An R2R3-MYB Transcription Factor OsMYBAS1 Promotes Seed Germination under Different Sowing Depths in Transgenic Rice
Abstract
:1. Introduction
2. Results
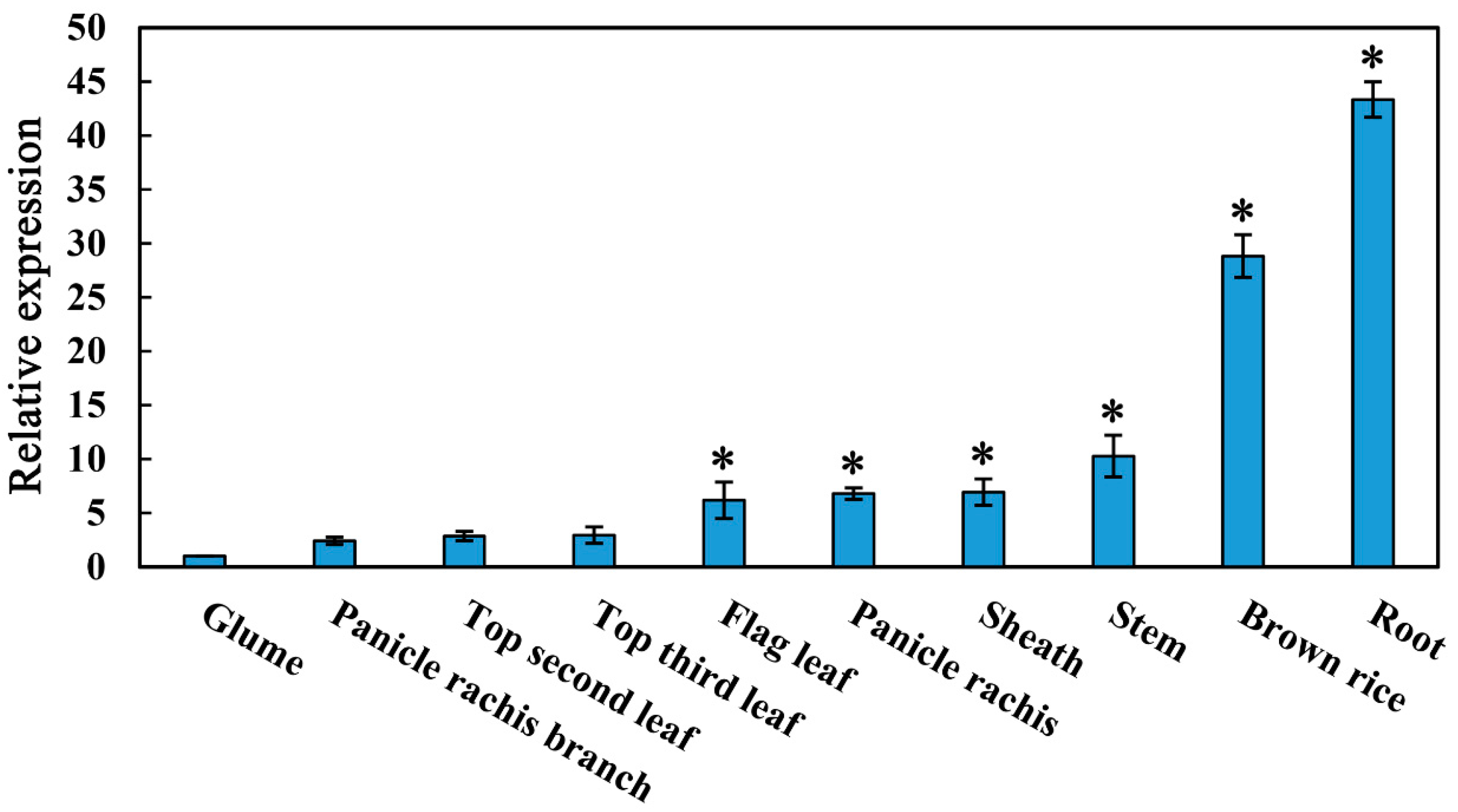
2.1. Expression Profile of OsMYBAS1
2.2. Subcellular Localization of OsMYBAS1
2.3. OsMYBAS1 Regulates Seed Germination under Deep-Sowing Condition
2.4. Antioxidant Capacity of OsMYBAS1-Overexpressing Rice
3. Discussion
4. Material and Methods
4.1. Plant Material, Growth Conditions and Treatments
4.2. Subcellular Localization
4.3. Vector Construction and Plant Transformation
4.4. qRT-PCR
4.5. Germination Rate, Root Length and Seedling Height Measurements
4.6. Determination of Peroxidase, Superoxide Dismutase and Catalase Activity and Malondialdehyde Content
4.7. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seck, P.A.; Diagne, A.; Mohanty, S.; Wopereis, M.C.S. Crops that feed the world 7: Rice. Food Secur. 2012, 4, 7–24. [Google Scholar] [CrossRef]
- Yang, W.J.; Xu, D.T.S.; Li, S.Y.; Tang, X.G.; Pan, S.G.; Chen, X.F.; Mo, Z.W. Emergence and seedling establishment of rice varieties at different sowing depths. J. Plant Growth Regul. 2021. [Google Scholar] [CrossRef]
- Wang, W.Q.; Peng, S.B.; Chen, Q.; Mei, J.H.; Dong, H.L.; Nie, L.X. Effects of pre-sowing seed treatments on establishment of dry direct-seeded early rice under chilling stress. AoB Plants 2016, 8, plw074. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.Y.; Hussain, S.; Zheng, M.M.; Peng, S.B.; Huang, J.J.; Cui, K.H.; Nie, L.X. Dry direct-seeded rice as an alternative to transplanted-flooded rice in Central China. Agron. Sustain. Dev. 2014, 35, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.X.; Du, J.; Zhou, Y.Z.; Zeng, Y.J.; Tan, X.M.; Pan, X.H.; Shi, Q.H.; Wu, Z.M.; Zeng, Y.H. Effects of different mechanical direct seeding methods on grain yield and lodging resistance of early Indica rice in South China. J. Integr. Agr. 2021, 20, 1204–1215. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, Q.S.; Kuai, J.; Zhao, L.; Hu, Z.; Wu, J.S.; Zhou, G.S. The effect of sowing depth and soil compaction on the growth and yield of rapeseed in rice straw returning field. Field Crop. Res. 2017, 203, 47–54. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, W.P.; Jiang, C.H.; Wang, X.N.; Xiong, H.Y.; Todorovska, E.G.; Yin, Z.G.; Chen, Y.F.; Wang, X.; Xie, J.Y.; et al. Genetic architecture and candidate genes for deep-sowing tolerance in rice revealed by non-syn GWAS. Front. Plant Sci. 2018, 9, 332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Li, C.N.