Transcriptional Controls for Early Bolting and Flowering in Angelica sinensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Total RNA Isolation and Illumina Sequencing
2.3. Sequence Filtration, Assembly and Unigene Expression Analysis
2.4. Basic Annotation of DEGs and Gene Cluster Analysis
2.5. qRT-PCR Validation
2.6. GA Quantification and Identification
2.7. Soluble Sugar Measurement
2.8. Statistical Analysis
3. Results
3.1. Global Gene Analysis
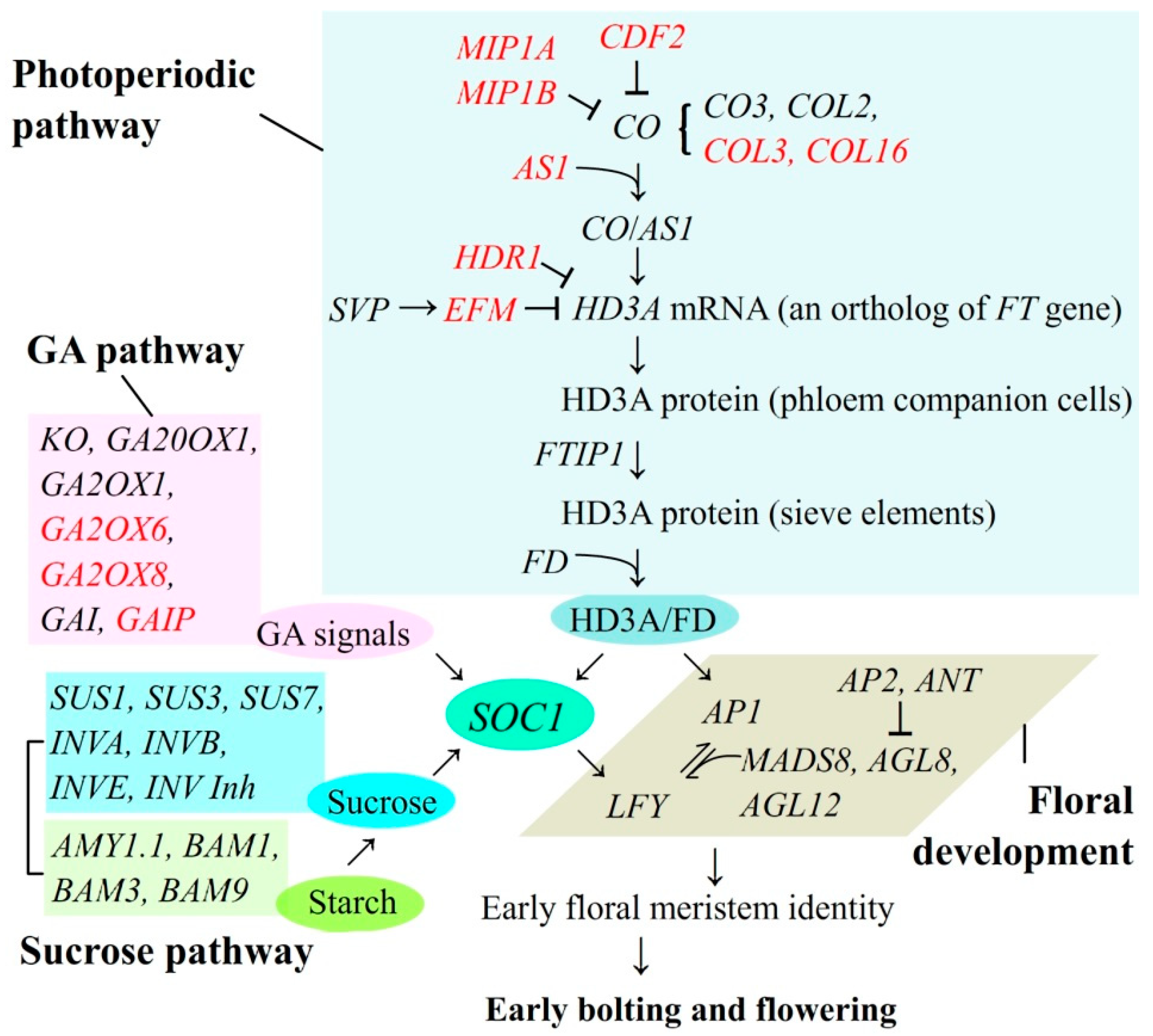
3.2. DEGs Linked with Bolting and Flowering
3.3. Flower-Regulating DEGs Inarticulately Expressed with EBF
3.4. Sucrose and GA Accumulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, M.F.; Liu, X.Z.; Wei, J.H.; Zhang, Z.; Chen, S.J.; Liu, Z.H.; Xing, H. Selection of high altitude planting area of Angelica sinensis based on biomass, bioactive compounds accumulation and antioxidant capacity. Chin. Tradit. Herbal Drugs 2020, 51, 474–481. [Google Scholar]
- Xu, X.Q.; Zhang, X.B.; Chen, J.; Zhao, W.L.; Jin, L. Study on ecological suitability of Angelica sinensis in Gansu Province. Chin. Tradit. Herbal Drugs 2020, 51, 3304–3307. [Google Scholar]
- Zhang, H.Y.; Bi, W.G.; Yu, Y.; Liao, W.B. Angelica sinensis (Oliv.) Diels in China: Distribution, cultivation, utilization and variation. Genet. Resour. Crop Evol. 2012, 59, 607–613. [Google Scholar] [CrossRef]
- Upton, R. American Herbal Pharmacopoeia and Therapeutic compendium: Dang Gui Root-Angelica sinensis (Oliv.); American Herbal Pharmacopoeia: Scotts Valley, CA, USA, 2003; pp. 1–41. [Google Scholar]
- Ma, J.P.; Guo, Z.B.; Jin, L.; Li, Y.D. Phytochemical progress made in investigations of Angelica sinensis (Oliv.) Diels. Chin. J. Nat. Med. 2015, 13, 241–249. [Google Scholar] [CrossRef]
- Wei, W.L.; Zeng, R.; Gu, C.M.; Qu, Y.; Huang, L.F. Angelica sinensis in China-A review of botanical profile, ethnopharmacology, phytochemistry and chemical analysis. J. Ethnopharmacol. 2016, 190, 116–141. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.W.; Hong, Y.H.; Chen, M.L.; Lin, B.F. Inhibitory effects of Angelica sinensis ethyl acetate extract and major compounds on NF-κB trans-activation activity and LPS-induced inflammation. J. Ethnopharmacol. 2010, 129, 244–249. [Google Scholar] [CrossRef]
- Wang, L.Y.; Tang, Y.P.; Liu, X.; Zhu, M.; Tao, W.W.; Li, W.X.; Duan, J.A. Effects of ferulic acid on antioxidant activity in Angelicae sinensis Radix, Chuan xiong Rhizoma, and their combination. Chin. J. Nat. Med. 2015, 13, 401–408. [Google Scholar]
