 | 17.273 | 18.567 | 6.903 | 2.833 |
| 93 | White | Magura-s | 0.303 | 0.0002 | 17.273 | 5.700 | 10.560 | 3.542 |
| 94 | White | AC.44603 | 0.216 | 0.0003 | 32.273 | 34.351 | 11.940 | 36.208 |
| 95 | Red | AC.44585 | 0.346 | 0.0009 | 85.227 | 48.295 | 29.963 | 48.917 |
| 96 | White | AC.44598 | 0.152 | 0.0002 | 20.000 | 18.321 | 8.955 | 24.542 |
| 97 | Red | AC.44592 | 0.216 | 0.0006 | 68.068 | 83.969 | 34.104 | 151.833 |
| 98 | Red | AC.44646 | 0.866 | 0.0004 | 65.000 | 83.461 | 44.813 | 211.625 |
| 99 | White | AC.44604 | 0.390 | 0.0003 | 19.886 | 12.276 | 12.201 | 35.375 |
| 100 | White | AC.44597 | 0.108 | 0.0004 | 32.955 | 38.168 | 16.567 | 16.208 |
| 101 | White | AC.44638 | 0.173 | 0.0003 | 25.795 | 15.776 | 10.672 | 27.250 |
| 102 | Red | AC.44595 | 0.519 | 0.0003 | 54.659 | 84.478 | 49.440 | 238.708 |
| 103 | Red | AC.44588 | 0.346 | 0.0002 | 54.545 | 83.715 | 35.149 | 186.417 |
| 104 | Red | AC.44591 | 0.087 | 0.0004 | 94.773 | 46.387 | 35.597 | 57.458 |
| 105 | Red | AC.44594 | 0.390 | 0.0002 | 62.273 | 86.005 | 29.507 | 114.750 |
| 106 | Red | AC.43737 | 0.411 | 0.0003 | 52.500 | 88.931 | 36.642 | 166.000 |
| 107 | White | AC.43660 | 0.563 | 0.0003 | 44.432 | 26.005 | 11.231 | 47.875 |
| 108 | White | AC.43732 | 0.108 | 0.0002 | 41.250 | 26.768 | 25.672 | 59.542 |
| 109 | White | AC.43661 | 0.238 | 0.0003 | 36.818 | 34.020 | 14.627 | 39.542 |
| 110 | Red | AC.43738 | 0.606 | 0.0002 | 75.341 | 86.768 | 40.933 | 174.958 |
| 111 | White | AC.43669 | 0.238 | 0.0002 | 45.000 | 27.405 | 15.746 | 18.083 |
| 112 | White | AC.43663 | 0.238 | 0.0003 | 22.045 | 25.496 | 9.478 | 24.750 |
| 113 | Red | AC.43658 | 0.130 | 0.0004 | 86.364 | 85.496 | 38.843 | 93.708 |
| 114 | White | AC.43662 | 0.130 | 0.0004 | 21.250 | 23.282 | 10.634 | 34.333 |
| 115 | Red | AC.43670 | 0.238 | 0.0004 | 80.455 | 88.550 | 59.142 | 287.042 |
| 116 | White | AC.43675 | 0.303 | 0.0003 | 48.636 | 16.768 | 11.716 | 39.750 |
| 117 | Red | AC.43676 | 0.281 | 0.0003 | 45.341 | 73.664 | 35.522 | 149.125 |
| Mean | 0.219 | 0.000 | 58.365 | 44.918 | 18.435 | 58.691 | ||
| CV % | 10.75 | 11.32 | 5.78 | 4.64 | 4.72 | 5.81 | ||
| LSD5% | 0.0448 | 0.00009 | 6.122 | 3.303 | 1.579 | 6.54 |
| Sl. No. | Accession No./ Vernacular Name of Germplasm Line | Inferred Ancestry Value at K = 3 | Group | Antioxidants Traits in Each Germplasm Line | ||
|---|---|---|---|---|---|---|
| Q1 | Q2 | Q3 | ||||
| 1 | AC5993 | 0.995 | 0.003 | 0.003 | SP1 | Very low |
| 2 | AC6170 | 0.992 | 0.006 | 0.002 | SP1 | Low |
| 3 | AC6023 | 0.981 | 0.017 | 0.002 | SP1 | High Peroxidase |
