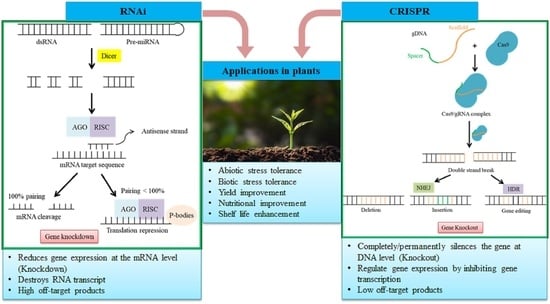
RNA Interference and CRISPR/Cas Gene Editing for Crop Improvement: Paradigm Shift towards Sustainable Agriculture
Abstract
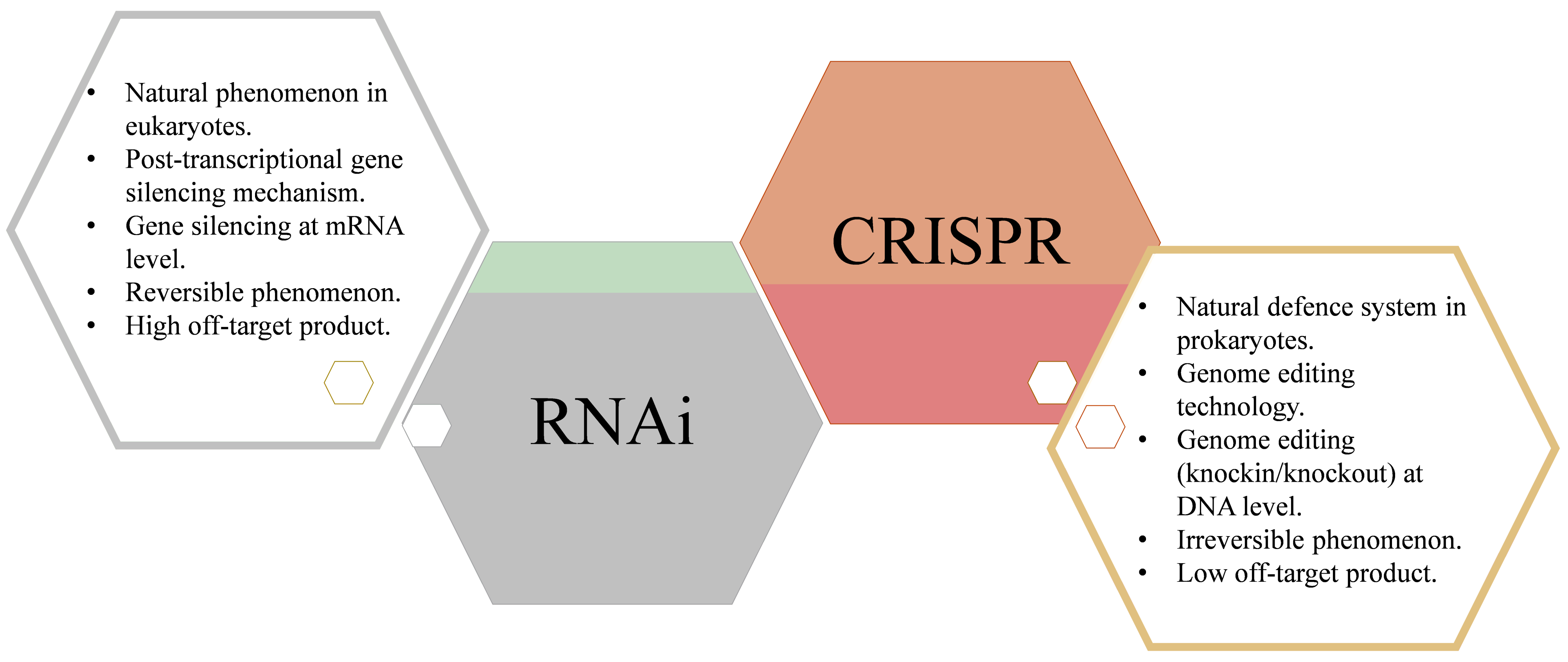
:1. Introduction
2. RNA Interference
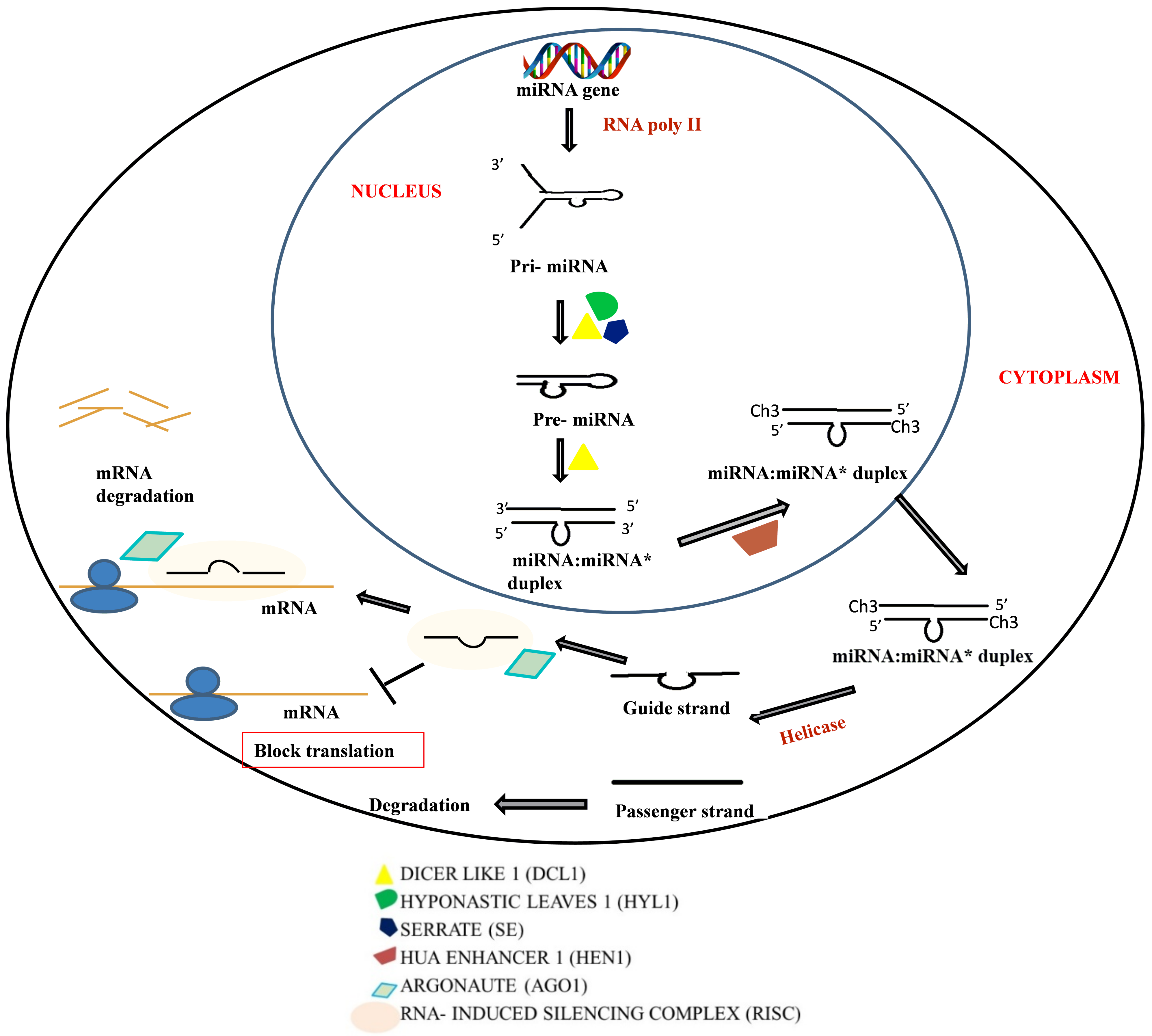
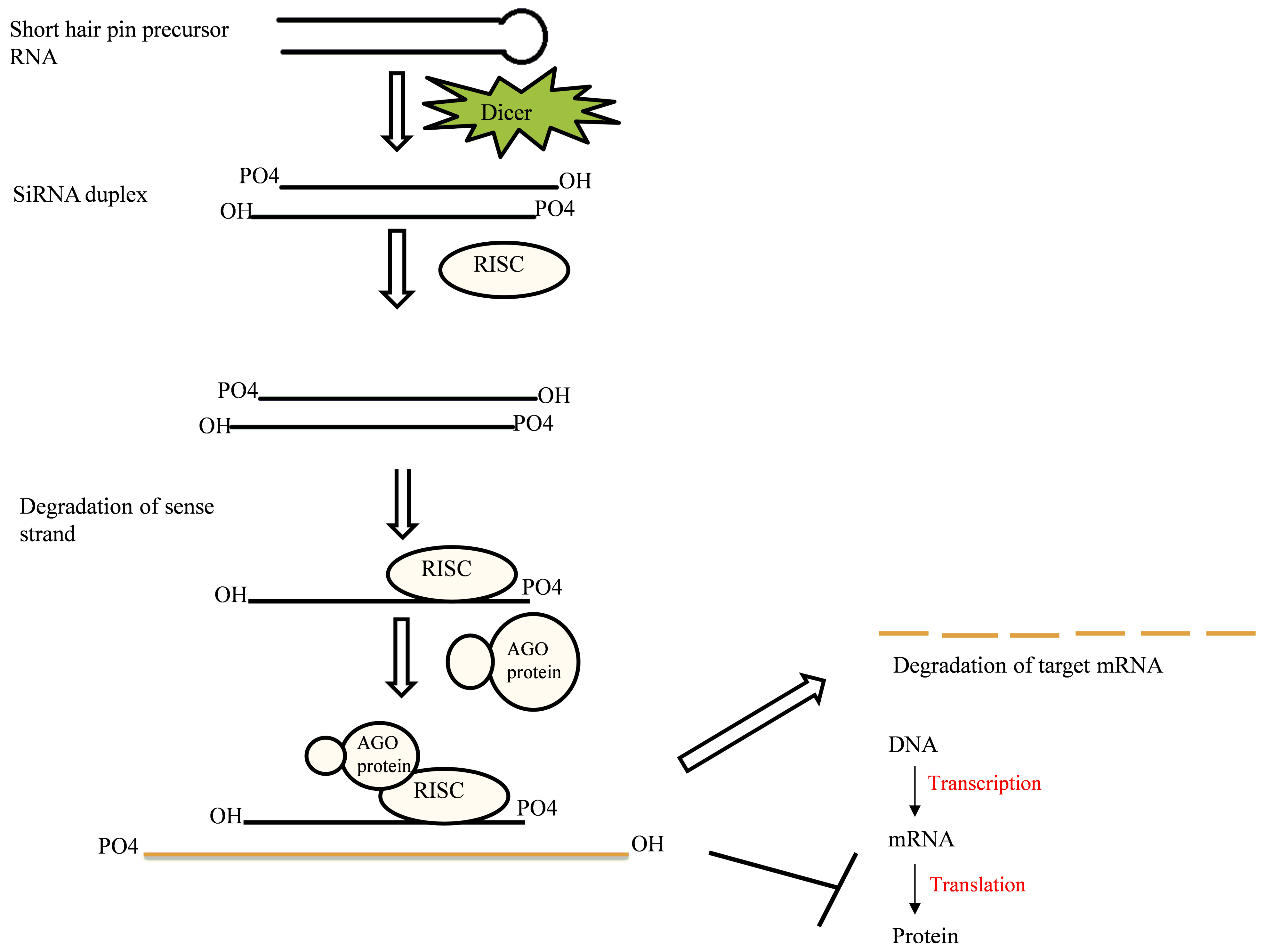
2.1. RNAi Mechanism
2.1.1. Components of RNAi Machinery
2.1.2. Mechanism of Action
2.2. Micro RNA (miRNA)
2.3. Small Interfering RNA (siRNA)
2.4. Role of RNAi in Crop Improvement
2.4.1. Biotic Stress Resistance
RNAi-Mediated Virus Resistance
RNAi-Mediated Bacterial Resistance
RNAi-Mediated Fungal Resistance
RNAi-Mediated Insects and Nematode Resistance
2.4.2. Abiotic Stress Tolerance
2.4.3. Seedless Fruit Development
2.4.4. Shelf-Life Enhancement
2.4.5. Male Sterile Plants Development
2.4.6. Flower Color Modification
2.4.7. Nutritional Improvement
2.4.8. Secondary Metabolite Production
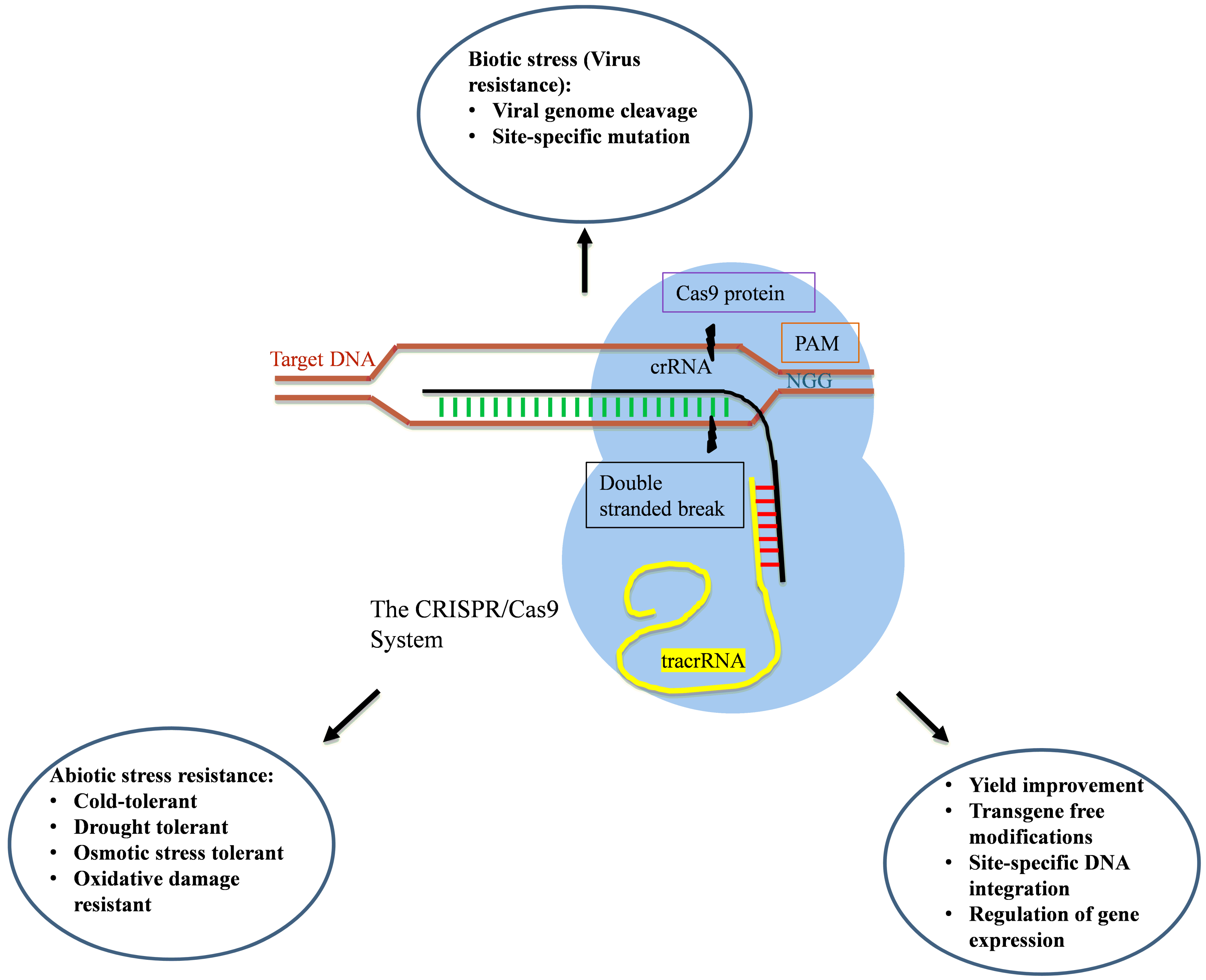
3. Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-Associated Protein (CRISPR/Cas)
3.1. Mechanism of Action