- Wang, B.H.; Ou-Yang, J.P. Pharmacological actions of sodium ferulate in cardiovascular system. Cardiovasc. Drug Rev. 2005, 23, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Li, M.F.; Kang, T.L.; Jin, L.; Wei, J.H. Research progress on bolting and flowering of Angelica sinensis and regulation pathways. Chin. Tradit. Herbal Drugs 2020, 51, 5894–5899. [Google Scholar]
- Huang, L.Q.; Jin, L. Suitable Technology for Production and Processing of Angelica Sinensis; Pharmaceutical Science and Technology Press: Beijing, China, 2018; pp. 1–14. [Google Scholar]
- Zhang, Z.; Liu, X.Z.; Bao, Y.J.; Li, J.; Cao, X.L.; Li, M.F. Selection of cropping rotations of Angelica sinensis based on biomass, bioactive compounds accumulation and antioxidant capacity. J. Gansu Agric. Univ. 2018, 53, 86–89. [Google Scholar]
- Wang, W.J. Analysis and control of early bolting characteristic of Angelica sinensis. J. Northwest Univ. (Nat. Sci. Ed.) 1977, 7, 32–39. [Google Scholar]
- Wang, W.J. Technology and principle of seedling frozen storage of Angelica sinensis. J. Chin. Med. Mat. 1979, 3, 1–4. [Google Scholar]
- Lin, H.M.; Wu, Y.A.; Cao, Z.F.; Lv, S.L.; Mao, X.J. Influence of sun shade cultivation on premature bolting in Angelica sinensis and growth environment factors. Chin. J. Exp. Trad. Med. Formula. 2010, 16, 79–83. [Google Scholar]
- Qiu, D.Y.; Lin, H.M.; Fang, Z.S.; Li, Y.D. Effects of seedlings with different root diameters on Angelica sinensis early bolting and physiological changes during the medicine formation period. Acta Pratac. Sin. 2010, 19, 100–105. [Google Scholar]
- Qiu, D.Y.; Lin, H.M.; Chen, Y.; Li, Y.D.; Guo, F.X. Effects of latitude, longitude and altitude on Angelica growth and early bolting in medicine formation period. Acta Agrestia Sin. 2010, 18, 838–843. [Google Scholar]
- Qi, J.T.; Lin, H.M.; Liu, X.Z. Preliminary study on the effect of nitrogen and phosphorus fertilizer on bolting rate of Angelica sinensis. J. Chin. Med. Mater. 2004, 27, 82–83. [Google Scholar]
- Li, M.S.; Yu, Z.Y. Research on Prevention of Early Bolting of Angelica sinensis. Chin. Tradit. Herbal Drugs 1979, 57, 34–38, 49. [Google Scholar]
- Taiz, L.; Zeiger, E. Plant Physiology, 5th ed.; Sinauer Associates, Inc., Publishers: Sunderland, MA, USA, 2010; pp. 559–590. [Google Scholar]
- Michaels, S.D.; Amasino, R.M. Memories of winter: Vernalization and the competence to flower. Plant Cell Environ. 2000, 23, 1145–1154. [Google Scholar] [CrossRef]
- Chen, L.; Cheng, J.C.; Castle, L.; Sung, Z.R. EMF genes regulate Arabidopsis inflorescence development. Plant Cell 1997, 9, 2011–2024. [Google Scholar] [PubMed]