| 4 | AC6172 | 0.961 | 0.037 | 0.002 | SP1 | Low |
| 5 | AC6027 | 0.009 | 0.002 | 0.989 | SP3 | Very low |
| 6 | AC9006 | 0.995 | 0.003 | 0.002 | SP1 | Very low |
| 7 | AC9021 | 0.986 | 0.011 | 0.003 | SP1 | Very low |
| 8 | AC9028 | 0.937 | 0.06 | 0.003 | SP1 | High Peroxidase |
| 9 | AC9030 | 0.994 | 0.005 | 0.001 | SP1 | High Catalase |
| 10 | AC9035 | 0.987 | 0.004 | 0.009 | SP1 | High Peroxidase |
| 11 | AC9038 | 0.998 | 0.001 | 0.001 | SP1 | Very low |
| 12 | AC9043 | 0.998 | 0.001 | 0.001 | SP1 | Low |
| 13 | AC9044 | 0.994 | 0.002 | 0.003 | SP1 | Very low |
| 14 | AC20920 | 0.995 | 0.002 | 0.003 | SP1 | High DPPH |
| 15 | AC20907 | 0.997 | 0.002 | 0.001 | SP1 | High DPPH |
| 16 | AC20845 | 0.997 | 0.002 | 0.001 | SP1 | High Peroxidase |
| 17 | AC20770 | 0.997 | 0.001 | 0.002 | SP1 | High DPPH |
| 18 | AC20627 | 0.998 | 0.001 | 0.001 | SP1 | High DPPH |
| 19 | AC20686 | 0.997 | 0.002 | 0.001 | SP1 | Very low |
| 20 | AC20664 | 0.996 | 0.002 | 0.001 | SP1 | Low |
| 21 | AC20614 | 0.996 | 0.001 | 0.003 | SP1 | High DPPH |
| 22 | Jhagrikarti | 0.99 | 0.009 | 0.002 | SP1 | Very low |
| 23 | Dadghani | 0.991 | 0.005 | 0.004 | SP1 | Very low |
| 24 | Shayam | 0.004 | 0.002 | 0.994 | SP3 | High Catalase |
| 25 | Basumati | 0.138 | 0.004 | 0.859 | SP3 | Low |
| 26 | Bharati | 0.997 | 0.001 | 0.002 | SP1 | High DPPH |
| 27 | Joha | 0.997 | 0.001 | 0.002 | SP1 | Very low |
| 28 | Adira-1 | 0.613 | 0.018 | 0.369 | A | Medium |
| 29 | Adira-2 | 0.997 | 0.002 | 0.001 | SP1 | High DPPH |
| 30 | Adira-3 | 0.622 | 0.003 | 0.376 | A | High DPPH |
| 31 | PK6 | 0.969 | 0.003 | 0.028 | SP1 | High DPPH |
| 32 | Vachaw | 0.946 | 0.002 | 0.053 | SP1 | High DPPH |
| 33 | Kozhivalan | 0.998 | 0.002 | 0.001 | SP1 | High DPPH |
| 34 | Marathondi | 0.515 | 0.022 | 0.463 | A | High DPPH |
| 35 | Ezhoml-2 | 0.998 | 0.001 | 0.001 | SP1 | High DPPH |
| 36 | Jyothi | 0.998 | 0.001 | 0.001 | SP1 | High DPPH |
| 37 | Kantakopura | 0.997 | 0.001 | 0.002 | SP1 | High DPPH |
| 38 | Kantakaamal | 0.693 | 0.081 | 0.227 | A | High DPPH |
| 39 | Kapanthi | 0.302 | 0.323 | 0.375 | A | High DPPH |
| 40 | Karpurkanti | 0.002 | 0.001 | 0.997 | SP3 | Very low |
| 41 | Kathidhan | 0.899 | 0.095 | 0.006 | SP1 | High DPPH |
| 42 | Kundadhan | 0.995 | 0.004 | 0.001 | SP1 | High TPC and DPPH |
| 43 | Champaeisia | 0.991 | 0.003 | 0.006 | SP1 | High DPPH |
| 44 | Latamahu | 0.996 | 0.002 | 0.002 | SP1 | Low |
| 45 | Latachaunri | 0.994 | 0.004 | 0.002 | SP1 | High DPPH and Peroxidase |
| 46 | AC10608 | 0.995 | 0.005 | 0.001 | SP1 | High Peroxidase |
| 47 | AC10187 | 0.945 | 0.052 | 0.002 | SP1 | High DPPH |