- Type I systems contain signature protein Cas3 that consists of both helicase and DNase domains for the degradation of target [188]. Recently, six subtypes of the type I system (Subtype I-A to I-F) have been identified to contain a variable number of Cas proteins. Aside from Cas proteins, the type I system also encodes for the CRISPR-associated complex for the antiviral defense (Cascade) complex, and Cas3 is also the part of this complex.
- Type II encodes three signature proteins, viz. Cas1, Cas2, and Cas9, and sometimes a fourth protein, i.e., Csn2 and Cas4. Cas9 is a multifunctional protein that plays a crucial role in the Type II system in adaptation to the degradation of the target along with trans-encoded small RNA (tracr RNA) [4,185,189,190]. Three subtypes of the type II system have been discovered, namely type II-A, type II-B, and type II-C [191,192].
- Type III is defined by the presence of Cas10, whose function is still unclear. Two subtypes of the type III system (type III-A and type III-B) have been identified [193].
- Adaption stage: The short pieces of DNA homologous to the genomic sequence of the invading virus or plasmid get incorporated at the leader side of the CRISPR locus. A new spacer unit is created by the duplication of repeats at every integration step. In type I and III CRISPR/Cas systems, the selection of proto-spacers occur by the recognition of PAMs present on or near the location of proto-spacers of the invading genetic element [183,194,195]. After the recognition, Cas1 and Cas2 proteins help in the integration of proto-spacers in between the repeat arrays of CRISPR.
- Expression stage: At this stage, the expression of the spacer takes place via transcription of the CRISPR locus and leads to the generation of a long transcript of pre-CRISPR RNA (pre-crRNA), which is processed into short crRNA by endoribonucleases. In the type I CRISPR/Cas system, pre-crRNA binds with the CRISPR-associated complex for the antiviral defense (Cascade) complex, processed into crRNA by cleavage through Cas6e and Cas6f. The crRNA produced has an 8-nt repeat fragment at the 5′ end and the fragment left forms the hairpin structure on the 3′ end. In the type II CRISPR/Cas system, a repeated fragment of pre-crRNA pairs with the trans-encoded small RNA (tracer RNA), which is further cleaved by RNase III in the presence of Cas9 [189]. Consequently, cleavage at a fixed distance in spacers may lead to maturation. The type III system uses the Cas6 protein for processing to crRNA, but afterward, crRNA is transferred to a different complex of Cas proteins, namely Csm in subtype III-A systems and Cmr in Subtype III-B. Further, cleavage occurs at the 3′ end in subtype III-B subsystems [196].
- Interference stage: After the expression, invading DNA or RNA is targeted and cleaved within proto-spacer sequences. The crRNA acts as a single guide RNA and guides the Cas protein towards the complementary target sequences of the invading genome of the virus or plasmid. In type I systems, the Cascade complex is guided by crRNA towards complementary target DNA, and invading DNA possibly cleaved by Cas3 protein. The Cas9 protein loaded with crRNA cleaves the target DNA in type II systems. The subtype of the type III system, III-A systems, target DNA [194] whereas III-B systems target RNA [196].