- Goodrich, J.; Puangsomlee, P.; Martin, M.; Long, D.; Meyerowitz, E.M.; Coupland, G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 1997, 386, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Piñeiro, M.; Gómez-Mena, C.; Schaffer, R.; Martínez-Zapater, J.M.; Coupland, G. EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 2003, 15, 1552–1562. [Google Scholar] [CrossRef] [Green Version]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Ruehl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Graeff, M.; Straub, D.; Eguen, T.; Dolde, U.; Rodrigues, V.; Brandt, R.; Wenkel, S. Microprotein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. Plos Genet. 2016, 12, e1005959. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.Q.; Zhang, J.W.; Ren, L.R.; Huang, H.Y.; Ma, Z.C.; Qi, J.T. Analysis on physiological and biochemical characteristics of bolting Angelica sinensis plant. Chin. Tradit. Herbal Drugs 2011, 42, 2326–2329. [Google Scholar]
- Yu, G.; Zhou, Y.; Yu, J.J.; Hu, X.Q.; Tang, Y.; Yan, H.; Duan, J.A. Transcriptome and digital gene expression analysis unravels the novel mechanism of early flowering in Angelica sinensis. Sci. Rep. 2019, 9, 10035. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, M.L.; Zhu, T.T.; Zhang, X.N.; Li, M.F.; Wei, J.H. Integrated transcriptomics and metabolites at different growth stages reveals the regulation mechanism of bolting and flowering of Angelica sinensis. Plant Biol. 2021, 23, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Mortazavi, A.; Williams, B.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Lee, J.; Oh, M.; Park, H.; Lee, I. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J. 2008, 55, 832–843. [Google Scholar] [CrossRef]
- Theissen, G.; Becker, A.; Di Rosa, A.; Kanno, A.; Kim, J.T.; Munster, T.; Winter, K.U.; Saedler, H. A short history of MADS-box genes in plants. Plant Mol. Biol. 2000, 42, 115–149. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.G.; Jang, S.; Chung, J.E.; Cho, Y.G.; An, G. Characterization of two rice MADS box genes that control flowering time. Mol. Cells 1997, 7, 559–566. [Google Scholar] [PubMed]
- Ferrandiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000, 127, 725–734. [Google Scholar] [CrossRef]
- Tapia-Lopez, R.; Garcia-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Perez-Ruiz, R.V.; Kim, S.H.; Acevedo, F.; Pelaz, S.; Alvarez-Buylla, E.R. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef] [Green Version]
- Sommer, H.; Beltrán, J.P.; Huijser, P.; Pape, H.; Lönnig, W.E.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef]
- Gregis, V.; Sessa, A.; Dorca-Fornell, C.; Kater, M.M. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 2009, 60, 626–637. [Google Scholar] [CrossRef]
