Effects on Capsicum annuum Plants Colonized with Trichoderma atroviride P. Karst Strains Genetically Modified in Taswo1, a Gene Coding for a Protein with Expansin-like Activity
Abstract
:1. Introduction
2. Results
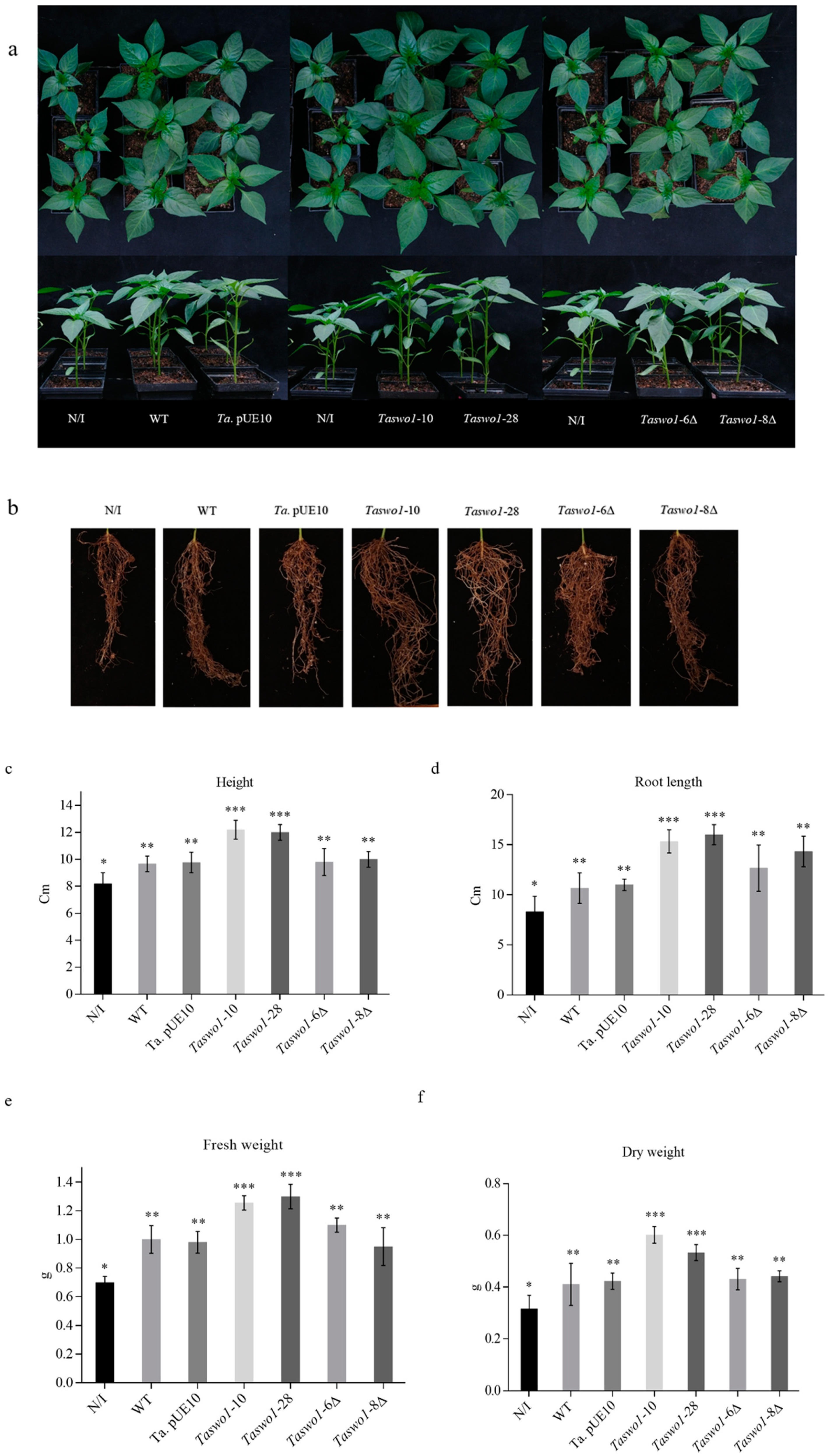
2.1. Overexpression of Taswo1 of T. atroviride Promotes Growth and Development of C. annuum and S. lycopersicum Plants
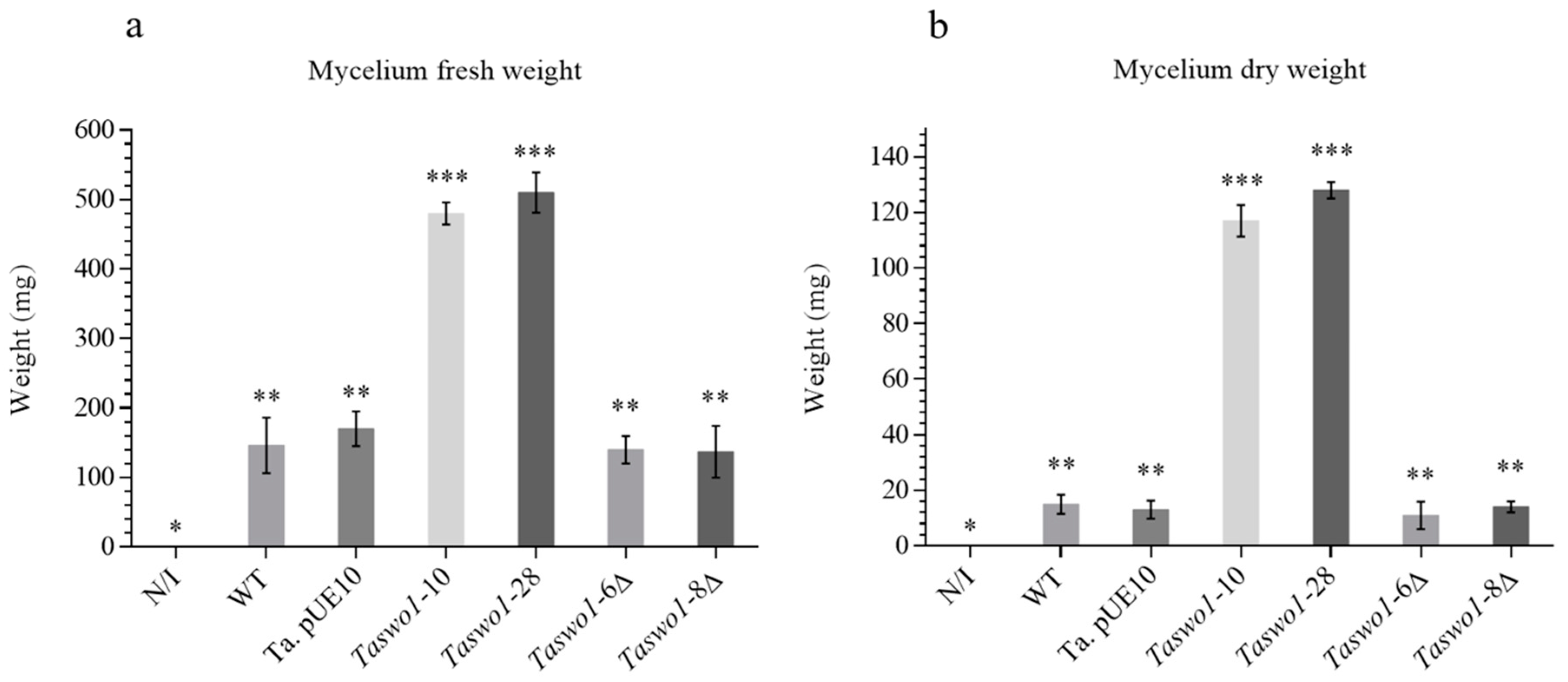
2.2. Root Colonization Assays by the T. atroviride Genetically Modified Strains
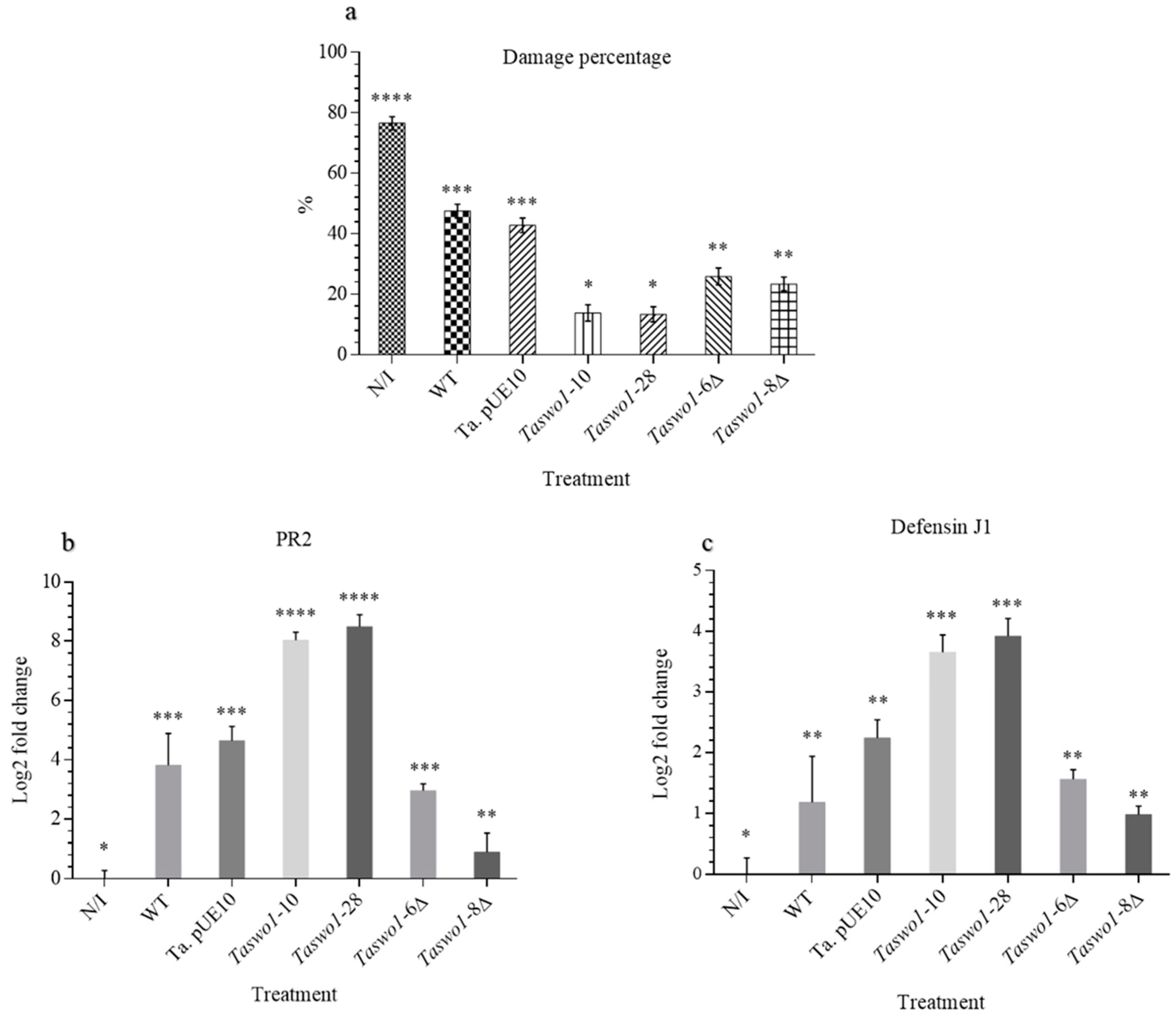