| 48 | AC10162 | 0.963 | 0.021 | 0.016 | SP1 | High DPPH |
| 49 | AC7282 | 0.002 | 0.001 | 0.997 | SP1 | High Peroxidase |
| 50 | AC7269 | 0.997 | 0.002 | 0.001 | SP1 | Very low |
| 51 | AC7134 | 0.785 | 0.006 | 0.208 | A | Low |
| 52 | AC7008 | 0.998 | 0.001 | 0.001 | SP1 | Low |
| 53 | AC9093 | 0.996 | 0.001 | 0.003 | SP1 | High Catalase |
| 54 | AC9090 | 0.993 | 0.003 | 0.004 | SP1 | Very low |
| 55 | AC9076A | 0.995 | 0.003 | 0.001 | SP1 | Low |
| 56 | AC9065 | 0.973 | 0.003 | 0.024 | SP1 | Low |
| 57 | AC9063 | 0.993 | 0.006 | 0.001 | SP1 | High TPC |
| 58 | AC9058 | 0.998 | 0.001 | 0.001 | SP1 | Low |
| 59 | AC9053A | 0.831 | 0.158 | 0.011 | SP1 | High Peroxidase |
| 60 | AC9050 | 0.994 | 0.001 | 0.004 | SP1 | Low |
| 61 | AC9005 | 0.994 | 0.004 | 0.002 | SP1 | Low |
| 62 | AC20389 | 0.945 | 0.035 | 0.02 | SP1 | Low |
| 63 | AC20371 | 0.993 | 0.006 | 0.001 | SP1 | Low |
| 64 | AC20423 | 0.996 | 0.003 | 0.001 | SP1 | Low |
| 65 | AC20362 | 0.977 | 0.018 | 0.005 | SP1 | Low |
| 66 | AC20328 | 0.986 | 0.008 | 0.006 | SP1 | Low |
| 67 | AC20317 | 0.996 | 0.002 | 0.002 | SP1 | Low |
| 68 | AC20282 | 0.889 | 0.104 | 0.007 | SP1 | High CUPRAC, Cata, TPC, DPPH and FRAP |
| 69 | AC20246 | 0.894 | 0.017 | 0.089 | SP1 | High CUPRAC, FRAP and DPPH |
| 70 | AC20347 | 0.943 | 0.055 | 0.002 | SP1 | Low |
| 71 | Palinadhan- | 0.334 | 0.214 | 0.452 | A | High DPPH |
| 72 | Chatuimuchi | 0.001 | 0.001 | 0.998 | SP3 | Very low |
| 73 | Uttarbangal | 0.904 | 0.094 | 0.002 | SP1 | Very low |
| 74 | Gochi | 0.941 | 0.053 | 0.006 | SP1 | High DPPH |
| 75 | Sugandha-2 | 0.002 | 0.001 | 0.998 | SP3 | Very low |
| 76 | Jhingesal | 0.997 | 0.002 | 0.001 | SP1 | High Catatalase |
| 77 | Cheruviripp | 0.996 | 0.003 | 0.001 | SP1 | High DPPH |
| 78 | Mahamaga | 0.985 | 0.013 | 0.002 | SP1 | High DPPH |
| 79 | Jaya | 0.991 | 0.008 | 0.001 | SP1 | Low |
| 80 | D1 | 0.944 | 0.033 | 0.023 | SP1 | High DPPH |
| 81 | PK21 | 0.985 | 0.014 | 0.001 | SP1 | High DPPH |
| 82 | Gandhakasal | 0.004 | 0.003 | 0.993 | SP3 | Very low |
| 83 | Sreyas | 0.995 | 0.002 | 0.003 | SP1 | High DPPH |
| 84 | Gondiachamp | 0.995 | 0.002 | 0.003 | SP1 | High DPPH |
| 85 | Chinamal | 0.981 | 0.001 | 0.017 | SP1 | Low |
| 86 | Magra | 0.995 | 0.002 | 0.003 | SP1 | Very low |
| 87 | Landi | 0.997 | 0.001 | 0.002 | SP1 | High DPPH |
| 88 | Lalgundi | 0.99 | 0.003 | 0.007 | SP1 | Very low |
| 89 | Balisaralak | 0.994 | 0.003 | 0.003 | SP1 | Very low |
| 90 | Laxmibilash | 0.426 | 0.002 | 0.572 | A | Low |
| 91 | Kaniar | 0.98 | 0.005 | 0.016 | SP1 | High DPPH |