3.2. Applications
3.2.1. Yield Improvement
3.2.2. Abiotic Stress Tolerance
3.2.3. Biotic Stress Tolerance
4. Conclusions and Future Prospects
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Pathak, K.; Gogoi, B. RNA interference (RNAi): Application in crop improvement: A review. Agric. Rev. 2016, 37, 245–249. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Pardo, B.; Gómez-González, B.; Aguilera, A. DNA Repair in Mammalian Cells. Cell. Mol. Life Sci. 2009, 66, 1039–1056. [Google Scholar] [CrossRef]
- Gill, R.A.; Scossa, F.; King, G.J.; Golicz, A.; Tong, C.; Snowdon, R.J.; Fernie, A.R.; Liu, S. On the Role of Transposable Elements in the Regulation of Gene Expression and Subgenomic Interactions in Crop Genomes. Crit. Rev. Plant Sci. 2021, 40, 157–189. [Google Scholar] [CrossRef]
- Mamta, B.; Rajam, M.V. RNA Interference: A Promising Approach for Crop Improvement. In Biotechnologies of Crop Improvement; Gosal, S., Wani, S., Eds.; Springer: Cham, Germany, 2018; Volume 2, pp. 41–65. [Google Scholar] [CrossRef]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell 1990, 2, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Johnson, N.; Axtell, M.J. Small RNA warfare: Exploring origins and function of trans-species microRNAs from the parasitic plant Cuscuta. Curr. Opin. Plant Biol. 2019, 50, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.K.; Banakar, P.; Rao, U. The status of RNAi-based transgenic research in plant nematology. Front. Microbiol. 2015, 5, 760. [Google Scholar] [CrossRef] [PubMed]
- Lunardon, A.; Johnson, N.; Hagerott, E.; Phifer, T.; Polydore, S.; Coruh, C.; Axtell, M.J. Integrated annotations and analyses of small RNA–producing loci from 47 diverse plants. Genome Res. 2020, 30, 497–513. [Google Scholar] [CrossRef]
- Lunardon, A.; Kariuki, S.M.; Axtell, M.J. Expression and processing of polycistronic artificial microRNAs and trans -acting siRNAs from transiently introduced transgenes in Solanum lycopersicum and Nicotiana benthamiana. Plant J. 2021, 106, 1087–1104. [Google Scholar] [CrossRef]
- Yang, X.; Sanchez, R.; Kundariya, H.; Maher, T.; Dopp, I.; Schwegel, R.; Virdi, K.; Axtell, M.J.; MacKenzie, S.A. Segregation of an MSH1 RNAi transgene produces heritable non-genetic memory in association with methylome reprogramming. Nat. Commun. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Romano, N.; Macino, G. Quelling: Transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 1992, 6, 3343–3353. [Google Scholar] [CrossRef]
- Wilson, R.C.; Doudna, J.A. Molecular Mechanisms of RNA Interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef] [Green Version]
- Cerutti, H.; Ma, X.; Msanne, J.; Repas, T. RNA-mediated silencing in algae: Biological roles and tools for analysis of gene function. Eukaryot. Cell 2011, 10, 1164–1172. [Google Scholar] [CrossRef] [Green Version]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nat. Cell Biol. 2000, 404, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nat. Cell Biol. 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Datta, S.K.; Datta, K. RNA interference in designing transgenic crops. GM Crop 2010, 1, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005, 579, 5822–5829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutvagner, G.; Simard, M. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redfern, A.D.; Colley, S.M.; Beveridge, D.J.; Ikeda, N.; Epis, M.R.; Li, X.; Foulds, C.E.; Stuart, L.M.; Barker, A.; Russell, V.J.; et al. RNA-induced silencing complex (RISC) Proteins PACT, TRBP, and Dicer are SRA binding nuclear receptor coregulators. Proc. Natl. Acad. Sci. USA 2013, 110, 6536–6541. [Google Scholar] [CrossRef] [Green Version]
- Dash, S.K.; Sushil, K.M.; N, M.H. RNA Interference a fine tuner of gene regulation: A review. Int. J. Biotechnol. Mol. Biol. Res. 2015, 6, 35–39. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; A Kolb, F.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single Processing Center Models for Human Dicer and Bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aalto, A.P.; E Pasquinelli, A. Small non-coding RNAs mount a silent revolution in gene expression. Curr. Opin. Cell Biol. 2012, 24, 333–340. [Google Scholar] [CrossRef] [Green Version]
- Kurihara, Y.; Takashi, Y.; Watanabe, Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 2006, 12, 206–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurihara, Y.; Watanabe, Y. From The Cover: Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Ji, L.; Huang, Q.; Vassylyev, D.G.; Chen, X.; Ma, J.-B. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nat. Cell Biol. 2009, 461, 823–827. [Google Scholar] [CrossRef]