2.3. Overexpressing and Mutant Strains Provide Protection against Fungal Infection to a Higher Extent Than the WT
2.4. Overexpression of Taswo1 Provides Abiotic Stress Protection in C. annuum Plants
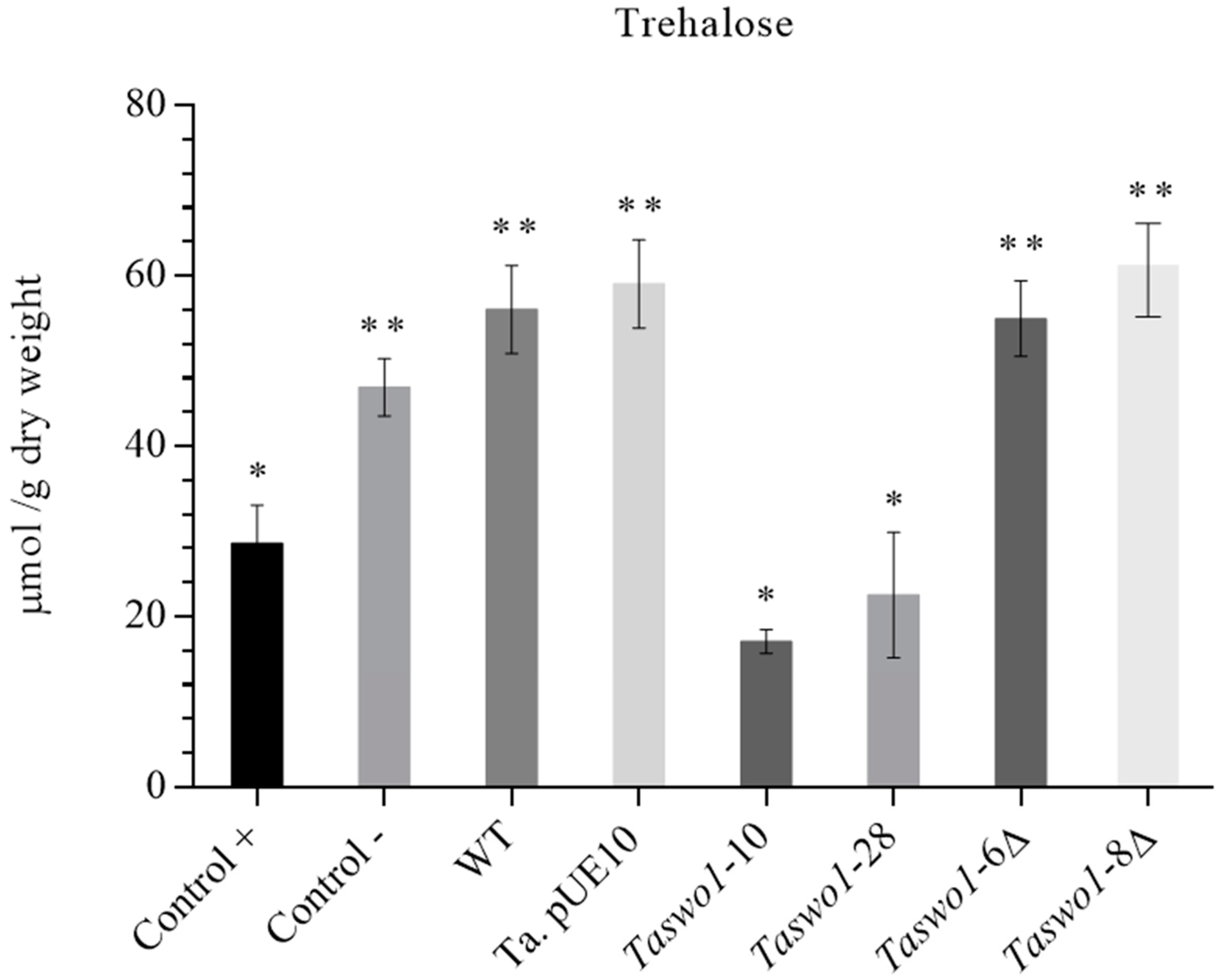
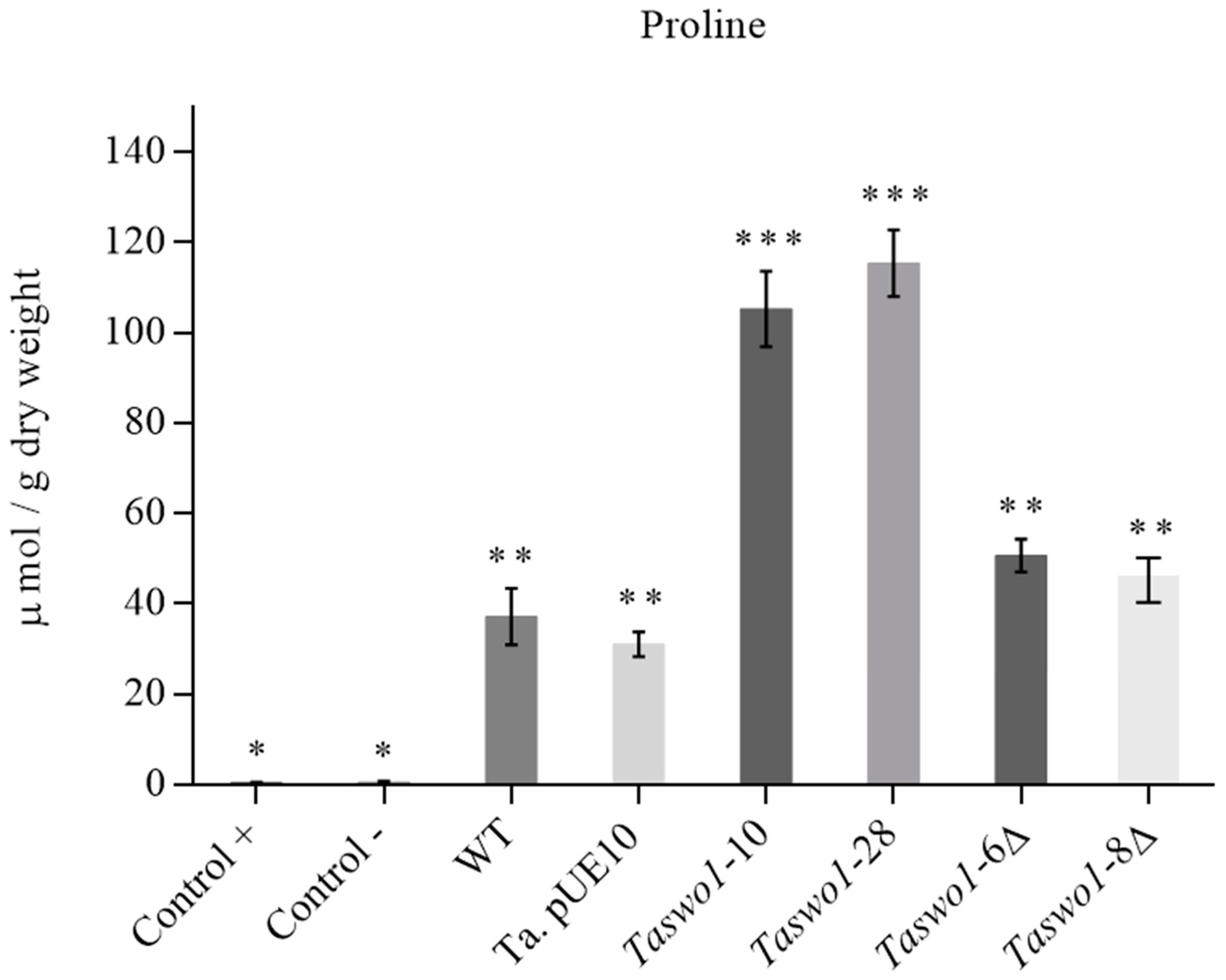
2.5. Osmolyte Accumulation under Drought Conditions in Plants Inoculated with Overexpressing and Mutant Strains
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahanty, T.; Bhattacharjee, S.; Goswami, M.; Bhattacharyya, P.; Das, B.; Ghosh, A.; Tribedi, P. Biofertilizers: A potential approach for sustainable agriculture development. Environ. Sci. Pollut. Res. 2016, 24, 3315–3335. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Kumar, R.; Yadav, S.; Kumari, P.; Singh, R.K.; Kumar, C.R. Effect of bio-control agents on soil borne pathogens: A review. J. Pharmacogn. Phytochem. 2018, 7, 406–411. [Google Scholar]
- Brotman, Y.; Briff, E.; Viterbo, A.; Chet, I. Role of Swollenin, an Expansin-like protein from Trichoderma, in plant root colonization. Plant Physiol. 2008, 147, 779–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma Species—Opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef]
- Brotman, Y.; Landau, U.; Cuadros-Inostroza, A.; Takayuki, T.; Fernie, A.R.; Chet, I.; Viterbo, A.; Willmitzer, L. Trichoderma-plant root colonization: Escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 2013, 9, e1003221. [Google Scholar] [CrossRef]
- Mukherjee, P.K. (Ed.) Trichoderma: Biology and Applications; CABI: Boston, MA, USA, 2013.