- Krogan, N.T.; Hogan, K.; Long, J.A. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 2012, 139, 4180–4190. [Google Scholar] [CrossRef] [Green Version]
- Elliott, R.C.; Betzner, A.S.; Huttner, E.; Oakes, M.P.; Tucker, W.Q.J.; Gerentes, D.; Perez, P.; Smyth, D.R. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 1996, 8, 155–168. [Google Scholar]
- Krizek, B.A.; Prost, V.; Macias, A. AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell 2000, 12, 1357–1366. [Google Scholar] [CrossRef]
- Smeekens, S.; Ma, J.; Hanson, J.; Rolland, F. Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 2010, 3, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Nunez, J.G.; Tiessen, A. Arabidopsis sucrose synthase 2 and 3 modulate metabolic homeostasis and direct carbon towards starch synthesis in developing seeds. Planta 2010, 232, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P.; Stitt, M. Sucrose synthase catalyzes a readily reversible-reaction in vivo in developing potato tubers and other plant tissues. Planta 1993, 189, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Tamoi, M.; Tabuchi, T.; Demuratani, M.; Otori, K.; Tanabe, N.; Maruta, T.; Shigeoka, S. Point mutation of a plastidic invertase inhibits development of the photosynthetic apparatus and enhances nitrate assimilation in sugar-treated Arabidopsis seedlings. J. Biol. Chem. 2010, 285, 15399–15407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas, W.A.; Pontis, H.G.; Salerno, G.L. New insights on sucrose metabolism: Evidence for an active A/N-Inv in chloroplasts uncovers a novel component of the intracellular carbon trafficking. Planta 2008, 227, 795–807. [Google Scholar] [CrossRef]
- Xiang, L.; Le Roy, K.; Bolouri-Moghaddam, M.R.; Vanhaecke, M.; Lammens, W.; Rolland, F.; Van den Ende, W. Exploring the neutral invertase-oxidative stress defence connection in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3849–3862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rausch, T.; Greiner, S. Plant protein inhibitors of invertases. Biochim. Biophys. Acta 2004, 1696, 253–261. [Google Scholar] [CrossRef]
- Fulton, D.C.; Stettler, M.; Mettler, T.; Vaughan, C.K.; Li, J.; Francisco, P.; Gil, M.; Reinhold, H.; Eicke, S.; Messerli, G.; et al. Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell 2008, 20, 1040–1058. [Google Scholar] [CrossRef] [Green Version]
- Edner, C.; Li, J.; Albrecht, T.; Mahlow, S.; Hejazi, M.; Hussain, H.; Kaplan, F.; Guy, C.; Smith, S.M.; Steup, M.; et al. Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial beta-amylases. Plant Physiol. 2007, 145, 17–28. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Gehan, J.P.; Sharkey, T.D. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005, 138, 2280–2291. [Google Scholar] [CrossRef] [Green Version]
