DNA Barcoding Medicinal Plant Species from Indonesia
Abstract
:1. Introduction
2. Results and Discussions
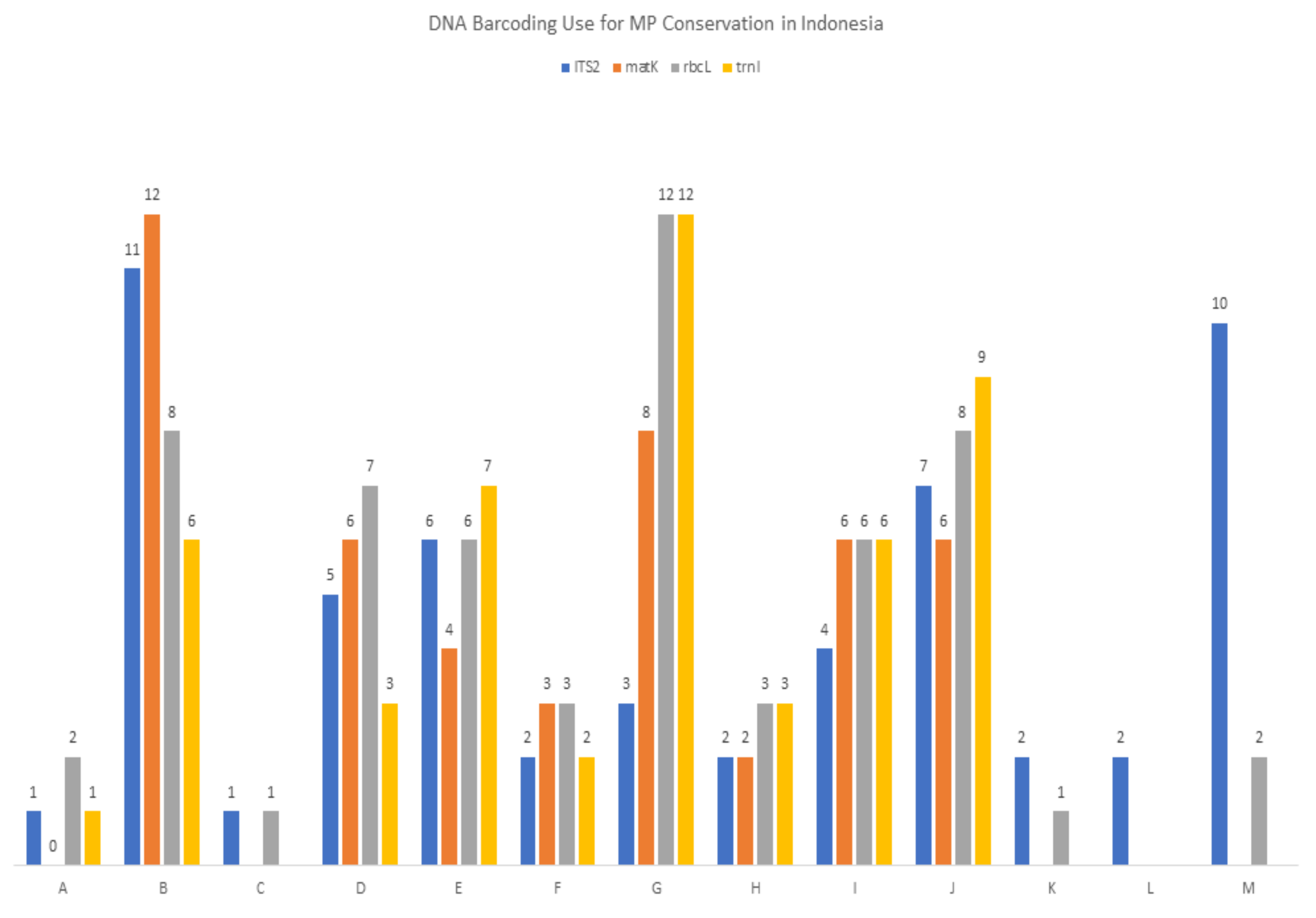
2.1. Understanding the Use of DNA Barcoding for Indonesian Medicinal Plants
2.2. Understanding the Effectiveness of Each DNA Barcoding Region Used for Indonesian Medicinal Plants Identification
2.3. Description of ITS2, matK, rbcL, and trnL Regions of Indonesian Medicinal Plants
2.4. Identification of Indonesian Medicinal Plants Using Sequences of Their ITS2, matK, rbcL, and trnL Regions
3. Materials and Methods
3.1. Plant Samples and Literature Survey
3.2. Molecular Analysis
3.3. Sequence Analyses and Data Interpretation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| DNA Barcoding Use for MP Conservation in Indonesia | ITS2 | matK | rbcL | trnL |
|---|---|---|---|---|
| A. new DNA barcoding and can strongly assist MP conservation | 1 | 1 | 2 | 1 |
| Anaxagorea javanica | 1 | |||
| Aquilaria hirta | 1 | |||
| Strongyleria pannea | 1 | 1 | 1 | |
| B. can strongly assist MP conservation | 11 | 12 | 8 | 6 |
| Alstonia scholaris | 1 | 1 | 1 | 1 |
| Alyxia reinwardtii | 1 | 1 | 1 | |
| Cymbidium aloifolium | 1 | 1 | 1 | |
| Dendrobium crumenatum | 1 | 1 | ||
| Dendrobium salaccense | 1 | 1 | 1 | |
| Euphorbia tirucalli | 1 | |||
| Ficus deltoidea | 1 | |||
| Galearia filiformis | 1 | 1 | 1 | |
| Kadsura scandens | 1 | |||
| Lunasia amara | 1 | 1 | 1 | |
| Nepenthes gracilis | 1 | |||
| Nepenthes reinwardtiana | 1 | 1 | ||
| Nervilia plicata | 1 | 1 | 1 | |
| Pangium edule | 1 | 1 | ||
| Parkia timoriana | 1 | |||
| Rauvolfia serpentina | 1 | 1 | 1 | 1 |
| C. new DNA barcoding and can assist MP conservation | 1 | 1 | ||
| Aglaonema commutatum | 1 | |||
| Meistera aculeata | 1 | |||
| D. can assist MP conservation | 5 | 6 | 7 | 3 |
| Alstonia macrophylla | 1 | 1 | ||
| Ancistrocladus tectorius | 1 | 1 | 1 | |
| Ardisia crenata | 1 | 1 | ||
| Dasymaschalon dasymaschalum | 1 | |||
| Justicia gendarussa | 1 | 1 | 1 | 1 |
| Orthosiphon aristatus | 1 | |||
| Phyllanthus oxyphyllus | 1 | 1 | ||
| Premna serratifolia | 1 | |||
| Toxicodendron succedaneum | 1 | 1 | 1 | 1 |
| Vitex glabrata | 1 | |||
| E. new to DNA bank data and new DNA barcoding and may strongly assist MP conservation | 6 | 4 | 6 | 7 |
| Amomum hochreutineri | 1 | 1 | 1 | |
| Dendrobium purpureum | 1 | 1 | 1 | 1 |
| Etlingera solaris | 1 | 1 | 1 | |
| Myristica succedanea | 1 | 1 | 1 | |
| Oberonia lycopodioides | 1 | 1 | 1 | 1 |
| Phanera fulva | 1 | 1 | ||
| Rhododendron macgregoriae | 1 | 1 | 1 | 1 |
| F. new DNA barcoding and may strongly assist MP conservation | 2 | 3 | 2 | 2 |
| Acriopsis liliifolia var. liliifolia | 1 | 1 | 1 | 1 |
| Anaxagorea javanica | 1 | 1 | ||
| Aquilaria hirta | 1 | 1 | 1 | |
| G. may strongly assist MP conservation | 3 | 8 | 12 | 12 |
| Alyxia reinwardtii | 1 | |||
| Cibotium barometz | 1 | |||
| Cymbidium aloifolium | 1 | |||
| Cymbidium ensifolium | 1 | 1 | ||
| Dendrobium crumenatum | 1 | |||
| Dendrobium salaccense | 1 | |||
| Euphorbia tirucalli | 1 | |||
| Ficus deltoidea | 1 | 1 | 1 | |
| Grammatophyllum speciosum | 1 | 1 | 1 | |
| Kadsura scandens | 1 | 1 | 1 | |
| Lunasia amara | 1 | |||
| Nepenthes ampullaria | 1 | 1 | 1 | |
| Nepenthes gracilis | 1 | 1 | ||
| Nepenthes mirabilis | 1 | 1 | 1 | 1 |
| Nepenthes reinwardtiana | 1 | 1 | ||
| Nervilia concolor | 1 | |||
| Pangium edule | 1 | |||
| Parkia timoriana | 1 | 1 | ||
| Smilax zeylanica | 1 | 1 | ||
| H. new to DNA bank data and new DNA barcoding and may assist MP conservation | 2 | 2 | 3 | 3 |
| Acalypha grandis | 1 | 1 | ||
| Ardisia complanata | 1 | 1 | 1 | 1 |
| Erycibe malaccensis | 1 | 1 | 1 | 1 |
| I. new DNA barcoding and may assist MP conservation | 4 | 6 | 7 | 6 |
| Aglaonema commutatum | 1 | 1 | ||
| Cinnamomum rhynchophyllum | 1 | 1 | 1 | |
| Decalobanthus mammosus | 1 | |||
| Hoya diversifolia | 1 | 1 | 1 | 1 |
| Meistera aculeata | 1 | |||
| Melicope lunu-ankenda | 1 | 1 | 1 | 1 |
| Psychotria montana | 1 | 1 | 1 | 1 |
| Spondias malayana | 1 | |||
| Ventilago madraspatana | 1 | 1 | 1 | |
| J. may assist MP conservation | 7 | 6 | 8 | 9 |
| Alstonia macrophylla | 1 | 1 | ||
| Ancistrocladus tectorius | 1 | |||
| Ardisia crenata | 1 | 1 | ||
| Benstonea affinis | 1 | 1 | 1 | |
| Dasymaschalon dasymaschalum | 1 | 1 | ||
| Millettia sericea | 1 | 1 | 1 | 1 |
| Orthosiphon aristatus | 1 | |||
| Phyllanthus oxyphyllus | 1 | 1 | ||
| Premna serratifolia | 1 | |||
| Smilax calophylla | 1 | |||
| Staurogyne elongata | 1 | 1 | 1 | 1 |
| Trevesia burckii | 1 | 1 | 1 | 1 |
| Vitex glabrata | 1 | 1 | 1 | |
| K. new to DNA bank data and new DNA barcoding, but sequences need to clarify further (K) | 2 | 1 | ||
| Acalypha grandis | 1 | |||
| Myristica succedanea | 1 | |||
| Phanera fulva | 1 | |||
| L. new DNA barcoding, but sequences need to clarify further | 2 | |||
| Aglaonema commutatum | 1 | |||
| Ventilago madraspatana | 1 | |||
| M. new DNA barcoding and may strongly assist MP conservation | 10 | 2 | ||
| Benstonea affinis | 1 | |||
| Cibotium barometz | 1 | |||
| Dasymaschalon dasymaschalum | 1 | |||
| Galearia filiformis | 1 | |||
| Grammatophyllum speciosum | 1 | |||
| Nervilia concolor | 1 | 1 | ||