- Druzhinina, I.S.; Seidl-Seiboth, V.; Herrera-Estrella, A.; Horwitz, B.A.; Kenerley, C.M.; Monte, E.; Mukherjee, P.K.; Zeilinger, S.; Grigoriev, I.; Kubicek, C.P. Trichoderma: The genomics of opportunistic success. Nat. Rev. Genet. 2011, 9, 749–759. [Google Scholar] [CrossRef] [PubMed]
- López-Bucio, J.; Pelagio-Flores, R.; Herrera-Estrella, A. Trichoderma as biostimulant: Exploiting the multilevel properties of a plant beneficial fungus. Sci. Hortic. 2015, 196, 109–123. [Google Scholar] [CrossRef]
- Valdespino, C.A.R.; Casas-Flores, S.; Olmedo-Monfil, V. Trichoderma as a model to study effector-like molecules. Front. Microbiol. 2019, 10, 1030. [Google Scholar] [CrossRef] [PubMed]
- Romera, F.J.; García, M.J.; Lucena, C.; Martinez-Medina, A.; Aparicio, M.A.; Ramos, J.; Alcántara, E.; Angulo, M.; Pérez-Vicente, R. Induced Systemic Resistance (ISR) and Fe deficiency responses in dicot plants. Front. Plant Sci. 2019, 10, 287. [Google Scholar] [CrossRef]
- Parisi, K.; Shafee, T.; Quimbar, P.; van der Weerden, N.L.; Bleackley, M.; Anderson, M.A. The evolution, function and mechanisms of action for plant defensins. Semin. Cell Dev. Biol. 2019, 88, 107–118. [Google Scholar] [CrossRef]
- Benítez, T.; Rincón, A.M.; Limón, M.C.; Codón, A.C. Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 2004, 7, 249–260. [Google Scholar]
- Altomare, C.; Norvell, W.A.; Björkman, T.; Harman, G.E. Solubilization of phosphates and micronutrients by the plant-growth-promoting and biocontrol fungus Trichoderma harzianum Rifai 1295-22. Appl. Environ. Microbiol. 1999, 65, 2926–2933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Björkman, T.; Blanchard, L.M.; Harman, G.E. Growth enhancement of shrunken-2 (sh2) sweet corn by Trichoderma harzianum 1295-22: Effect of Environmental Stress. J. Am. Soc. Hortic. Sci. 1998, 123, 35–40. [Google Scholar] [CrossRef]
- Marina, M.A.S.; Silva-Flores, M.A.; Uresti-Rivera, E.E.; Castro-Longoria, E.; Herrera-Estrella, A.; Casas-Flores, S. Colonization of Arabidopsis roots by Trichoderma atroviride promotes growth and enhances systemic disease resistance through jasmonic acid/ethylene and salicylic acid pathways. Eur. J. Plant Pathol. 2011, 131, 15–26. [Google Scholar] [CrossRef]
- Ahmad, J.S.; Baker, R. Implications of rhizosphere competence of Trichoderma harzianum. Can. J. Microbiol. 1988, 34, 229–234. [Google Scholar] [CrossRef]
- Strakowska, J.; Blaszczyk, L.; Chelkowski, J. The significance of cellulolytic enzymes produced by Trichoderma in opportunistic lifestyle of this fungus. J. Basic Microbiol. 2014, 54, S2–S13. [Google Scholar] [CrossRef]
- Zaidi, N.W.; Dar, M.H.; Singh, S.; Singh, U.S. Trichoderma species as abiotic stress relievers in plants. In Biotechnology and Biology of Trichoderma; Vijai, G.G., Monika, S., Alfredo, H.-E., Upadhyay, R.S., Irina, D., Maria, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 515–525. [Google Scholar] [CrossRef]
- Hidangmayum, A.; Dwivedi, P. Plant responses to Trichoderma Spp. and their tolerance to abiotic stresses: A review. J. Pharm. Phytochem. 2018, 7, 758–766. [Google Scholar]
- Zhang, F.; Wang, Y.; Liu, C.; Chen, F.; Ge, H.; Tian, F.; Yang, T.; Ma, K.; Zhang, Y. Trichoderma harzianum mitigates salt stress in cucumber via multiple responses. Ecotoxicol. Environ. Saf. 2018, 170, 436–445. [Google Scholar] [CrossRef]
- Pelagio-Flores, R.; Esparza-Reynoso, S.; Garnica-Vergara, A.; López-Bucio, J.; Herrera-Estrella, A. Trichoderma-induced acidification is an early trigger for changes in Arabidopsis root growth and determines fungal phytostimulation. Front. Plant Sci. 2017, 8, 822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mona, S.A.; Hashem, A.; Abd_Allah, E.; Alqarawi, A.; Soliman, D.W.K.; Wirth, S.; Egamberdieva, D. Increased resistance of drought by Trichoderma harzianum fungal treatment correlates with increased secondary metabolites and proline content. J. Integr. Agric. 2017, 16, 1751–1757. [Google Scholar] [CrossRef]