- Li, J.; Yang, Z.; Yu, B.; Liu, J.; Chen, X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005, 15, 1501–1507. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Liu, Q.; ASmith, N.; Liang, G.; Wang, M.B. RNA silencing in plants: Mechanisms, technologies and applications in horticultural crops. Curr. Genom. 2016, 17, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Pareek, M.; Yogindran, S.; Mukherjee, S.K.; Rajam, M.V. Plant MicroRNAs: Biogenesis, Functions, and Applications. In Plant Biology and Biotechnology; Springer: New Delhi, India, 2015; pp. 639–661. [Google Scholar] [CrossRef]
- Mello, C.C.; Conte, D. Revealing the world of RNA interference. Nat. Cell Biol. 2004, 431, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nat. Cell Biol. 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef] [Green Version]
- Saurabh, S.; Vidyarthi, A.S.; Prasad, D. RNA interference: Concept to reality in crop improvement. Planta 2014, 239, 543–564. [Google Scholar] [CrossRef] [Green Version]
- Aoki, K.; Moriguchi, H.; Yoshioka, T.; Okawa, K.; Tabara, H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007, 26, 5007–5019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halic, M.; Moazed, D. Dicer-Independent Primal RNAs Trigger RNAi and Heterochromatin Formation. Cell 2010, 140, 504–516. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-C.; Li, L.; Gu, W.; Xue, Z.; Crosthwaite, S.K.; Pertsemlidis, A.; Lewis, Z.; Freitag, M.; Selker, E.U.; Mello, C.C.; et al. Diverse Pathways Generate MicroRNA-like RNAs and Dicer-Independent Small Interfering RNAs in Fungi. Mol. Cell 2010, 38, 803–814. [Google Scholar] [CrossRef] [Green Version]
- Ye, R.; Chen, Z.; Lian, B.; Rowley, J.; Xia, N.; Chai, J.; Li, Y.; He, X.-J.; Wierzbicki, A.T.; Qi, Y. A Dicer-Independent Route for Biogenesis of siRNAs that Direct DNA Methylation in Arabidopsis. Mol. Cell 2016, 61, 222–235. [Google Scholar] [CrossRef] [Green Version]
- Tester, M.; Langridge, P. Breeding Technologies to Increase Crop Production in a Changing World. Science 2010, 327, 818–822. [Google Scholar] [CrossRef]
- Sharma, V.K.; Basu, S.; Chakraborty, S. RNAi mediated broad-spectrum transgenic resistance in Nicotiana benthamiana to chilli-infecting begomoviruses. Plant Cell Rep. 2015, 34, 1389–1399. [Google Scholar] [CrossRef]
- Rupp, J.; Cruz, L.F.; Trick, H.; Fellers, J.P. RNAi-Mediated, Stable Resistance to Triticum mosaic virus in Wheat. Crop Sci. 2016, 56, 1602–1610. [Google Scholar] [CrossRef]
- Ahmed, M.M.S.; Bian, S.; Wang, M.; Zhao, J.; Zhang, B.; Liu, Q.; Zhang, C.; Tang, S.; Gu, M.; Yu, H. RNAi-mediated resistance to rice black-streaked dwarf virus in transgenic rice. Transgenic Res. 2017, 26, 197–207. [Google Scholar] [CrossRef]
- Hameed, A.; Tahir, M.N.; Asad, S.; Bilal, R.; Van Eck, J.; Jander, G.; Mansoor, S. RNAi-Mediated Simultaneous Resistance Against Three RNA Viruses in Potato. Mol. Biotechnol. 2017, 59, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Niu, L.; Zhang, W.; Yang, J.; Xing, G.; He, H.; Guo, D.; Du, Q.; Qian, X.; Yao, Y.; et al. RNAi-mediated SMV P3 cistron silencing confers significantly enhanced resistance to multiple Potyvirus strains and isolates in transgenic soybean. Plant Cell Rep. 2018, 37, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Hada, A.; Subramanyam, K.; Theboral, J.; Misra, S.; Ganapathi, A.; Malathi, V.G. RNAi-mediated resistance to yellow mosaic viruses in soybean targeting coat protein gene. Acta Physiol. Plant 2018, 40, 1–12. [Google Scholar] [CrossRef]
- Senthilraja, C.; Reddy, M.G.; Rajeswaran, J.; Kokiladevi, E.; Velazhahan, R. RNA interference-mediated resistance to Tobacco streak virus in transgenic peanut. Australas. Plant Pathol. 2018, 47, 227–230. [Google Scholar] [CrossRef]
- Malathi, P.; Muzammil, S.A.; Krishnaveni, D.; Balachandran, S.; Mangrauthia, S.K. Coat protein 3 of Rice tungro spherical virus is the key target gene for development of RNAi mediated tungro disease resistance in rice. Agri Gene 2019, 12, 100084. [Google Scholar] [CrossRef]
- Gao, L.; Luo, J.; Ding, X.; Wang, T.; Hu, T.; Song, P.; Zhai, R.; Zhang, H.; Zhang, K.; Li, K.; et al. Soybean RNA interference lines silenced for eIF4E show broad potyvirus resistance. Mol. Plant Pathol. 2019, 21, 303–317. [Google Scholar] [CrossRef]