- Blázquez, M.A.; Weigel, D. Integration of floral inductive signals in Arabidopsis. Nature 2000, 404, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Sudhakar, D.; Peng, J.; Richards, D.E.; Christou, P.; Harberd, N.P. Expression of Arabidopsis GAI in transgenic rice represses multiple gibberellin responses. Plant Cell 2001, 13, 1791–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukazawa, J.; Teramura, H.; Murakoshi, S.; Nasuno, K.; Nishida, N.; Ito, T.; Yoshida, M.; Kamiya, Y.; Yamaguchi, S.; Takahashi, Y. DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell 2014, 26, 2920–2938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrone, D.; Chen, X.; Coates, R.M.; Peters, R.J. Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem. J. 2010, 431, 337–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rieu, I.; Ruiz-Rivero, O.; Fernandez-Garcia, N.; Griffiths, J.; Powers, S.J.; Gong, F.; Linhartova, T.; Eriksson, S.; Nilsson, O.; Thomas, S.G.; et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008, 53, 488–504. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.K.; Yun, C.H.; Lee, J.H.; Jang, Y.H.; Park, H.Y.; Kim, J.K. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 2008, 228, 355–365. [Google Scholar] [CrossRef]
- Liu, L.; Liu, C.; Hou, X.; Xi, W.; Shen, L.; Tao, Z.; Wang, Y.; Yu, H. FTIP1 is an essential regulator required for florigen transport. PloS Biol. 2012, 10, e1001313. [Google Scholar] [CrossRef] [Green Version]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005, 309, 1056–1059. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Lee, I.; Lee, S.Y.; Imaizumi, T.; Hong, J.C. CONSTANS and ASYMMETRIC LEAVES 1 complex is involved in the induction of FLOWERING LOCUS T in photoperiodic flowering in Arabidopsis. Plant J. 2012, 69, 332–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Zhang, Z.; Wu, J.; Cui, X.; Feng, D.; Wang, K.; Xu, M.; Zhou, L.; Han, X.; Gu, X.; et al. The Oryza sativa regulator HDR1 associates with the kinase OsK4 to control photoperiodic flowering. PLoS Genet. 2016, 12, e1005927. [Google Scholar] [CrossRef] [PubMed]
- van der Hoeven, C.; Dietz, A.; Landsmann, J. Variability of organ-specific gene expression in transgenic tobacco plants. Transgenic Res. 1994, 3, 159–166. [Google Scholar] [CrossRef]