| Nervilia plicata | 1 | |||
| Pangium edule | 1 | |||
| Parkia timoriana | 1 | |||
| Smilax calophylla | 1 | |||
| Smilax zeylanica | 1 |
| No. | Species [38] | Author | Fam. | Region | Max Score | Total Score | Query Cover | E Value | Per. Ident | Best Matched Species | Sum. | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Justicia gendarussa | Burm.f. | Acanth. | ITS2 | 562 | 562 | 0.73 | 5.00E-156 | 0.9968 | Justicia gendarussa | c | |
| matK | 1330 | 1330 | 0.96 | 0 | 0.9986 | Justicia gendarussa | c | |||||
| rbcL | 1055 | 1055 | 0.97 | 0 | 1 | Justicia gendarussa | c | |||||
| trnL | 1487 | 1487 | 0.92 | 0 | 0.9975 | Justicia gendarussa | c | |||||
| 2 | Staurogyne elongata | (Nees) Kuntze | Acanth. | ITS2 | 597 | 597 | 0.89 | 1.00E-166 | 0.9526 | Ophiorrhiziphyllon macrobotryum | a ** | |
| matK | 1273 | 1273 | 0.97 | 0 | 0.9821 | Staurogyne concinnula | a * | |||||
| rbcL | 939 | 939 | 0.91 | 0 | 0.9923 | Staurogyne concinnula | a * | |||||
| trnL | 1013 | 1427 | 0.99 | 0 | 0.9732 | Staurogyne trinitensis | a * | |||||
| 3 | Pangium edule | Reinw. | Achari. | ITS2 | 163 | 163 | 0.15 | 1.00E-35 | 0.9286 | Celastraceae sp. | i | |
| matK | 1387 | 1387 | 1 | 0 | 0.9974 | Pangium edule | c | |||||
| rbcL | 972 | 972 | 0.91 | 0 | 1 | Pangium edule | c | |||||
| trnL | 1158 | 1741 | 0.98 | 0 | 0.982 | Ryparosa kurrangii | a * | |||||
| 4 | Spondias malayana | Kosterm. | Anacardi. | ITS2 | 636 | 636 | 1 | 3.00E-178 | 0.9332 | Spondias tuberosa | a * | |
| 5 | Toxicodendron succedaneum | (L.) Kuntze | Anacardi. | ITS2 | 660 | 660 | 0.75 | 0 | 1 | Toxicodendron succedaneum | c | |
| matK | 1452 | 1452 | 0.99 | 0 | 1 | Toxicodendron succedaneum | c | |||||
| rbcL | 1038 | 1038 | 0.97 | 0 | 1 | Toxicodendron succedaneum | c | |||||
| trnL | 1598 | 1598 | 1 | 0 | 1 | Toxicodendron succedaneum | c | 1/7 is a * | ||||
| 6 | Ancistrocladus tectorius | (Lour.) Merr. | Ancistroclad. | ITS2 | 774 | 774 | 1 | 0 | 0.9953 | Ancistrocladus benomensis | c | 1/3 is a * |
| matK | 1387 | 1387 | 1 | 0 | 0.9987 | Ancistrocladus heyneanus | a * | |||||
| rbcL | 1053 | 1053 | 1 | 0 | 1 | Ancistrocladus tectorius | c | |||||
| trnL | 1663 | 1663 | 1 | 0 | 0.9903 | Ancistrocladus tectorius | c | |||||
| 7 | Anaxagorea javanica | Blume | Annon. | matK | 1502 | 1502 | 0.97 | 0 | 0.9928 | Anaxagorea luzonensis | a * | |
| rbcL | 1013 | 1013 | 0.94 | 0 | 1 | Anaxagorea luzonensis | a * | |||||
| trnL | 1423 | 1423 | 1 | 0 | 1 | Anaxagorea javanica | c | |||||
| 8 | Dasymaschalon dasymaschalum | (Blume) I.M.Turner | Annon. | ITS2 | 237 | 237 | 0.38 | 3.00E-58 | 0.9474 | Acer palmatum | i | |
| matK | 1382 | 1382 | 1 | 0 | 0.9947 | Dasymaschalon clusiflorum | a * | |||||
| rbcL | 1020 | 1020 | 0.97 | 0 | 1 | Desmos dasymaschalus | c | |||||
| trnL | 1565 | 1565 | 0.95 | 0 | 0.9965 | Dasymaschalon megalanthum | a * | |||||
| 9 | Alstonia macrophylla | Wall. Ex. G.Don | Apocyn. | ITS2 | 763 | 763 | 0.98 | 0 | 0.9976 | Alstonia scholaris | a * | |
| matK | 1386 | 1386 | 1 | 0 | 0.9987 | Alstonia macrophylla | c | |||||
| rbcL | 857 | 857 | 1 | 0 | 0.9876 | Alstonia scholaris | c | 13/14 is a * with the same coverage | ||||
| trnL | 1557 | 1557 | 1 | 0 | 0.9908 | Alstonia scholaris | a * | |||||
| 10 | Alstonia scholaris | (L.) R. Br. | Apocyn. | ITS2 | 457 | 457 | 0.62 | 3.00E-124 | 0.9772 | Alstonia scholaris | c | |
| matK | 1380 | 1380 | 1 | 0 | 0.9987 | Alstonia yunnanensis | c | 1/9 a is a * with same coverage | ||||
| rbcL | 1051 | 1051 | 1 | 0 | 0.9983 | Alstonia scholaris | c | |||||
| trnL | 1589 | 1589 | 1 | 0 | 0.9977 | Alstonia scholaris | c | 1/2 is a * | ||||
| 11 | Alyxia reinwardtii | Blume | Apocyn. | ITS2 | 614 | 614 | 0.8 | 1.00E-171 | 0.9912 | Alyxia reinwardtii | c | |
| matK | 1317 | 1317 | 0.95 | 0 | 0.9972 | Alyxia reinwardtii | c | |||||
| rbcL | 1020 | 1020 | 0.96 | 0 | 1 | Alyxia reinwardtii | c | 1/2 is a * with higher coverage | ||||
| trnL | 1524 | 1524 | 0.98 | 0 | 0.9929 | Alyxia grandis | a * | |||||
| 12 | Hoya diversifolia | Blume | Apocyn. | ITS2 | 507 | 507 | 0.63 | 3.00E-139 | 1 | Hoya glabra | a * | |
| matK | 1347 | 1347 | 1 | 0 | 1 | Hoya vitellinoides | a * | |||||
| rbcL | 1051 | 1051 | 0.99 | 0 | 1 | Hoya pottsii | a * | |||||
| trnL | 1539 | 1539 | 0.98 | 0 | 0.9988 | Hoya sp. | a * | |||||
| 13 | Rauvolfia serpentina | (L.) Benth. ex Kurz | Apocyn. | ITS2 | 617 | 617 | 0.73 | 1.00E-172 | 1 | Rauvolfia serpentina | c | |
| matK | 1380 | 1380 | 0.99 | 0 | 1 | Rauvolfia serpentina | c | |||||
| rbcL | 1057 | 1057 | 0.99 | 0 | 1 | Rauvolfia serpentina | c | |||||
| trnL | 1395 | 1395 | 0.89 | 0 | 0.9873 | Rauvolfia serpentina | c | |||||
| 14 | Aglaonema commutatum | Schott | Ar. | ITS2 | 501 | 805 | 0.59 | 2.00E-137 | 0.9964 | Thunbergia coccinea | i | |
| matK | 1384 | 1384 | 1 | 0 | 0.9974 | Aglaonema crispum | a * | |||||
| rbcL | 1022 | 1022 | 0.97 | 0 | 1 | Aglaonema commutatum | c | |||||
| trnL | 1650 | 1650 | 1 | 0 | 0.9989 | Aglaonema crispum | a * | |||||
| 15 | Trevesia burckii | R.Br. | Arali. | ITS2 | 745 | 745 | 0.95 | 0 | 0.988 | Trevesia palmata | a * | |
| matK | 1393 | 1393 | 1 | 0 | 1 | Trevesia palmata | a * | |||||