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Smita, S.; Katiyar, A.; Chinnusamy, V.; Pandey, D.M.; Bansal, K.C. Transcriptional regulatory network analysis of MYB transcription factor family genes in rice. Front. Plant Sci. 2015, 6, 1157. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.H.; Bao, X.X.; Zhi, Y.L.; Wu, Q.; Guo, Y.R.; Yin, X.H.; Zeng, L.Q.; Li, J.; Zhang, J.; He, W.L.; et al. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef] [Green Version]
- Klempnauer, K.H.; Gonda, T.J.; Bishop, J.M. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: The architecture of a transduced oncogene. Cell 1982, 31, 453–463. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Petersont, P.A.; Saedler, H. The regulatory Cl locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Vannini, C.; Locatelli, F.; Bracale, M.; Magnani, E.; Marsoni, M.; Osnato, M.; Mattana, M.; Baldoni, E.; Coraggio, I. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 2004, 37, 115–127. [Google Scholar] [CrossRef]
- Pasquali, G.; Biricolti, S.; Locatelli, F.; Baldoni, E.; Mattana, M. Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep. 2008, 27, 1677–1686. [Google Scholar] [CrossRef]
- Hu, Z.Y.; Yamauchi, T.; Yang, J.H.; Jikumaru, Y.; Tsuchida-Mayama, T.; Ichikawa, H.; Takamure, I.; Nagamura, Y.; Tsutsumi, N.; Yamaguchi, S.; et al. Strigolactone and cytokinin act antagonistically in regulating rice mesocotyl elongation in darkness. Plant Cell Physiol. 2014, 55, 30–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamiru, M.; Abe, A.; Utsushi, H.; Yoshida, K.; Takagi, H.; Fujisaki, K.; Undan, J.R.; Rakshit, S.; Takaichi, S.; Jikumaru, Y.; et al. The tillering phenotype of the rice plastid terminal oxidase (PTOX) loss-of-function mutant is associated with strigolactone deficiency. New Phytol. 2014, 202, 116–131. [Google Scholar] [CrossRef]
- Du, L.G.; Jiang, H.Y.; Zhao, G.W.; Ren, J.Y. Gene cloning of ZmMYB59 transcription factor in maize and its expression during seed germination in response to deep-sowing and exogenous hormones. Plant Breed. 2017, 136, 834–844. [Google Scholar] [CrossRef]