- Tao, S.Q.; Li, J.; Gu, X.G.; Wang, Y.A.; Xia, Q.; Qin, B.; Zhu, L. Quantitative analysis of ATP sulfurylase and selenocysteine methyltransferase gene expression in different organs of tea plant (Camellia sinensis). Amer. J. Plant Sci. 2012, 3, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Che, P.; Lall, S.; Nettleton, D.; Howell, S.H. Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 2006, 141, 620–637. [Google Scholar] [CrossRef] [Green Version]



| Bolted | Unbolted | |
|---|---|---|
| Unfiltered data | ||
| Data of reads number (million) | 60.73 | 52.48 |
| Reads length | 150 | 150 |
| GC (%) | 44.69 | 45.12 |
| Data of reads number×read length (million) | 9110 | 7872 |
| Q20 (%) | 98.50 | 98.47 |
| Q30 (%) | 95.25 | 95.18 |
| Filtered data 1 | ||
| Data of reads number (million) | 60.66 | 52.41 |
| Data of reads number×read length (million) | 9098 | 7862 |
| Q20 (%) | 98.56 | 98.53 |
| Q30 (%) | 95.34 | 95.26 |
| Mapped data 2 | ||
| Data of unique mapped reads (million) | 44.70 | 37.40 |
| Data of multiple mapped reads (million) | 7.80 | 6.40 |
| Mapping ratio (%) | 86.56 | 83.57 |
| Compiled data | ||
| Total number of unigenes | 72,502 | |
| Total Length (bp) (million) | 64.14 | |
| N50 (bp) | 1534 | |
| Max length (bp) | 15,601 | |
| Min length (bp) | 201 | |
| Average Length (bp) | 884 | |
| GC content (%) | 41.17 | |
| BLASTx Searching against Specific Platforms | Values | Percentage (%) |
|---|---|---|
| NR | 44,708 | 61.66 |
| SwissProt | 30,471 | 42.03 |
| KOG | 22,959 | 31.67 |
| KEGG | 18,056 | 24.90 |
| GO | 12,473 | 17.20 |
| Gene Name | Gene ID | Protein Name | log2 Ratio (BRPKM/UBRPKM) |
|---|---|---|---|
| Floral development (8) | |||
| Genes favoring flowering | |||
| SOC1 | XP_017245180.1 | MADS-box protein SOC1 | 1.06 |
| MADS8 | XP_017257209.1 | MADS-box transcription factor 8 | 7.21 |
| AGL8 | XP_017244085.1 | Agamous-like MADS-box protein AGL8 | 4.16 |
| AGL12 | XP_017218759.1 | Agamous-like MADS-box protein AGL12 | 3.42 |
| DEFA | XP_017253634.1 | Floral homeotic protein DEFICIENS | 1.11 |
| AP1 | AGX01569.1 | Floral homeotic protein APETALA 1 | 4.29 |
| Genes disfavoring flowering | |||
| AP2 | XP_017231882.1 | Floral homeotic protein APETALA 2 | −6.14 |
| ANT | XP_017254585.1 | AP2-like ethylene-responsive transcription factor ANT | −3.27 |
| Sucrose pathway (11) | |||
| Genes favoring flowering | |||
| SUS1 | XP_017219197.1 | Sucrose synthase isoform 1 | 1.31 |
| SUS3 | XP_017225961.1 | Sucrose synthase 3 | 1.40 |
| SUS7 | XP_017244457.1 | Sucrose synthase 7 | −2.70 |
| INVA | CAA76145.1 | Alkaline/neutral invertase A, mitochondrial | 1.41 |
| INVB | XP_017254796.1 | Probable alkaline/neutral invertase B | 1.22 |
| INVE | XP_017258042.1 | Alkaline/neutral invertase E, chloroplastic | 1.09 |
| AMY1.1 | XP_017218607.1 | Alpha-amylase | 1.03 |