| rbcL | 1048 | 1048 | 0.98 | 0 | 0.9982 | Brassaiopsis gracilis | a * | |||||
| trnL | 1668 | 1668 | 0.99 | 0 | 0.9989 | Brassaiopsis ciliata | a * | |||||
| 16 | Cibotium barometz | (L.) J.Sm. | Ciboti. | ITS2 | 348 | 858 | 0.75 | 3.00E-91 | 0.9896 | Cucumis sativus | i | |
| rbcL | 965 | 965 | 0.94 | 0 | 0.9872 | Cyathea chinensis | a ** | |||||
| 17 | Decalobanthus mammosus | (Lour.) A.R.Simoes & Staples | Convolvul. | rbcL | 1031 | 1031 | 0.97 | 0 | 0.9982 | Merremia peltata | a * | |
| 18 | Erycibe malaccensis | C.B.Clarke | Convolvul. | ITS2 | 466 | 466 | 0.95 | 5.00E-127 | 0.8631 | Erycibe obtusifolia | a * | |
| matK | 1389 | 1389 | 1 | 0 | 1 | Erycibe cochinchinensis | a * | |||||
| rbcL | 1033 | 1033 | 0.96 | 0 | 1 | Erycibe sp. | a * | |||||
| trnL | 1347 | 1347 | 0.93 | 0 | 0.9881 | Erycibe coccinea | a * | |||||
| 19 | Rhododendron macgregoriae | F.Muell. | Eric. | ITS2 | 723 | 723 | 1 | 0 | 0.9658 | Rhododendron groenlandicum | a * | |
| matK | 1369 | 1369 | 1 | 0 | 0.9908 | Rhododendron javanicum | a * | |||||
| rbcL | 1027 | 1027 | 0.98 | 0 | 0.9912 | Rhododendron simsii | a * | |||||
| trnL | 1629 | 1629 | 0.96 | 0 | 0.9955 | Rhododendron javanicum | a * | |||||
| 20 | Acalypha grandis | Benth. | Euphorbi. | ITS2 | 272 | 272 | 0.35 | 1.00E-68 | 0.9808 | Acer tataricum subsp. theiferum | i | |
| rbcL | 1062 | 1062 | 0.99 | 0 | 1 | Acalypha grisebachiana | a * | |||||
| trnL | 1729 | 1729 | 1 | 0 | 0.9886 | Acalypha hispida | a * | |||||
| 21 | Euphorbia tirucalli | L. | Euphorbi. | ITS2 | 617 | 617 | 0.71 | 1.00E-172 | 1 | Euphorbia tirucalli | c | 1/12 I with higher coverage |
| rbcL | 1046 | 1046 | 0.98 | 0 | 1 | Euphorbia rauhii | a * | |||||
| 22 | Millettia sericea | (Vent.) Benth. | Fab. | ITS2 | 712 | 712 | 0.94 | 0 | 0.9571 | Millettia pulchra | a * | |
| matK | 1332 | 1332 | 0.97 | 0 | 0.988 | Millettia pulchra | a * | |||||
| rbcL | 1042 | 1042 | 0.97 | 0 | 0.9982 | Dahlstedtia pinnata | a * | |||||
| trnL | 1543 | 1543 | 1 | 0 | 0.9819 | Millettia pinnata | a * | |||||
| 23 | Parkia timoriana | (DC.) Merr. | Fab. | ITS2 | 593 | 593 | 0.71 | 2.00E-165 | 0.9909 | Parkia timoriana | c | |
| matK | 1376 | 1376 | 0.98 | 0 | 0.996 | Parkia biglandulosa | a * | |||||
| rbcL | 1000 | 1000 | 0.95 | 0 | 0.9927 | Magnoliophyta sp. | i | |||||
| trnL | 1814 | 1814 | 0.99 | 0 | 0.999 | Parkia biglandulosa | a * | |||||
| 24 | Phanera fulva | (Korth.) Benth. | Fab. | ITS2 | 475 | 475 | 0.68 | 7.00E-130 | 0.9477 | Bauhinia sp. | a * | |
| rbcL | 1016 | 1016 | 0.96 | 0 | 0.9982 | Embryophyte environmental | i | |||||
| trnL | 1404 | 1404 | 0.78 | 0 | 0.9974 | Phanera vahlii | a ** | |||||
| 25 | Orthosiphon aristatus | (Blume) Miq. | Lami. | ITS2 | 562 | 562 | 0.69 | 5.00E-156 | 1 | Orthosiphon aristatus | c | |
| rbcL | 1042 | 1042 | 0.98 | 0 | 1 | Clerodendranthus spicatus | a ** | |||||
| 26 | Premna serratifolia | L. | Lami. | ITS2 | 422 | 422 | 0.99 | 9.00E-114 | 0.8495 | Premna microphylla | a * | |
| rbcL | 1040 | 1040 | 0.97 | 0 | 1 | Premna serratifolia | c | 2/3 is a * with higher and lower coverage | ||||
| 27 | Vitex glabrata | Gaertn. | Lami. | ITS2 | 651 | 651 | 0.91 | 0 | 0.9558 | Vitex carvalhoi | a * | |
| matK | 1587 | 1587 | 1 | 0 | 0.9988 | Vitex glabrata | c | |||||
| rbcL | 1050 | 1050 | 1 | 0 | 0.9982 | Vitex doniana | a * | |||||
| trnL | 1411 | 1411 | 0.94 | 0 | 0.9923 | Vitex triflora | a * | |||||
| 28 | Cinnamomum rhynchophyllum | Miq. | Laur. | matK | 1375 | 1375 | 0.99 | 0 | 0.9987 | Cinnamomum camphora | a * | |
| rbcL | 1055 | 1055 | 1 | 0 | 1 | Cinnamomum dubium | a * | |||||
| trnL | 1587 | 1587 | 1 | 0 | 1 | Cinnamomum pittosporoides | a * | |||||
| 29 | Ficus deltoidea | Jack | Mor. | ITS2 | 616 | 616 | 0.78 | 4.00E-172 | 1 | Ficus deltoidea | c | |
| matK | 1380 | 1380 | 1 | 0 | 0.996 | Ficus cf. | a * | |||||
| rbcL | 1051 | 1051 | 0.98 | 0 | 0.9983 | Ficus benjamina | a * | |||||
| trnL | 1664 | 1664 | 0.99 | 0 | 0.9967 | Ficus carica | a * | |||||
| 30 | Myristica succedanea | Blume | Myristic. | ITS2 | 185 | 185 | 0.17 | 2.00E-42 | 0.9231 | Rhodohypoxis milloides | i | |
| matK | 1476 | 1476 | 0.92 | 0 | 0.9988 | Myristica fragrans | a * | |||||
| rbcL | 1057 | 1057 | 1 | 0 | 1 | Horsfieldia amygdalina | a * | 4/11 is a ** | ||||
| trnL | 1371 | 1371 | 0.83 | 0 | 0.9987 | Myristica iners | a * | |||||
| 31 | Nepenthes ampullaria | Jack | Nepenth. | matK | 1375 | 1375 | 0.99 | 0 | 0.9973 | Nepenthes mapuluensis | a * | |
| rbcL | 1042 | 1042 | 1 | 0 | 1 | Nepenthes mirabilis | a * | |||||
| trnL | 1648 | 1648 | 1 | 0 | 0.9956 | Nepenthes mirabilis | a * | |||||
| 32 | Nepenthes gracilis | Korth. | Nepenth. | matK | 1371 | 1371 | 1 | 0 | 0.9973 | Nepenthes gracilis | c | |
| rbcL | 1046 | 1046 | 1 | 0 | 1 | Nepenthes mirabilis | a * | |||||
| trnL | 961 | 961 | 0.57 | 0 | 0.9962 | Nepenthes ampullaria | a * | |||||
| 33 | Nepenthes mirabilis | (Lour.) Druce | Nepenth. | ITS2 | 857 | 857 | 1 | 0 | 0.9979 | Nepenthes reinwardtiana | a * | |
| matK | 1371 | 1371 | 1 | 0 | 0.9973 | Nepenthes mapuluensis | a * | |||||
| rbcL | 1038 | 1038 | 1 | 0 | 0.9965 | Nepenthes graciliflora | a * | |||||
| trnL | 959 | 959 | 0.57 | 0 | 0.9943 | Nepenthes sanguinea | a * | |||||