- Seidl, V.; Marchetti, M.; Schandl, R.; Allmaier, G.; Kubicek, C.P.; Marchetti-Deschmann, M. Epl1, the major secreted protein of Hypocrea atroviridis on glucose, is a member of a strongly conserved protein family comprising plant defense response elicitors. FEBS J. 2006, 273, 4346–4359. [Google Scholar] [CrossRef] [PubMed]
- Gaderer, R.; Lamdan, N.L.; Frischmann, A.; Sulyok, M.; Krska, R.; Horwitz, B.A.; Seidl-Seiboth, V. Sm2, a paralog of the Trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of Trichoderma with maize. BMC Microbiol. 2015, 15, 2. [Google Scholar] [CrossRef] [Green Version]
- Guzmán-Guzmán, P.; Alemán-Duarte, M.I.; Delaye, L.; Herrera-Estrella, A.; Olmedo-Monfil, V. Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 2017, 18, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendoza, A.M.; Zaid, R.; Lawry, R.; Hermosa, R.; Monte, E.; Horwitz, B.A.; Mukherjee, P.K. Molecular dialogues between Trichoderma and roots: Role of the fungal secretome. Fungal Biol. Rev. 2018, 32, 62–85. [Google Scholar] [CrossRef]
- Saloheimo, M.; Paloheimo, M.; Hakola, S.; Pere, J.; Swanson, B.; Nyyssönen, E.; Bhatia, A.; Ward, M.; Penttilä, M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant Expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 2002, 269, 4202–4211. [Google Scholar] [CrossRef]
- Chen, X.-A.; Ishida, N.; Todaka, N.; Nakamura, R.; Maruyama, J.-I.; Takahashi, H.; Kitamoto, K. Promotion of efficient saccharification of crystalline cellulose by Aspergillus fumigatus swo1. Appl. Environ. Microbiol. 2010, 76, 2556–2561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, K.; Wang, S.; Lai, G.; Liu, G.; Xing, M. Characterization of a novel swollenin from Penicillium oxalicum in facilitating enzymatic saccharification of cellulose. BMC Biotechnol. 2013, 13, 42. [Google Scholar] [CrossRef] [Green Version]
- Yao, Q.; Sun, T.-T.; Liu, W.-F.; Chen, G.-J. Gene cloning and heterologous expression of a novel endoglucanase, swollenin, from Trichoderma pseudokoningii S38. Biosci. Biotechnol. Biochem. 2008, 72, 2799–2805. [Google Scholar] [CrossRef]
- Reithner, B.; Ibarra-Laclette, E.; Mach, R.L.; Herrera-Estrella, A. Identification of mycoparasitism-related genes in Trichoderma atroviride. Appl. Environ. Microbiol. 2011, 77, 4361–4370. [Google Scholar] [CrossRef] [Green Version]
- Balcázar-López, E.; Méndez-Lorenzo, L.H.; Batista-García, R.A.; Esquivel-Naranjo, U.; Ayala, M.; Kumar, V.V.; Savary, O.; Cabana, H.; Herrera-Estrella, A.; Folch-Mallol, J.L. Xenobiotic compounds degradation by heterologous expression of a Trametes sanguineus laccase in Trichoderma atroviride. PLoS ONE 2016, 11, e0147997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andberg, M.; Penttilä, M.; Saloheimo, M. Swollenin from Trichoderma reesei exhibits hydrolytic activity against cellulosic substrates with features of both endoglucanases and cellobiohydrolases. Bioresour. Technol. 2015, 181, 105–113. [Google Scholar] [CrossRef]
- Cripps-Guazzone, N. Rhizosphere Competence of Selected Trichoderma Species. Ph.D. Thesis, Lincoln University, Lincoln, New Zealand, 2014. [Google Scholar]
- Berg, G.; Zachow, C.; Lottmann, J.; Götz, M.; Costa, R.; Smalla, K. Impact of plant species and site on rhizosphere-associated fungi antagonistic to Verticillium dahliae kleb. Appl. Environ. Microbiol. 2005, 71, 4203–4213. [Google Scholar] [CrossRef] [Green Version]
- Guillén, D.; Sánchez, S.; Rodríguez-Sanoja, R. Carbohydrate-binding domains: Multiplicity of biological roles. Appl. Microbiol. Biotechnol. 2009, 85, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Degenkolb, T.; Nielsen, K.F.; Dieckmann, R.; Branco-Rocha, F.; Chaverri, P.; Samuels, G.J.; Thrane, U.; Von Döhren, H.; Vilcinskas, A.; Brückner, H. Peptaibol, secondary-metabolite, and hydrophobin pattern of commercial biocontrol agents formulated with species of the Trichoderma harzianum complex. Chem. Biodivers. 2015, 12, 662–684. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Llano, Y.; Rodríguez-Pupo, E.C.; Druzhinina, I.S.; Chenthamara, K.; Cai, F.; Gunde-Cimerman, N.; Zalar, P.; Gostinčar, C.; Kostanjšek, R.; Folch-Mallol, J.L.; et al. Stress reshapes the physiological response of halophile fungi to salinity. Cells 2020, 9, 525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viterbo, A.; Chet, I. TasHyd1, a new hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol. Plant Pathol. 2006, 7, 249–258. [Google Scholar] [CrossRef]