| BAM1 | XP_017219233.1 | Beta-amylase 1, chloroplastic | 1.62 |
| BAM3 | XP_017236738.1 | Beta-amylase 3, chloroplastic | 1.05 |
| BAM9 | XP_017219710.1 | Inactive beta-amylase 9 | 1.30 |
| Genes disfavoring flowering | |||
| INV Inh | KZV43516.1 | Invertase inhibitor | −1.83 |
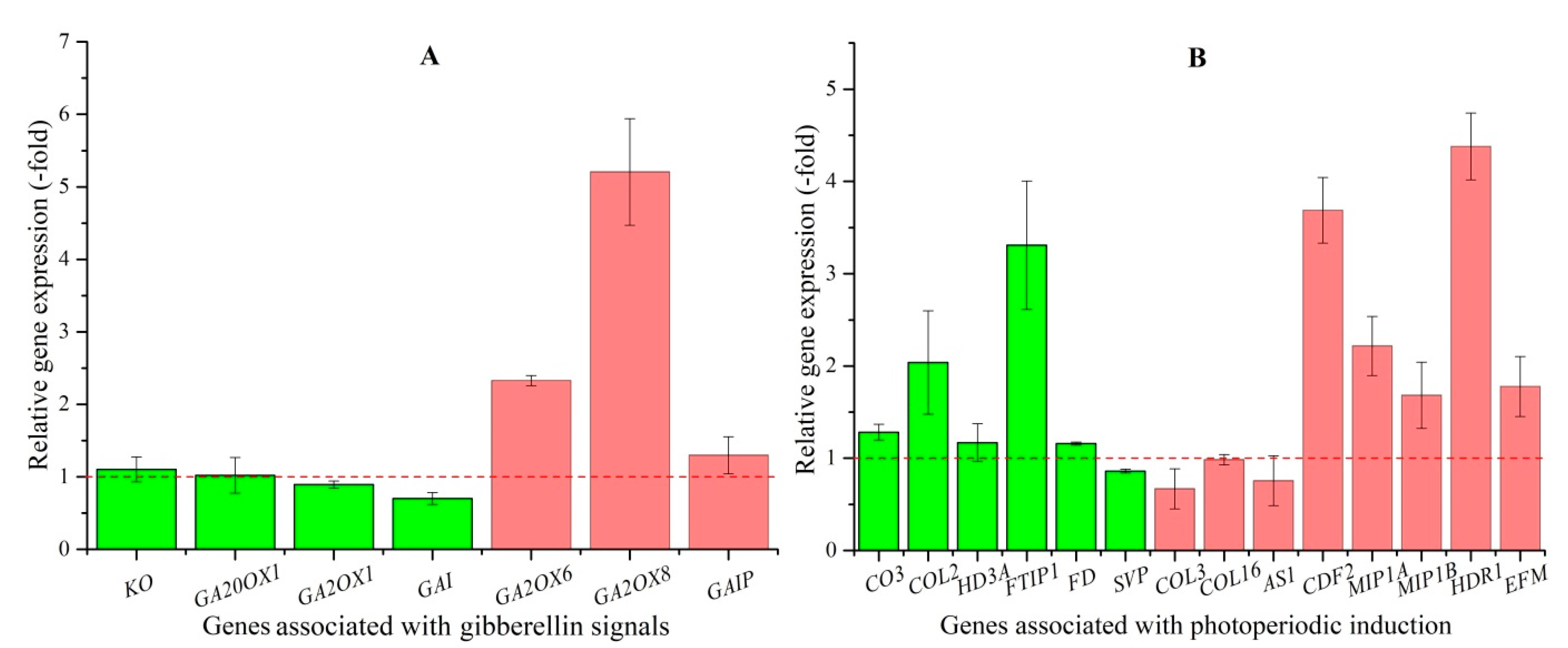
| GA pathway (7) | |||
| Genes favoring flowering | |||
| KO | XP_017253618.1 | Ent-kaurene oxidase, chloroplastic | 2.04 |
| GA20OX1 | XP_017239190.1 | Gibberellin 20 oxidase 1 | 1.77 |
| Genes disfavoring flowering | |||
| GA2OX1 | API85599.1 | Gibberellin 2-beta-dioxygenase 1 | −1.41 |
| GAI | XP_017238853.1 | DELLA protein GAI | −3.49 |
| GA2OX6 | XP_017243791.1 | Gibberellin 2-beta-dioxygenase 6 | 2.53 |
| GA2OX8 | XP_017220109.1 | Gibberellin 2-beta-dioxygenase 8 | 1.65 |
| GAIP | XP_017217018.1 | DELLA protein GAIP | 2.15 |
| Photoperiodic induction (14) | |||
| Genes favoring flowering | |||
| CO3 | XP_017232180.1 | Zinc finger protein CO3 | 2.58 |
| COL2 | XP_017231361.1 | Zinc finger protein CONSTANS-LIKE 2 | 3.5 |
| HD3A | XP_017216959.1 | Protein HEADING DATE 3A | 13.41 |
| FTIP1 | XP_019421416.1 | FT-interacting protein 1 | 2.13 |
| FD | XP_017256913.1 | Protein FD | 3.26 |
| HDR1 | XP_019170400.1 | Protein HEADING DATE REPRESSOR 1 | 2.12 |
| SVP | XP_017245967.1 | MADS-box protein SVP | −1.25 |
| Genes disfavoring flowering | |||
| COL3 | XP_017221909.1 | Zinc finger protein CONSTANS-LIKE 3 | −2.79 |
| COL16 | XP_017244294.1 | Zinc finger protein CONSTANS-LIKE 16 | −1.33 |
| AS1 | XP_017249788.1 | Transcription factor AS1 | −2.23 |
| CDF2 | XP_017221059.1 | Cyclic dof factor 2 | 3.03 |
| MIP1A | XP_017253198.1 | B-box domain protein 30 | 2.58 |
| MIP1B | XP_017253198.1 | B-box domain protein 31 | 3.50 |
| EFM | XP_017241902.1 | EARLY FLOWERING MYB PROTEIN | 1.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Li, J.; Wei, J.; Paré, P.W. Transcriptional Controls for Early Bolting and Flowering in Angelica sinensis. Plants 2021, 10, 1931. https://doi.org/10.3390/plants10091931
Li M, Li J, Wei J, Paré PW. Transcriptional Controls for Early Bolting and Flowering in Angelica sinensis. Plants. 2021; 10(9):1931. https://doi.org/10.3390/plants10091931
Chicago/Turabian StyleLi, Mengfei, Jie Li, Jianhe Wei, and Paul W. Paré. 2021. "Transcriptional Controls for Early Bolting and Flowering in Angelica sinensis" Plants 10, no. 9: 1931. https://doi.org/10.3390/plants10091931
APA StyleLi, M., Li, J., Wei, J., & Paré, P. W. (2021). Transcriptional Controls for Early Bolting and Flowering in Angelica sinensis. Plants, 10(9), 1931. https://doi.org/10.3390/plants10091931