| 34 | Nepenthes reinwardtiana | Miq. | Nepenth. | ITS2 | 861 | 861 | 1 | 0 | 0.9979 | Nepenthes reinwardtiana | c | |
| matK | 1376 | 1376 | 1 | 0 | 0.996 | Nepenthes reinwardtiana | c | |||||
| rbcL | 1042 | 1042 | 0.98 | 0 | 0.9965 | Nepenthes mirabilis | a * | |||||
| trnL | 948 | 948 | 0.57 | 0 | 0.9924 | Nepenthes alba | a * | |||||
| 35 | Acriopsis liliifolia var. liliifolia | (J.Koenig) Ormerod | Orchid. | ITS2 | 394 | 394 | 0.94 | 2.00E-105 | 0.8428 | Cymbidium ensifolium | a ** | |
| matK | 1408 | 1408 | 1 | 0 | 0.9987 | Acriopsis sp. | a * | |||||
| rbcL | 911 | 911 | 1 | 0 | 0.9824 | Acriopsis sp. | a * | |||||
| trnL | 824 | 1591 | 0.91 | 0 | 0.9265 | Cymbidium erythraeum | a ** | |||||
| 36 | Cymbidium aloifolium | (L.) Sw. | Orchid. | ITS2 | 468 | 468 | 0.61 | 1.00E-127 | 0.9884 | Cymbidium aloifolium | c | |
| matK | 1386 | 1386 | 1 | 0 | 0.9987 | Cymbidium aloifolium | c | 1/5 is a * | ||||
| rbcL | 1048 | 1048 | 0.98 | 0 | 0.9982 | Cymbidium aloifolium | c | 1/4 is a * | ||||
| trnL | 989 | 989 | 0.79 | 0 | 0.953 | Cymbidium wadae | a * | |||||
| 37 | Cymbidium ensifolium | (L.) Sw. | Orchid. | ITS2 | 387 | 387 | 0.66 | 4.00E-103 | 0.9072 | Cymbidium goeringii | a * | |
| matK | 1293 | 1293 | 0.99 | 0 | 0.9889 | Cymbidium longibracteatum | a * | |||||
| 38 | Dendrobium crumenatum | Sw. | Orchid. | ITS2 | 577 | 577 | 0.7 | 2.00E-160 | 0.9968 | Dendrobium crumenatum | c | |
| matK | 1400 | 1400 | 0.99 | 0 | 0.9961 | Dendrobium crumenatum | c | |||||
| rbcL | 1038 | 1038 | 0.97 | 0 | 0.9982 | Dendrobium pseudotenellum | a * | |||||
| 39 | Dendrobium purpureum | Roxb. | Orchid. | ITS2 | 481 | 537 | 0.86 | 2.00E-131 | 0.9005 | Dendrobium calcaratum | a * | |
| matK | 1360 | 1360 | 1 | 0 | 0.9947 | Dendrobium faciferum | a * | |||||
| rbcL | 1042 | 1042 | 0.98 | 0 | 0.9965 | Dendrobium aggregatum | a * | |||||
| trnL | 562 | 998 | 0.98 | 8.00E-156 | 0.9814 | Dendrobium chrysanthum | a * | |||||
| 40 | Dendrobium salaccense | (Blume) Lindl. | Orchid. | ITS2 | 627 | 627 | 0.79 | 2.00E-175 | 0.9914 | Dendrobium haemoglossum | a * | |
| matK | 1382 | 1382 | 0.99 | 0 | 0.9987 | Dendrobium salaccense | c | |||||
| rbcL | 1031 | 1031 | 1 | 0 | 1 | Dendrobium salaccense | c | 2/3 is a * | ||||
| trnL | 1328 | 1328 | 0.81 | 0 | 0.9959 | Dendrobium salaccense | c | |||||
| 41 | Grammatophyllum speciosum | Blume | Orchid. | ITS2 | 809 | 38152 | 1 | 0 | 1 | Raphanus raphanistrum subsp. landra | i | |
| matK | 1378 | 1378 | 0.99 | 0 | 0.996 | Grammatophyllum papuanum | a * | |||||
| rbcL | 1037 | 1037 | 0.97 | 0 | 0.9947 | Cymbidium faberi | a ** | |||||
| trnL | 568 | 1103 | 0.93 | 2.00E-157 | 0.9905 | Cymbidium serratum | a ** | |||||
| 42 | Nervilia concolor | (Blume) Schltr. | Orchid. | ITS2 | 828 | 828 | 1 | 0 | 1 | Cucumis sativus | i | |
| rbcL | 1062 | 1062 | 0.99 | 0 | 1 | Nepenthes mirabilis | i | |||||
| trnL | 1585 | 1585 | 1 | 0 | 0.9834 | Nervilia mekongensis | a * | |||||
| 43 | Nervilia plicata | (Andrews) Schltr. | Orchid. | ITS2 | 721 | 721 | 0.88 | 0 | 0.9741 | Syzygium megacarpum | i | |
| matK | 1413 | 1413 | 0.97 | 0 | 0.9987 | Nervilia plicata | c | |||||
| rbcL | 1005 | 1005 | 0.94 | 0 | 1 | Nervilia plicata | c | 1/4 is a * with higher coverage | ||||
| trnL | 1663 | 1663 | 0.99 | 0 | 0.9967 | Nervilia plicata | c | |||||
| 44 | Oberonia lycopodioides | (J.Koenig) Ormerod | Orchid. | ITS2 | 398 | 398 | 0.88 | 1.00E-106 | 0.8765 | Oberonia caulescens | a * | |
| matK | 1205 | 1205 | 0.93 | 0 | 0.9732 | Oberonia mucronata | a * | |||||
| rbcL | 922 | 922 | 1 | 0 | 0.9921 | Ancistrochilus sp. | a ** | |||||
| trnL | 592 | 1078 | 0.91 | 2.00E-164 | 0.8734 | Liparis loeselii | a ** | |||||
| 45 | Strongyleria pannea | (Lindl.) Schuit., Y.P.Ng & H.A.Pedersen | Orchid. | ITS2 | 431 | 431 | 0.59 | 2.00E-116 | 0.959 | Mycaranthes pannea | c | |
| matK | 1375 | 1375 | 1 | 0 | 0.996 | Mycaranthes pannea | c | |||||
| rbcL | 1055 | 1055 | 1 | 0 | 0.9965 | Mycaranthes pannea | c | |||||
| 46 | Galearia filiformis | (Blume) Boerl. | Pand. | ITS2 | 433 | 433 | 0.99 | 4.00E-117 | 0.8552 | Populus nigra | i | |
| matK | 1393 | 1393 | 1 | 0 | 1 | Galearia filiformis | c | |||||
| rbcL | 1042 | 1042 | 0.98 | 0 | 1 | Galearia filiformis | c | |||||
| trnL | 1744 | 1744 | 1 | 0 | 0.9969 | Galearia filiformis | c | |||||
| 47 | Benstonea affinis | (Kurz) Callm. & Buerki | Pandan. | ITS2 | 124 | 124 | 0.24 | 6.00E-24 | 0.8611 | Magnolia henryi | i | |
| matK | 1397 | 1397 | 0.91 | 0 | 0.9935 | Pandanus oblatus | a * | |||||
| rbcL | 1057 | 1057 | 1 | 0 | 1 | Pandanus adinobotrys | a * | |||||
| trnL | 1705 | 1705 | 1 | 0 | 0.9989 | Pandanus baptistii | a * | |||||
| 48 | Phyllanthus oxyphyllus | Miq. | Phyllanth. | ITS2 | 621 | 621 | 0.74 | 9.00E-174 | 0.9971 | Phyllanthus oxyphyllus | c | 1/2 is a * with higher coverage |
| matK | 1375 | 1375 | 1 | 0 | 0.9973 | Phyllanthus oxyphyllus | c | |||||
| rbcL | 1059 | 1059 | 1 | 0 | 1 | Phyllanthus emblica | a * | |||||
| trnL | 989 | 989 | 0.58 | 0 | 0.9945 | Phyllanthus emblica | a * | |||||
| 49 | Ardisia complanata | Wall. | Primul. | ITS2 | 667 | 667 | 0.78 | 0 | 0.9973 | Ardisia dasyrhizomatica | a * | |
| matK | 1574 | 1574 | 1 | 0 | 0.9931 | Ardisia mamillata | a * | |||||