- Solheim, B. Possible Role of Lectins in Binding Rhizobia to Host Roots; Bog-Hansen, T.C., Spengler, G.A., Eds.; University of Bern: Bern, Switzerland, 1983; pp. 539–548. [Google Scholar] [CrossRef]
- Inbar, J.; Chet, I. The role of lectins in recognition and adhesion of the mycoparasitic fungus Trichoderma spp. to its host. In Toward Anti-Adhesion Therapy for Microbial Diseases; Advances in Experimental Medicine and Biology; Kahane, I., Ofek, I., Eds.; Springer: Boston, MA, USA, 1996; Volume 408, pp. 229–231. [Google Scholar] [CrossRef]
- Chacón, M.R.; Rofríguez-Galán, O. Microscopic and transcriptome analyses of early colonization of tomato roots by Trichoderma harzianum. Int. Microbiol. 2007, 10, 19–27. [Google Scholar] [CrossRef]
- Yedidia, I.; Benhamou, N.; Chet, I. Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol. 1999, 65, 1061–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoresh, M.; Harman, G.E. Differential expression of maize chitinases in the presence or absence of Trichoderma harzianum strain T22 and indications of a novel exo- endo-heterodimeric chitinase activity. BMC Plant Biol. 2010, 10, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yedidia, I.; Shoresh, M.; Kerem, Z.; Benhamou, N.; Kapulnik, Y.; Chet, I. Concomitant induction of systemic resistance to Pseudomonas syringae Pv. lachrymans in cucumber by Trichoderma asperellum (T-203) and accumulation of phytoalexins. Appl. Environ. Microbiol. 2003, 69, 7343–7353. [Google Scholar] [CrossRef] [Green Version]
- De Meyer, G.; Bigirimana, J.; Elad, Y.; Höfte, M. Induced systemic resistance in Trichoderma harzianum T39 biocontrol of Botrytis cinerea. Eur. J. Plant Pathol. 1998, 104, 279–286. [Google Scholar] [CrossRef]
- Howell, C.R. Mechanisms Employed by Trichoderma species in the biological control of plant diseases: The history and evolution of current concepts. Plant Dis. 2003, 87, 4–10. [Google Scholar] [CrossRef] [Green Version]
- Arias, A.C.R.; Espitia, H.M.Z. Defensinas de plantas y su uso potencial como controladores de plagas en la agricultura. Acta Biol. Colomb. 2010, 15, 33–46. [Google Scholar]
- Georgelis, N.; Nikolaidis, N.; Cosgrove, D.J. Bacterial expansins and related proteins from the world of microbes. Appl. Microbiol. Biotechnol. 2015, 99, 3807–3823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luti, S.; Sella, L.; Quarantin, A.; Pazzagli, L.; Baccelli, I. Twenty years of research on cerato-platanin family proteins: Clues, conclusions, and unsolved issues. Fungal Biol. Rev. 2019, 34, 13–24. [Google Scholar] [CrossRef]
- Ebaccelli, I. Cerato-platanin family proteins: One function for multiple biological roles? Front. Plant Sci. 2015, 5, 769. [Google Scholar] [CrossRef]
- Baccelli, I.; Luti, S.; Bernardi, R.; Scala, A.; Pazzagli, L. Cerato-platanin shows expansin-like activity on cellulosic materials. Appl. Microbiol. Biotechnol. 2013, 98, 175–184. [Google Scholar] [CrossRef]
- Crutcher, F.K.; Diez, M.E.M.; Ding, S.; Liu, J.; Horwitz, B.A.; Mukherjee, P.K.; Kenerley, C.M. A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by Trichoderma virens. Fungal Biol. 2015, 119, 476–486. [Google Scholar] [CrossRef]
- Navazio, L.; Baldan, B.; Moscatiello, R.; Zuppini, A.; Woo, S.L.; Mariani, P.; Lorito, M. Calcium-mediated perception and defense responses activated in plant cells by metabolite mixtures secreted by the biocontrol fungus Trichoderma atroviride. BMC Plant Biol. 2007, 7, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moscatiello, R.; Sello, S.; Ruocco, M.; Barbulova, A.; Cortese, E.; Nigris, S.; Baldan, B.; Chiurazzi, M.; Mariani, P.; Lorito, M.; et al. The Hydrophobin HYTLO1 secreted by the biocontrol fungus Trichoderma longibrachiatum triggers a NAADP-mediated calcium signalling pathway in Lotus japonicus. Int. J. Mol. Sci. 2018, 19, 2596. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Delaney, T.P. Arabidopsis SON1 Is an F-Box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 2002, 14, 1469–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Yao, W.; Wang, L.; Ma, F.; Tong, W.; Wang, C.; Bao, R.; Jiang, C.; Yang, Y.; Zhang, J.; et al. Overexpression of VpEIFP1, a novel F-box/Kelch-repeat protein from wild Chinese Vitis pseudoreticulata, confers higher tolerance to powdery mildew by inducing thioredoxin z proteolysis. Plant Sci. 2017, 263, 142–155. [Google Scholar] [CrossRef]