- Tzean, Y.; Chang, H.-H.; Tu, T.-C.; Hou, B.H.; Chen, H.-M.; Chiu, Y.-S.; Chou, W.-Y.; Chang, L.; Yeh, H.-H. Engineering Plant Resistance to Tomato Yellow Leaf Curl Thailand Virus Using a Phloem-Specific Promoter Expressing Hairpin RNA. Mol. Plant-Microbe Interact. 2020, 33, 87–97. [Google Scholar] [CrossRef]
- Escobar, M.A.; Civerolo, E.L.; Summerfelt, K.R.; Dandekar, A.M. RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc. Natl. Acad. Sci. USA 2001, 98, 13437–13442. [Google Scholar] [CrossRef] [Green Version]
- Katiyar-Agarwal, S.; Morgan, R.; Dahlbeck, D.; Borsani, O.; Villegas, A.; Zhu, J.-K.; Staskawicz, B.J.; Jin, H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 2006, 103, 18002–18007. [Google Scholar] [CrossRef] [Green Version]
- Enrique, R.; Siciliano, F.; Favaro, M.A.; Gerhardt, N.; Roeschlin, R.; Rigano, L.; Castagnaro, A.; Vojnov, A.; Sendin, L.; Marano, M.R. Novel demonstration of RNAi in citrus reveals importance of citrus callose synthase in defence against Xanthomonas citri subsp. citri. Plant Biotechnol. J. 2011, 9, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Sanju, S.; Siddappa, S.; Thakur, A.; Shukla, P.K.; Srivastava, N.; Pattanayak, D.; Sharma, S.K.; Singh, B.P. Host-mediated gene silencing of a single effector gene from the potato pathogen Phytophthora infestans imparts partial resistance to late blight disease. Funct. Integr. Genom. 2015, 15, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Song, X.-S.; Xiao-Li, Q.; Cao, L.-H.; Sun, K.; Qiu, X.-L.; Xu, Y.-B.; Yang, P.; Huang, T.; Zhang, J.-B.; et al. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 2015, 13, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
- Ghag, S.B.; Shekhawat, U.K.S.; Ganapathi, T.R. Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 2014, 12, 541–553. [Google Scholar] [CrossRef]
- Andrade, C.M.; Tinoco, M.L.P.; Rieth, A.F.; Maia, F.C.O.; Aragão, F.J.L. Host-induced gene silencing in the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Pathol. 2016, 65, 626–632. [Google Scholar] [CrossRef] [Green Version]
- Bharti, P.; Jyoti, P.; Kapoor, P.; Sharma, V.; Shanmugam, V.; Yadav, S.K. Host-induced silencing of pathogenicity genes enhances resistance to Fusarium oxysporum wilt in tomato. Mol. Biotechnol. 2017, 59, 343–352. [Google Scholar] [CrossRef]
- Zhu, L.; Zhu, J.; Liu, Z.; Wang, Z.; Zhou, C.; Wang, H. Host-induced gene silencing of rice blast fungus Magnaporthe oryzae pathogenicity genes mediated by the brome mosaic virus. Genes 2017, 8, 241. [Google Scholar] [CrossRef]
- Tiwari, I.M.; Jesuraj, A.; Kamboj, R.; Devanna, B.N.; Botella, J.; Sharma, T.R. Host Delivered RNAi, an efficient approach to increase rice resistance to sheath blight pathogen (Rhizoctonia solani). Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Majumdar, R.; Rajasekaran, K.; Sickler, C.; Lebar, M.; Musungu, B.M.; Fakhoury, A.M.; Payne, G.A.; Geisler, M.; Carter-Wientjes, C.; Wei, Q.; et al. The pathogenesis-related maize seed (PRms ) gene plays a role in resistance to Aspergillus flavus infection and aflatoxin contamination. Front. Plant Sci. 2017, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Pareek, M.; Rajam, M.V. RNAi-mediated silencing of MAP kinase signalling genes (Fmk1, Hog1, and Pbs2) in Fusarium oxysporum reduces pathogenesis on tomato plants. Fungal Biol. 2017, 121, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.K.; Majumdar, R.; Rajasekaran, K.; Chen, Z.-Y.; Wei, Q.; Sickler, C.M.; Lebar, M.D.; Cary, J.W.; Frame, B.R.; Wang, K. RNA interference based silencing of the alpha-amylase (amy1) gene in Aspergillus flavus decreases fungal growth and aflatoxin production in maize kernels. Planta 2018, 247, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, M.; Adachi, A.; Sato, Y.; Doke, N.; Kondo, T.; Yoshioka, H. RNAi of the sesquiterpene cyclase gene for phytoalexin production impairs pre- and post-invasive resistance to potato blight pathogens. Mol. Plant Pathol. 2019, 20, 907–922. [Google Scholar] [CrossRef]
- Wang, L.; Wang, H.; He, S.; Meng, F.; Zhang, C.; Fan, S.; Wu, J.; Zhang, S.; Xu, P. GmSnRK1. 1, a sucrose nonfermenting-1 (SNF1)-Related Protein Kinase, Promotes Soybean Resistance to Phytophthora sojae. Front. Plant Sci. 2019, 10, 996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.; Mukherjee, S.K.; Rajam, M.V. Silencing of the ornithine decarboxylase gene of Fusarium oxysporum f. sp. lycopersici by host-induced RNAi confers resistance to Fusarium wilt in tomato. Plant Mol. Biol. Rep. 2020, 38, 419–429. [Google Scholar] [CrossRef]