| rbcL | 1031 | 1031 | 0.99 | 0 | 0.9965 | Ardisia crenata | a * | |||||
| trnL | 1483 | 1483 | 1 | 0 | 0.9951 | Ardisia dasyrhizomatica | a * | |||||
| 50 | Ardisia crenata | Sims | Primul. | ITS2 | 617 | 617 | 0.74 | 1.00E-172 | 0.997 | Ardisia villosa | a * | |
| matK | 1404 | 1404 | 0.88 | 0 | 0.9987 | Ardisia crenata | c | |||||
| rbcL | 1048 | 1048 | 1 | 0 | 1 | Ardisia cornudentata subsp. morrisonensis | c | 1/2 is a * | ||||
| trnL | 1476 | 1476 | 0.99 | 0 | 0.9988 | Ardisia affinis | a * | |||||
| 51 | Ventilago madraspatana | Boerl. | Rhamn. | ITS2 | 206 | 316 | 0.45 | 1.00E-48 | 0.9444 | Hibiscus panduriformis | i | |
| matK | 1347 | 1347 | 0.96 | 0 | 0.9973 | Ventilago leiocarpa | a * | |||||
| rbcL | 1022 | 1022 | 0.96 | 0 | 0.9947 | Ventilago leiocarpa | a * | |||||
| trnL | 1574 | 1574 | 1 | 0 | 0.9722 | Ventilago kurzii | a * | |||||
| 52 | Psychotria montana | Blume | Rubi. | ITS2 | 398 | 398 | 1 | 8.00E-107 | 0.9744 | Psychotria camerunensis | a * | |
| matK | 1376 | 1376 | 0.99 | 0 | 0.996 | Psychotria asiatica | a * | |||||
| rbcL | 1029 | 1029 | 0.96 | 0 | 1 | Psychotria adenophylla | a * | |||||
| trnL | 1504 | 1504 | 0.96 | 0 | 0.9826 | Psychotria asiatica | a * | |||||
| 53 | Lunasia amara | Blanco | Rut. | ITS2 | 579 | 579 | 0.74 | 6.00E-161 | 0.9654 | Lunasia amara | c | |
| matK | 1243 | 1243 | 0.88 | 0 | 0.9971 | Lunasia amara | c | |||||
| rbcL | 1026 | 1026 | 0.97 | 0 | 0.9947 | Flindersia brayleyana | a ** | |||||
| trnL | 1668 | 1668 | 0.95 | 0 | 0.9946 | Lunasia amara | c | |||||
| 54 | Melicope lunu-ankenda | (Gaertn.) T.G. Hartley | Rut. | ITS2 | 787 | 787 | 1 | 0 | 0.9823 | Melicope pteleifolia | a * | |
| matK | 1408 | 1408 | 1 | 0 | 0.9987 | Melicope pteleifolia | a * | |||||
| rbcL | 1031 | 1031 | 0.98 | 0 | 0.9965 | Melicope pteleifolia | a * | |||||
| trnL | 1168 | 1168 | 1 | 0 | 0.9953 | Melicope grisea | a * | |||||
| 55 | Kadsura scandens | (Blume) Blume | Schisandr. | ITS2 | 558 | 558 | 0.69 | 7.00E-155 | 0.9967 | Kadsura scandens | c | |
| matK | 1376 | 1376 | 1 | 0 | 0.9947 | Kadsura philippinensis | a * | |||||
| rbcL | 1050 | 1050 | 0.99 | 0 | 1 | Kadsura cf. | a * | |||||
| trnL | 1635 | 1635 | 0.99 | 0 | 0.986 | Kadsura matsudae | a * | |||||
| 56 | Smilax calophylla | Wall. ex A.DC. | Smilac. | ITS2 | 821 | 821 | 1 | 0 | 0.9933 | Phaseolus vulgaris | I | |
| rbcL | 1048 | 1048 | 0.98 | 0 | 0.9982 | Smilax cocculoides | a * | |||||
| 57 | Smilax zeylanica | L. | Smilac. | ITS2 | 274 | 274 | 0.35 | 3.00E-69 | 0.9809 | Acer tataricum subsp. theiferum | i | |
| matK | 1371 | 1371 | 1 | 0 | 1 | Smilax ovalifolia | a * | |||||
| rbcL | 1044 | 1044 | 0.98 | 0 | 1 | Smilax ocreata | a * | |||||
| 58 | Aquilaria hirta | Ridl. | Thymelae. | ITS2 | 702 | 702 | 0.82 | 0 | 0.9948 | Aquilaria microcarpa | a * | |
| matK | 1402 | 1402 | 1 | 0 | 0.9974 | Aquilaria malaccensis | a * | |||||
| rbcL | 1057 | 1057 | 0.99 | 0 | 1 | Rauvolfia serpentina | c | |||||
| trnL | 987 | 987 | 0.67 | 0 | 0.9945 | Aquilaria microcarpa | a * | |||||
| 59 | Amomum hochreutineri | Valeton | Zingiber. | ITS2 | 616 | 616 | 0.79 | 4.00E-172 | 0.9884 | Sundamomum hastilabium | a ** | |
| rbcL | 1044 | 1044 | 0.98 | 0 | 1 | Amomum villosum var. xanthioides | a * | |||||
| trnL | 1568 | 1568 | 0.98 | 0 | 0.9931 | Amomum fulviceps | a * | |||||
| 60 | Etlingera solaris | (Blume) R.M.Sm. | Zingiber. | ITS2 | 656 | 656 | 0.89 | 0 | 0.9764 | Hornstedtia conica | a ** | |
| rbcL | 1053 | 1053 | 0.99 | 0 | 1 | Alpinia arundelliana | a ** | |||||
| trnL | 1622 | 1622 | 0.99 | 0 | 0.9955 | Etlingera yunnanensis | a ** | |||||
| 61 | Meistera aculeata | (Roxb.) Skornick. & M.F. Newman | Zingiber. | ITS2 | 592 | 592 | 0.72 | 7.00E-165 | 1 | Amomum aculeatum | c | |
| rbcL | 1020 | 1020 | 0.96 | 0 | 1 | Amomum dallachyi | a * |
References
- Miller, S.E.; Hausmann, A.; Hallwachs, W.; Janzen, D.H. Advancing taxonomy and bioinventories with DNA barcodes. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CBOL Plant Working Group A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [CrossRef] [PubMed] [Green Version]
- Dick, C.W.; Webb, C.O. Chapter 18 Plant DNA Barcodes, Taxonomic Management, and Species Discovery in Tropical Forests. In DNA Barcodes: Methods and Protocols; John Kress, W., Erickson, D.L., Eds.; Springer Science+Business Media: Berlin/Heidelberg, Germany, 2012; Volume 858, pp. 379–393. ISBN 9781617795916. [Google Scholar]
- Eurlings, M.C.M.; Lens, F.; Pakusza, C.; Peelen, T.; Wieringa, J.J.; Gravendeel, B. Forensic Identification of Indian Snakeroot (Rauvolfia serpentina Benth. ex Kurz) Using DNA Barcoding. J. Forensic Sci. 2013, 58, 822–830. [Google Scholar] [CrossRef] [Green Version]
- Kress, W.J.; Garcı-Robledo, C.; Uriarte, M.; Erickson, D.L. DNA barcodes for ecology, evolution, and conservation. Trends Ecol. Evol. 2014, 30, 25–35. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007, 35, e14. [Google Scholar] [CrossRef] [Green Version]
- Fazekas, A.J.; Kuzmina, M.L.; Newmaster, S.G.; Hollingsworth, P.M. DNA Barcoding Methods for Land Plants. Methods Mol. Biol. 2012, 858, 223–252. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L. A Two-Locus Global DNA Barcode for Land Plants: The Coding rbcL Gene Complements the Non-Coding trnH- psbA Spacer Region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [Green Version]