- Chen, Y.H.; Yang, X.Y.; He, K.; Liu, M.H.; Li, J.; Gao, Z.F.; Lin, Z.Q.; Zhang, Y.F.; Wang, X.X.; Qiu, X.M.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Niu, Q.W.; Wu, H.W.; Liu, J.; Ye, J.; Yu, N.; Chua, N.H. Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 2015, 84, 404–416. [Google Scholar] [CrossRef]
- El-Kereamy, A.; Bi, Y.M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 2012, 7, e52030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, X.Y.; Xu, Y.Y.; Ma, Q.B.; Xu, W.Y.; Wang, T.; Xue, Y.B.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.D.; Jiang, Y.L.; Liang, Y.N.; Chen, L.; Chen, W.J.; Cheng, B.J. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 2019, 137, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Mu, R.L.; Cao, Y.R.; Liu, Y.F.; Lei, G.; Zou, H.F.; Liao, Y.; Wang, H.W.; Zhang, W.K.; Ma, B.; Du, J.Z.; et al. An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res. 2009, 19, 1291–1304. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.J.; Zhang, L.; Wang, J.C.; Li, C.S.; Zeng, X.; Xie, S.P.; Zhang, Y.Z.; Liu, S.S.; Hu, S.L.; Wang, J.H.; et al. Quantitative trait locus analysis for deep-sowing germination ability in the maize IBM Syn10 DH population. Front. Plant Sci. 2017, 8, 813. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.S.; Sasaki, K.; Kang, J.W.; Sato, T.; Song, W.Y.; Ahn, S.N. Mesocotyl elongation is essential for seedling emergence under deep-seeding condition in rice. Rice 2017, 10, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno, H.; Banayo, N.P.M.C.; Bueno, C.S.; Kashiwagi, J.I.; Nakashima, T.; Corales, A.M.; Garcia, R.; Sandhu, N.; Kumar, A.; Kato, Y. Longer mesocotyl contributes to quick seedling establishment, improved root anchorage, and early vigor of deep-sown rice. Field Crop. Res. 2018, 228, 84–92. [Google Scholar] [CrossRef]
- Lu, Q.; Zhang, M.C.; Niu, X.J.; Wang, C.H.; Xu, Q.; Feng, Y.; Wang, S.; Yuan, X.P.; Yu, H.Y.; Wang, Y.P.; et al. Uncovering novel loci for mesocotyl elongation and shoot length in India rice through genome-wide association mapping. Planta 2016, 243, 645–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, Y.; Kim, J.H.; Nguyen, H.N.; Jikumaru, Y.; Kamiya, Y.; Hong, S.W.; Lee, H. A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J. Exp. Bot. 2013, 64, 3911–3922. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.; Zhang, Y.M.; Zhang, M.S.; Li, T.; Dirk, L.M.; Downie, B.; Zhao, T.Y. ZmGOLS2, a target of transcription factor ZmDREB2A, offers similar protection against abiotic stress as ZmDREB2A. Plant Mol. Biol. 2016, 90, 157–170. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fu, H.; Cao, D.D.; Hu, W.M.; Guan, Y.J.; Fu, Y.Y.; Fang, Y.F.; Hu, J. Studies on optimum harvest time for hybrid rice seed. J. Sci. Food Agric. 2017, 97, 1124–1133. [Google Scholar] [CrossRef]