- Glazebrook, J.; Weigel, D. Arabidopsis: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2002. [Google Scholar]
- Herrera-Estrella, A.; Goldman, G.; Van Montagu, M. Notes high-efficiency transformation system for the biocontrol agents, Trichoderma spp. Mol. Microbiol. 1990, 4, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Viterbo, A.; Harel, M.; Horwitz, B.A.; Chet, I.; Mukherjee, P.K. Trichoderma mitogen-activated protein kinase signaling is involved in induction of plant systemic resistance. Appl. Environ. Microbiol. 2005, 71, 6241–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atriztán-Hernández, K.; Moreno-Pedraza, A.; Winkler, R.; Markow, T.; Herrera-Estrella, A. Trichoderma atroviride from predator to prey: Role of the mitogen-activated protein kinase Tmk3 in fungal chemical defense against fungivory by Drosophila melanogaster larvae. Appl. Environ. Microbiol. 2019, 85, e01825–e01918. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Elad, Y. Production of ethylene by tissues of tomato, pepper, French-bean and cucumber in response to infection by Botrytis cinerea. Physiol. Mol. Plant Pathol. 1990, 36, 277–287. [Google Scholar] [CrossRef]


| Strain | Growth Rate (cm/day) | Std Dev. |
|---|---|---|
| WT | 1.61 | ±0.15 |
| Ta. pUE10 (BMH-0063) | 1.7 | ±0.01 |
| Taswo1-28 (BMH-0064) | 1.6 | ±0.05 |
| Taswo1-10 (BMH-0065) | 1.7 | ±0.1 |
| Taswo1-6 (BMH-0066) | 1.5 | ±0.1 |
| Taswo1-8 (BMH-0067) | 1.6 | ±0.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Cruz, R.; Mehta, R.; Atriztán-Hernández, K.; Martínez-Villamil, O.; del Rayo Sánchez-Carbente, M.; Sánchez-Reyes, A.; Lira-Ruan, V.; González-Chávez, C.A.; Tabche-Barrera, M.L.; Bárcenas-Rodríguez, R.C.; et al. Effects on Capsicum annuum Plants Colonized with Trichoderma atroviride P. Karst Strains Genetically Modified in Taswo1, a Gene Coding for a Protein with Expansin-like Activity. Plants 2021, 10, 1919. https://doi.org/10.3390/plants10091919
Sánchez-Cruz R, Mehta R, Atriztán-Hernández K, Martínez-Villamil O, del Rayo Sánchez-Carbente M, Sánchez-Reyes A, Lira-Ruan V, González-Chávez CA, Tabche-Barrera ML, Bárcenas-Rodríguez RC, et al. Effects on Capsicum annuum Plants Colonized with Trichoderma atroviride P. Karst Strains Genetically Modified in Taswo1, a Gene Coding for a Protein with Expansin-like Activity. Plants. 2021; 10(9):1919. https://doi.org/10.3390/plants10091919
Chicago/Turabian StyleSánchez-Cruz, Ricardo, Richa Mehta, Karina Atriztán-Hernández, Olivia Martínez-Villamil, María del Rayo Sánchez-Carbente, Ayixon Sánchez-Reyes, Verónica Lira-Ruan, Carlos Alberto González-Chávez, María Luisa Tabche-Barrera, Roberto Carlos Bárcenas-Rodríguez, and et al. 2021. "Effects on Capsicum annuum Plants Colonized with Trichoderma atroviride P. Karst Strains Genetically Modified in Taswo1, a Gene Coding for a Protein with Expansin-like Activity" Plants 10, no. 9: 1919. https://doi.org/10.3390/plants10091919
APA StyleSánchez-Cruz, R., Mehta, R., Atriztán-Hernández, K., Martínez-Villamil, O., del Rayo Sánchez-Carbente, M., Sánchez-Reyes, A., Lira-Ruan, V., González-Chávez, C. A., Tabche-Barrera, M. L., Bárcenas-Rodríguez, R. C., Batista-García, R. A., Herrera-Estrella, A., Balcázar-López, E., & Folch-Mallol, J. L. (2021). Effects on Capsicum annuum Plants Colonized with Trichoderma atroviride P. Karst Strains Genetically Modified in Taswo1, a Gene Coding for a Protein with Expansin-like Activity. Plants, 10(9), 1919. https://doi.org/10.3390/plants10091919