- Sucher, N.; Hennell, J.; Carles, M. Plant DNA Fingerprinting and Barcoding; Humana Press: New York, NY, USA, 2012; Volume 862, ISBN 978-1-61779-608-1. [Google Scholar]
- Ratnasingham, S.; Hebert, P.D.N. BARCODING BOLD: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [Green Version]
- Newmaster, S.G.; Grguric, M.; Shanmughanandhan, D.; Ramalingam, S.; Ragupathy, S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013, 11, 222. [Google Scholar] [CrossRef] [Green Version]
- Schindel, D.E.; Miller, S.E. DNA barcoding a useful tool for taxonomists. Nature 2005, 435, 17. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for iFdentifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Techen, N.; Parveen, I.; Pan, Z.; Khan, I.A. DNA barcoding of medicinal plant material for identification. Curr. Opin. Biotechnol. 2014, 25, 103–110. [Google Scholar] [CrossRef]
- Huda, M.K.; Price, A.; Wilcock, C.C. Identification of Medicinal Orchids of Bangladesh: DNA Barcoding Vs. Traditional Taxonomy. J. Orchid Soc. India 2017, 31, 33–40. [Google Scholar]
- Hartvig, I.; Czako, M.; Kjær, E.D.; Nielsen, L.R.; Theilade, I. The use of DNA barcoding in identification and conservation of rosewood (Dalbergia spp.). PLoS ONE 2015, 10, e0138231. [Google Scholar] [CrossRef] [Green Version]
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef]
- Vassou, S.L.; Nithaniyal, S.; Raju, B.; Parani, M. Creation of reference DNA barcode library and authentication of medicinal plant raw drugs used in Ayurvedic medicine. BMC Complement. Altern. Med. 2016, 16, 186. [Google Scholar] [CrossRef] [Green Version]
- Ferri, G.; Corradini, B.; Ferrari, F.; Santunione, A.L.; Palazzoli, F.; Alu’, M. Forensic botany II, DNA barcode for land plants: Which markers after the international agreement? Forensic. Sci. Int. Genet. 2015, 15, 131–136. [Google Scholar] [CrossRef]
- Paranaiba, R.T.F.; Carvalho, C.B.V.; Freitas, J.M.; Fassio, L.H.; Botelho, É.D.; Neves, D.B.J.; Silva, R.C.; Aguiar, S.M.; Naaum, A. Forensic botany and forensic chemistry working together: Application of plant DNA barcoding as a complement to forensic chemistry—A case study in Brazil. Genome 2019, 62, 11–18. [Google Scholar] [CrossRef]
- Sass, C.; Little, D.P.; Stevenson, D.W.; Specht, C.D. DNA barcoding in the Cycadales: Testing the potential of proposed barcoding markers for species identification of Cycads. PLoS ONE 2007, 2, e1154. [Google Scholar] [CrossRef]
- Mahadani, P.; Sharma, G.D.; Ghosh, S.K. Identification of ethnomedicinal plants (Rauvolfioideae: Apocynaceae) through DNA barcoding from northeast India. Pharmacogn. Mag. 2013, 9, 255–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abubakar, B.M.; Salleh, F.M.; Omar, M.S.S.; Wagiran, A. Assessing product adulteration of Eurycoma longifolia (Tongkat Ali) herbal medicinal product using DNA barcoding and HPLC analysis. Pharm. Biol. 2018, 56, 368–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabelin, V.L.D.; Alejandro, G.J.D. Efficiency of matK, rbcL, trnH-psbA, and trnL-F (cpDNA) to molecularly authenticate Philippine ethnomedicinal Apocynaceae through DNA barcoding. Pharmacogn. Mag. 2016, 12, S384–S388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, L.; Qiu, X.H.; Huang, J.; Xu, W.; Bai, J.Q.; Zhang, J.; Su, H.; Xu, C.M.; Huang, Z.H. Constructing a DNA barcode reference library for southern herbs in China: A resource for authentication of southern Chinese medicine. PLoS ONE 2018, 13, e0201240. [Google Scholar] [CrossRef]
- Chao, Z.; Zeng, W.; Liao, J.; Liu, L.; Liang, Z.; Li, X. DNA barcoding Chinese medicinal Bupleurum. Phytomedicine 2014, 21, 1767–1773. [Google Scholar] [CrossRef]
- Suba, M.D.L.; Arriola, A.H.; Jonathan, G.; Alejandro, D. Evaluation of cpDNA Barcodes in Selected Medicinal Plants of Mt. Arayat National Park, Pampanga, the Philippines. J. Young Pharm. 2019, 11, 130–135. [Google Scholar] [CrossRef] [Green Version]
- Vavilov, N.I. Theoretical Basis for Plant Breeding, Vol. Moscow. Origin and Geography of Cultivated Plants. In The Phytogeographical Basis for Plant Breeding (D. Love, Transl.); Löve, D., Ed.; Cambridge University Press: Cambridge, UK, 1935; pp. 316–366. ISBN 0521404274. [Google Scholar]
- Ministry of National Development Planning. Indonesian Biodiversity Strategy and Action Plan 2015–2020; Indonesian Government: Jakarta, Indonesia, 2016. [Google Scholar]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- De Padua, L.S.; Bunyapraphatsara, N.; Lemmens, R.H.M.J. (Eds.) Medicinal and Poisonous Plants PROSEA; Plant Resources of South-East Asia 12(1); Backhuys Pub.: Leiden, The Netherlands, 1999. [Google Scholar]
- Von Rintelen, K.; Arida, E.; Häuser, C. A review of biodiversity-related issues and challenges in megadiverse Indonesia and other Southeast Asian countries. Res. Ideas Outcomes 2017, 3, e20860. [Google Scholar] [CrossRef] [Green Version]
- POWO. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew.. 2022. Available online: http://www.plantsoftheworldonline.org/ (accessed on 6 February 2022).