| 92 | Kanakchampa | 0.976 | 0.003 | 0.02 | SP1 | Very low |
| 93 | Magura-S | 0.895 | 0.001 | 0.104 | SP1 | Very low |
| 94 | AC44603 | 0.017 | 0.981 | 0.001 | SP2 | Low |
| 95 | AC44585 | 0.004 | 0.984 | 0.012 | SP2 | High Peroxidase |
| 96 | AC44598 | 0.007 | 0.987 | 0.006 | SP2 | Low |
| 97 | AC44592 | 0.995 | 0.003 | 0.003 | SP2 | High DPPH |
| 98 | AC44646 | 0.001 | 0.997 | 0.001 | SP2 | High Cata, DPPH, FRAP and CUPRAC |
| 99 | AC44604 | 0.001 | 0.998 | 0.001 | SP2 | Medium |
| 100 | AC44597 | 0.013 | 0.98 | 0.007 | SP2 | Medium |
| 101 | AC44638 | 0.002 | 0.997 | 0.001 | A | Low |
| 102 | AC44595 | 0.001 | 0.284 | 0.715 | SP2 | High CUPRAC, FRAP and DPPH |
| 103 | AC44588 | 0.005 | 0.994 | 0.002 | SP2 | High CUPRAC, DPPH and FRAP |
| 104 | AC44591 | 0.002 | 0.997 | 0.001 | SP2 | Low |
| 105 | AC44594 | 0.002 | 0.998 | 0.001 | SP2 | High DPPH |
| 106 | AC43737 | 0.01 | 0.988 | 0.002 | SP2 | High DPPH and CUPRAC |
| 107 | AC43660 | 0.002 | 0.997 | 0.001 | SP2 | High Catalase, DPPH, FRAP, CUPRAC |
| 108 | AC43732 | 0.003 | 0.996 | 0.001 | SP2 | High Catalase, DPPH and CUPRAC |
| 109 | AC43661 | 0.001 | 0.998 | 0.001 | SP2 | Low |
| 110 | AC43738 | 0.004 | 0.995 | 0.001 | SP2 | High Catalase and CUPRAC |
| 111 | AC43669 | 0.002 | 0.997 | 0.001 | SP2 | High DPPH |
| 112 | AC43663 | 0.003 | 0.994 | 0.003 | SP2 | High DPPH |
| 113 | AC43658 | 0.001 | 0.997 | 0.002 | SP2 | High DPPH |
| 114 | AC43662 | 0.001 | 0.998 | 0.001 | SP2 | Low |
| 115 | AC43670 | 0.003 | 0.981 | 0.016 | SP2 | High DPPH and CUPRAC |
| 116 | AC43675 | 0.002 | 0.805 | 0.193 | SP2 | High DPPH |
| 117 | AC43676 | 0.002 | 0.986 | 0.012 | SP2 | Medium |
| Source of Variation | AMOVA for the Four Subpopulations at K = 3 | |||
|---|---|---|---|---|
| df. | Mean Sum of Squares | Variance Components | Percentage Variation | |
| Among populations | 3 | 0.641 | 0.003 | 1% |
| Among individuals (accessions) within population | 113 | 0.514 | 0.022 | 4% |
| Within individuals (accessions) | 117 | 0.470 | 0.470 | 95% |
| Total | 233 | 0.495 | ||
| F-Statistics | Value | p-value | ||
| FST | 0.006 | 0.121 | ||
| FIS | 0.045 | 0.003 | ||
| FIT | 0.051 | 0.001 | ||
| FST max. | 0.014 | |||
| F’ST | 0.460 | |||
| Sl. No. | Antioxidant Enzymes | Marker | Position (cM) | GLM | MLM | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker_F | Marker_p | Marker_R2 | q-Value | Marker_F | Marker_p | Marker_R2 | q-Value | ||||
| 1 | Catalase | RM1341 | 80.2 | 9.99747 | 0.00204 | 0.08016 | 0.00564 | 7.8566 | 0.006 | 0.07179 | 0.00994 |