| Plant Material | 0 cm Sowing-Depth | 4 cm Sowing-Depth | ||||
|---|---|---|---|---|---|---|
| Germination Rate (%) | Root Length (cm) | Seedling Height (cm) | Germination Rate (%) | Root Length (cm) | Seedling Height (cm) | |
| WT | 96.7 ± 3.3 a | 7.9 ± 0.5 ab | 11.7 ± 0.3 a | 55.0 ± 3.8 b | 2.9 ± 0.6 abc | 4.8 ± 0.1 c |
| osmybas1-1 | 97.7 ± 1.5 a | 11.2 ± 1.4 a | 9.4 ± 0.8 b | 23.0 ± 3.2 c | 2.1 ± 0.6 bc | 3.0 ± 0.3 d |
| osmybas1-2 | 96.7 ± 1.8 a | 7.5 ± 1.1 b | 9.0 ± 0.2 b | 20.0 ± 2.5 c | 1.5 ± 0.4 c | 2.0 ± 0.1 d |
| OE-1 | 97.3 ± 1.5 a | 9.0 ± 0.3 ab | 12.0 ± 0.4 a | 79.3 ± 1.8 a | 4.5 ± 0.5 ab | 8.3 ± 0.9 a |
| OE-2 | 95.3 ± 2.6 a | 6.7 ± 1.4 b | 12.8 ± 0.5 a | 86.3 ± 2.0 a | 3.9 ± 0.8 a | 6.8 ± 0.1 b |
| Plant | SOD (U/g) | POD (nmol FW/min) | CAT (nmol FW/min) | MDA (nmol/g) |
|---|---|---|---|---|
| WT | 731.3 ± 3.8c | 23.4 ± 0.9a | 1.0 ± 0.1a | 415.8 ± 0.1a |
| OE-1 | 814.6 ± 0.8b | 26.1 ± 0.2a | 1.1 ± 0.4a | 224.2 ± 0.2b |
| OE-2 | 861.8 ± 0.1a | 26.1 ± 0.4a | 1.4 ± 0.1a | 207.1 ± 0.7c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Wu, R.; Shen, T.; Li, Z.; Li, C.; Wu, B.; Jiang, H.; Zhao, G. An R2R3-MYB Transcription Factor OsMYBAS1 Promotes Seed Germination under Different Sowing Depths in Transgenic Rice. Plants 2022, 11, 139. https://doi.org/10.3390/plants11010139
Wang X, Wu R, Shen T, Li Z, Li C, Wu B, Jiang H, Zhao G. An R2R3-MYB Transcription Factor OsMYBAS1 Promotes Seed Germination under Different Sowing Depths in Transgenic Rice. Plants. 2022; 11(1):139. https://doi.org/10.3390/plants11010139
Chicago/Turabian StyleWang, Xiaomin, Rong Wu, Tongshu Shen, Zhenan Li, Chengyong Li, Bangkui Wu, Hongye Jiang, and Guangwu Zhao. 2022. "An R2R3-MYB Transcription Factor OsMYBAS1 Promotes Seed Germination under Different Sowing Depths in Transgenic Rice" Plants 11, no. 1: 139. https://doi.org/10.3390/plants11010139
APA StyleWang, X., Wu, R., Shen, T., Li, Z., Li, C., Wu, B., Jiang, H., & Zhao, G. (2022). An R2R3-MYB Transcription Factor OsMYBAS1 Promotes Seed Germination under Different Sowing Depths in Transgenic Rice. Plants, 11(1), 139. https://doi.org/10.3390/plants11010139