- IUCN. The IUCN Red List of Threatened Species. Version 2020–2022. 2020. Available online: https://www.iucnredlist.org/ (accessed on 13 June 2020).
- UNEP-WCMC (Comps.). The Checklist of CITES Species Website. CITES Secretariat, 25 Geneva, Switzerland. Compiled by UNEP-WCMC, Cambridge, UK.. 2020. Available online: http://checklist.cites.org (accessed on 13 June 2020).
- The National Development Planning Agency. Indonesian Biodiversity Strategy and Action Plan National Document. BAPPENAS; National Development Planning Agency of Republic Indonesia: Jakarta, Indonesia, 2003. [Google Scholar]
- Harvey-Brown, Y. Aquilaria hirta. The IUCN Red List of Threatened Species 2018: E.T34561A. Available online: https://doi.org/10.2305/IUCN.UK.2018-1.RLTS.T34561A2853368.en (accessed on 17 January 2022).
- Olander, S.B. Etlingera solaris. The IUCN Red List of Threatened Species 2019: E.T117324858A124282372. Available online: https://doi.org/10.2305/IUCN.UK.2019-3.RLTS.T117324858A124282372.en (accessed on 28 January 2022).
- IUCN. IUCN Red List Categories and Criteria: Version 3, 2nd ed.; IUCN: Gland, Switzerland; Cambridge, UK, 2012; pp. iv + 32. [Google Scholar]
- Yu, J.; Xue, J.H.; Zhou, S.L. New universal matK primers for DNA barcoding angiosperms. J. Syst. Evol. 2011, 49, 176–181. [Google Scholar] [CrossRef]
- Amandita, F.Y.; Rembold, K.; Vornam, B.; Rahayu, S.; Siregar, I.Z.; Kreft, H.; Finkeldey, R. DNA barcoding of flowering plants in Sumatra, Indonesia. Ecol. Evol. 2019, 9, 1858–1868. [Google Scholar] [CrossRef] [Green Version]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Ming, R.; Yu, Q. Comparative Analysis of GC Content Variations in Plant Genomes. Trop. Plant Biol. 2016, 9, 136–149. [Google Scholar] [CrossRef]
- Wilkie, P.; Poulsen, A.D.; Harris, D.; Forrest, L.L. The collection and storage of plant material for DNA extraction: The Teabag Method. Gard. Bull. Singapore 2013, 65, 231–234. [Google Scholar] [CrossRef] [Green Version]
- Till, B.J.; Jankowicz-Cieslak, J.; Huynh, O.A.; Beshir, M.M.; Laport, R.G.; Hofinger, B.J. Low-Cost Methods for Molecular Characterisation of Mutant Plants: Tissue Desiccation, DNA Extraction and Mutation Discovery: Protocols; Springer Nature: Berlin/Heidelberg, Germany, 2015; ISBN 9783319162591. [Google Scholar]
- Maurin, O.; Epitawalage, N.; Eiserhardt, W.; Forest, F.; Fulcher, T.; Baker, W. A Visual Guide to Collecting Plant Tissues for DNA; Royal Botanic Gardens Kew: Richmond, UK, 2017; pp. 1–2. [Google Scholar]
- Cahyaningsih, R.; Magos Brehm, J.; Nigel, B. Setting the Priority Medicinal Plants for Conservation in Indonesia; Springer Netherlands: Amsterdam, The Netherlands, 2021; Volume 6, pp. 2019–2050. ISBN 0123456789. [Google Scholar]
- Costion, C.; Ford, A.; Cross, H.; Crayn, D.; Harrington, M.; Lowe, A. Plant dna barcodes can accurately estimate species richness in poorly known floras. PLoS ONE 2011, 6, 4–11. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Song, J.; Cao, Y.; Sun, Q.; Yao, H.; Wu, Q.; Chao, J.; Zhou, J.; Xue, W.; Duan, J. Application of the ITS2 Region for Barcoding Medicinal Plants of Selaginellaceae in Pteridophyta. PLoS ONE 2013, 8, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef]
- Casiraghi, M.; Labra, M.; Ferri, E.; Galimberti, A.; de Mattia, F. DNA barcoding: A six-question tour to improve users’ awareness about the method. Brief. Bioinform. 2010, 11, 440–453. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Phillips, J.D.; Gillis, D.J.; Hanner, R.H. Incomplete estimates of genetic diversity within species: Implications for DNA barcoding. Ecol. Evol. 2019, 9, 2996–3010. [Google Scholar] [CrossRef] [PubMed]



| No. | Species | Author | Family. | N/I | Important Sp. | Sp. No. per Genus | BOLD (NCBI)Database |
|---|---|---|---|---|---|---|---|
| 1 | Justicia gendarussa | Burm.f. | Acanthaceae | N | No | 921 | yes |
| 2 | Staurogyne elongata | (Nees) Kuntze | Acanthaceae | N | No | 148 | yes |
| 3 | Pangium edule | Reinw. | Achariaceae | N | Yes (P) | 1 | yes |
| 4 | Spondias malayana | Kosterm. | Anacardiaceae | N | No | 19 | no (yes) |
| 5 | Toxicodendron succedaneum | (L.) Kuntze | Anacardiaceae | I | No | 27 | yes |
| 6 | Ancistrocladus tectorius | (Lour.) Merr. | Ancistrocladaceae | N | No | 21 | yes |
| 7 | Anaxagorea javanica | Blume | Annonaceae | N | Yes (P) | 25 | no (yes) |
| 8 | Dasymaschalon dasymaschalum | (Blume) I.M.Turner | Annonaceae | N | No | 27 | yes |
| 9 | Alstonia macrophylla | Wall. Ex. G.Don | Apocynaceae | N | No | 44 | yes |
| 10 | Alstonia scholaris | (L.) R. Br. | Apocynaceae | N | Yes (P) | yes | |
| 11 | Alyxia reinwardtii | Blume | Apocynaceae | N | Yes (P) | 106 | yes |
| 12 | Hoya diversifolia | Blume | Apocynaceae | N | No | 521 | no (yes) |
| 13 | Rauvolfia serpentina | (L.) Benth. ex Kurz | Apocynaceae | N | Yes (II) | 74 | yes |
| 14 | Aglaonema commutatum | Schott | Araceae | N | No | 22 | no (yes) |
| 15 | Trevesia burckii | R.Br. | Araliaceae | N | No | 8 | yes (yes) |
| 16 | Cibotium barometz | (L.) J.Sm. | Cibotiaceae | N | Yes (II) | 10 | yes |
| 17 | Decalobanthus mammosus | (Lour.) A.R.Simoes & Staples | Convolvulaceae | I | No | 13 | no (yes) |
| 18 | Erycibe malaccensis | C.B. Clarke | Convolvulaceae | N | No | 70 | no (no) |
| 19 | Rhododendron macgregoriae | F. Muell. | Ericaceae | N | Yes (E) | 1057 | no (no) |
| 20 | Acalypha grandis | Benth. | Euphorbiaceae | N | No | 428 | no (no) |
| 21 | Euphorbia tirucalli | L. | Euphorbiaceae | I | Yes (II) | 1976 | yes |
| 22 | Millettia sericea | (Vent.) Benth. | Fabaceae | N | No | 187 | yes |
| 23 | Parkia timoriana | (DC.) Merr. | Fabaceae | N | No | 40 | yes |
| 24 | Phanera fulva | (Korth.) Benth. | Fabaceae | N | Yes (E) | 90 | no (no) |
| 25 | Orthosiphon aristatus | (Blume) Miq. | Lamiaceae | N | No | 44 | yes |
| 26 | Premna serratifolia | L. | Lamiaceae | N | No | 131 | yes |
| 27 | Vitex glabrata | Gaertn. | Lamiaceae | N | No | 203 | yes |