| 2 | Catalase | RM3231 | 32.7 | 10.55577 | 0.00154 | 0.08424 | 0.00564 | 8.16013 | 0.00514 | 0.07457 | 0.009638 |
| 3 | DPPH | RM247 | 32.3 | 10.72025 | 0.00142 | 0.08855 | 0.00966 | 10.07384 | 0.00196 | 0.09162 | 0.006125 |
| 4 | DPPH | RM3701 | 45.3 | 11.90813 | 7.99 × 10−4 | 0.09738 | 0.00564 | 11.09733 | 0.00118 | 0.10093 | 0.006125 |
| 5 | DPPH | RM13600 | 110.2 | 9.40651 | 0.00273 | 0.0779 | 0.00652 | 6.88723 | 0.00994 | 0.06264 | 0.00994 |
| 6 | FRAP | RM247 | 32.2 | 9.40651 | 0.00273 | 0.0779 | 0.00617 | 7.14597 | 0.00868 | 0.06551 | 0.00994 |
| 7 | FRAP | RM3701 | 45.3 | 9.11231 | 0.00317 | 0.06781 | 0.00617 | 8.98419 | 0.00338 | 0.08236 | 0.007243 |
| 8 | FRAP | RM309 | 74.5 | 12.35495 | 6.44 × 10−4 | 0.08946 | 0.00617 | 7.35763 | 0.00777 | 0.06745 | 0.00994 |
| 9 | CUPRAC | RM3701 | 45.3 | 14.56812 | 2.26 × 10−4 | 0.10344 | 0.00771 | 9.65365 | 0.00241 | 0.08678 | 0.006125 |
| 10 | CUPRAC | RM235 | 101.8 | 9.11231 | 0.00317 | 0.06781 | 0.00564 | 10.03931 | 0.00199 | 0.09024 | 0.006125 |
| 11 | CUPRAC | RM148 | 142.3 | 12.35495 | 6.44 × 10−4 | 0.08946 | 0.00966 | 7.03523 | 0.0092 | 0.06324 | 0.00994 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanghamitra, P.; Barik, S.R.; Bastia, R.; Mohanty, S.P.; Pandit, E.; Behera, A.; Mishra, J.; Kumar, G.; Pradhan, S.K. Detection of Genomic Regions Controlling the Antioxidant Enzymes, Phenolic Content, and Antioxidant Activities in Rice Grain through Association Mapping. Plants 2022, 11, 1463. https://doi.org/10.3390/plants11111463
Sanghamitra P, Barik SR, Bastia R, Mohanty SP, Pandit E, Behera A, Mishra J, Kumar G, Pradhan SK. Detection of Genomic Regions Controlling the Antioxidant Enzymes, Phenolic Content, and Antioxidant Activities in Rice Grain through Association Mapping. Plants. 2022; 11(11):1463. https://doi.org/10.3390/plants11111463
Chicago/Turabian StyleSanghamitra, Priyadarsini, Saumya Ranjan Barik, Ramakrushna Bastia, Shakti Prakash Mohanty, Elssa Pandit, Abhisarika Behera, Jyotirmayee Mishra, Gaurav Kumar, and Sharat Kumar Pradhan. 2022. "Detection of Genomic Regions Controlling the Antioxidant Enzymes, Phenolic Content, and Antioxidant Activities in Rice Grain through Association Mapping" Plants 11, no. 11: 1463. https://doi.org/10.3390/plants11111463
APA StyleSanghamitra, P., Barik, S. R., Bastia, R., Mohanty, S. P., Pandit, E., Behera, A., Mishra, J., Kumar, G., & Pradhan, S. K. (2022). Detection of Genomic Regions Controlling the Antioxidant Enzymes, Phenolic Content, and Antioxidant Activities in Rice Grain through Association Mapping. Plants, 11(11), 1463. https://doi.org/10.3390/plants11111463