| 28 | Cinnamomum rhynchophyllum | Miq. | Lauraceae | N | No | 241 | no (yes) |
| 29 | Ficus deltoidea | Jack | Moraceae | N | Yes (P) | 874 | yes |
| 30 | Myristica succedanea | Blume | Myristicaceae | N | Yes (E) | 175 | no (no) |
| 31 | Nepenthes ampullaria | Jack | Nepenthaceae | N | Yes (P, II) | 165 | yes |
| 32 | Nepenthes gracilis | Korth. | Nepenthaceae | N | Yes (P, II) | yes | |
| 33 | Nepenthes mirabilis | (Lour.) Druce | Nepenthaceae | N | Yes (P, II) | yes | |
| 34 | Nepenthes reinwardtiana | Miq. | Nepenthaceae | N | Yes (P, E, II) | yes | |
| 35 | Acriopsis liliifolia var. liliifolia | (J.Koenig) Ormerod | Orchidaceae | N | Yes (P, II) | 10 | no (yes) |
| 36 | Cymbidium aloifolium | (L.) Sw. | Orchidaceae | N | Yes (P, II) | 74 | yes |
| 37 | Cymbidium ensifolium | (L.) Sw. | Orchidaceae | I | Yes (II) | yes | |
| 38 | Dendrobium crumenatum | Sw. | Orchidaceae | N | Yes (P, II) | 1547 | yes |
| 39 | Dendrobium purpureum | Roxb. | Orchidaceae | N | Yes (P, E, II) | no (no) | |
| 40 | Dendrobium salaccense | (Blume) Lindl. | Orchidaceae | N | Yes (P, II) | yes | |
| 41 | Grammatophyllum speciosum | Blume | Orchidaceae | N | Yes (P, II) | 13 | yes |
| 42 | Nervilia concolor | (Blume) Schltr. | Orchidaceae | N | Yes (P, II) | 77 | yes |
| 43 | Nervilia plicata | (Andrews) Schltr. | Orchidaceae | N | Yes (P, II) | yes | |
| 44 | Oberonia lycopodioides | (J.Koenig) Ormerod | Orchidaceae | N | Yes (P, II) | 305 | no (no) |
| 45 | Strongyleria pannea | (Lindl.) Schuit., Y.P.Ng & H.A.Pedersen | Orchidaceae | N | Yes (P, II) | 4 | no (yes) |
| 46 | Galearia filiformis | (Blume) Boerl. | Pandaceae | N | Yes (E) | 5 | yes |
| 47 | Benstonea affinis | (Kurz) Callm. & Buerki | Pandanaceae | N | No | 61 | yes |
| 48 | Phyllanthus oxyphyllus | Miq. | Phyllanthaceae | N | No | 1016 | yes |
| 49 | Ardisia complanata | Wall. | Primulaceae | N | No | 719 | no (no) |
| 50 | Ardisia crenata | Sims | Primulaceae | I | No | yes | |
| 51 | Ventilago madraspatana | Boerl. | Rhamnaceae | N | No | 41 | no (yes) |
| 52 | Psychotria montana | Blume | Rubiaceae | N | No | 1531 | no (yes) |
| 53 | Lunasia amara | Blanco | Rutaceae | N | Yes (P) | 1 | yes |
| 54 | Melicope lunu-ankenda | (Gaertn.) T.G. Hartley | Rutaceae | N | No | 241 | no (yes) |
| 55 | Kadsura scandens | (Blume) Blume | Schisandraceae | N | Yes (P) | 17 | yes |
| 56 | Smilax calophylla | Wall. ex A.DC. | Smilacaceae | N | No | 262 | yes |
| 57 | Smilax zeylanica | L. | Smilacaceae | N | Yes (P) | yes | |
| 58 | Aquilaria hirta | Ridl. | Thymelaeaceae | N | Yes (P, VU) | 21 | no (yes) |
| 59 | Amomum hochreutineri | Valeton | Zingiberaceae | N | Yes (E) | 102 | no (no) |
| 60 | Etlingera solaris | (Blume) R.M.Sm. | Zingiberaceae | N | Yes (E, VU) | 143 | no (no) |
| 61 | Meistera aculeata | (Roxb.) Skornick. & M.F. Newman | Zingiberaceae | N | No | 41 | no (yes) |
| Observed Parameter | ITS2 (%) | matK * (%) | rbcL (%) | trnL (%) |
|---|---|---|---|---|
| No PCR amplicon obtained | 1.64 | 27.87 | 1.64 | 16.39 |
| Mixed sequences—no use | 8.20 | 0 | 1.64 | 3.28 |
| Sequence provided | 90.16 | 72.13 | 96.72 | 80.33 |
| Assembled consensus sequence | 88.52 | 65.57 | 96.72 | 73.77 |
| Unidirectional sequence | 1.64 | 6.56 | 0 | 6.56 |
| Identification Measure | Region | |||
|---|---|---|---|---|
| ITS2 (%) | matK * (%) | rbcL (%) | trnL (%) | |
| Correct identification at species level | 29.51 | 31.15 | 29.51 | 16.39 |
| Correct identification at genus level | 32.79 | 47.54 | 52.46 | 55.74 |
| Correct identification at family level | 6.56 | 0 | 9.84 | 8.20 |
| Incorrect identification | 22.95 | 0 | 4.92 | 0 |
| Region | Observation | Value (%) | Related Species |
|---|---|---|---|
| ITS2 | Overall average | 1.29503 | |
| Minimum distance | 0.00440 | Nepenthes reinwardtiana and Nervilia concolor *** | |
| Maximum distance | 2.70903 | Erycibe malaccensis and Acalypha grandis *** | |
| matK | Overall average | 1.12567 | |
| Minimum distance | 0.00615 | Nepenthes mirabilis and N. ampullaria * | |
| Maximum distance | 2.62368 | Nepenthes reinwardtiana and Parkia timoriana *** | |
| rbcL | Overall average | 1.19148 | |
| Minimum distance | 0.00350 | Amomum hochreutineri and Etlingera solaris ** | |
| Maximum distance | 2.62587 | Phyllanthus oxyphyllus and Galearia filiformis *** | |
| trnL | Overall average | 1.11310 | |
| Minimum distance | 0.02887 | Alstonia scholaris and Rauvolfia serpentina ** | |
| Maximum distance | 2.59858 | Millettia sericea and Cymbidium aloifolium *** |
| Gene Region | Name | Sequence | Reference |
|---|---|---|---|
| rbcL | rbcLa-F | ATGTCACCACAAACAGAGACTAAAGC | [50] |
| rbcLa-R | GTAAAATCAAGTCCACCRCG | ||
| matK | matK472F | CCCRTYCATCTGGAAATCTTGGTTC | [41] |
| matK1248R | GCTRTRATAATGAGAAAGATTTCTGC | ||
| matKa | matKxF | TAATTTACGATCAATTCATTC | [23] |
| matK5R | GTTCTAGCACAAGAAAGTCG | ||
| ITS2 | ITS2F | ATGCGATACTTGGTGTGAAT | [51] |
| ITS3R | GACGCTTCTCCAGACTACAAT | ||
| trnL | trnL-F | ATTTGAACTGGTGACACGAG | [7] |
| trnL-c | CGAAATCGGTAGACGCTACG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cahyaningsih, R.; Compton, L.J.; Rahayu, S.; Magos Brehm, J.; Maxted, N. DNA Barcoding Medicinal Plant Species from Indonesia. Plants 2022, 11, 1375. https://doi.org/10.3390/plants11101375
Cahyaningsih R, Compton LJ, Rahayu S, Magos Brehm J, Maxted N. DNA Barcoding Medicinal Plant Species from Indonesia. Plants. 2022; 11(10):1375. https://doi.org/10.3390/plants11101375
Chicago/Turabian StyleCahyaningsih, Ria, Lindsey Jane Compton, Sri Rahayu, Joana Magos Brehm, and Nigel Maxted. 2022. "DNA Barcoding Medicinal Plant Species from Indonesia" Plants 11, no. 10: 1375. https://doi.org/10.3390/plants11101375
APA StyleCahyaningsih, R., Compton, L. J., Rahayu, S., Magos Brehm, J., & Maxted, N. (2022). DNA Barcoding Medicinal Plant Species from Indonesia. Plants, 11(10), 1375. https://doi.org/10.3390/plants11101375