- Mamta; Reddy, K.R.K.; Rajam, M.V. Targeting chitinase gene of Helicoverpa armigera by host-induced RNA interference confers insect resistance in tobacco and tomato. Plant Mol. Biol. 2016, 90, 281–292. [Google Scholar] [CrossRef]
- Malik, H.J.; Raza, A.; Amin, I.; Scheffler, J.A.; Scheffler, B.E.; Brown, J.K.; Mansoor, S. RNAi-mediated mortality of the whitefly through transgenic expression of double- stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, A.B.; Monteiro, T.R.; Cabral, G.; Aragão, F.J.L. RNAi-mediated resistance to whitefly (Bemisia tabaci) in genetically engineered lettuce (Lactuca sativa). Transgenic Res. 2017, 26, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.; Bhattacharya, R. Host-mediated RNA interference targeting a cuticular protein gene impaired fecundity in the green peach aphid Myzus persicae. Pest Manag. Sci. 2018, 74, 2059–2068. [Google Scholar] [CrossRef]
- Shin, Y.H.; Lee, S.H.; Park, Y.-D. Development of mite (Tetranychus urticae)-resistant transgenic Chinese cabbage using plant-mediated RNA interference. Hortic. Environ. Biotechnol. 2020, 61, 305–315. [Google Scholar] [CrossRef]
- Dutta, T.K.; Papolu, P.K.; Banakar, P.; Choudhary, D.; Sirohi, A.; Rao, U. Tomato transgenic plants expressing hairpin construct of a nematode protease gene conferred enhanced resistance to root-knot nematodes. Front. Microbiol. 2015, 6, 260. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, K.; Lu, Q.; Du, J.; Wang, Z.; Wang, D.; Sun, B.; Li, H. Transgenic Nicotiana benthamiana plants expressing a hairpin RNAi construct of a nematode Rs-cps gene exhibit enhanced resistance to Radopholus similis. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Chukwurah, P.N.; Poku, S.A.; Yokoyama, A.; Fukuda, H.; Shishido, M.; Nakamura, I. Expression of Meloidogyne incognita PolA1 hairpin RNA reduced nematode multiplication in transgenic tomato. Plant Biotechnol. Rep. 2019, 13, 591–601. [Google Scholar] [CrossRef]
- Hu, Y.; You, J.; Li, C.; Pan, F.; Wang, C. The Heterodera glycines effector Hg16B09 is required for nematode parasitism and suppresses plant defense response. Plant Sci. 2019, 289, 110271. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Li, J.; Vodkin, L.O.; Todd, T.C.; Finer, J.J.; Trick, H.N. Host-derived gene silencing of parasite fitness genes improves resistance to soybean cyst nematodes in stable transgenic soybean. Theor. Appl. Genet. 2019, 132, 2651–2662. [Google Scholar] [CrossRef] [Green Version]
- Joshi, I.; Kumar, A.; Kohli, D.; Singh, A.K.; Sirohi, A.; Subramaniam, K.; Chaudhury, A.; Jain, P.K. Conferring root-knot nematode resistance via host-delivered RNAi-mediated silencing of four Mi-msp genes in Arabidopsis. Plant Sci. 2020, 298, 110592. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, P.; Xia, Y.; Wei, P.; Li, W.; Zhang, W.; Chen, X.; Cao, P.; Xu, Y.; Jin, L.; et al. Downregulation of the lycopene e-cyclase gene confers tolerance to salt and drought stress in Nicotiana tabacum. Acta Physiol. Plant 2015, 37, 1–15. [Google Scholar] [CrossRef]
- Cui, P.; Liu, H.; Islam, F.; Li, L.; Farooq, M.A.; Ruan, S.; Zhou, W. OsPEX11, a peroxisomal Biogenesis Factor 11, contributes to salt stress tolerance in Oryza sativa. Front. Plant Sci. 2016, 7, 1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.A.; Jung, H.-E.; Hong, J.K.; Hermand, V.; McClung, C.R.; Lee, Y.-H.; Kim, J.Y.; Lee, S.I.; Jeong, M.-J.; Kim, J.; et al. Reduction of GIGANTEA expression in transgenic Brassica rapa enhances salt tolerance. Plant Cell Rep. 2016, 35, 1943–1954. [Google Scholar] [CrossRef] [PubMed]
- Szechyńska-Hebda, M.; Czarnocka, W.; Hebda, M.; Karpiński, S. PAD4, LSD1 and EDS1 regulate drought tolerance, plant biomass production, and cell wall properties. Plant Cell Rep. 2016, 35, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Ninghui, C.; Wu, Q.; Peng, Z.; Sprague, S.A.; Wang, W.; Park, J.; Akhunov, E.; Jagadish, K.S.V.; Nakata, P.; Cheng, N.; et al. Silencing of OsGRXS17 in rice improves drought stress tolerance by modulating ROS accumulation and stomatal closure. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Huang, L.; Wang, M.; Cui, Y.; Xia, X. OsDSR-1, a calmodulin-like gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Mol. Breed. 2017, 37, 75. [Google Scholar] [CrossRef]
- Jin, Y.; Pan, W.; Zheng, X.; Cheng, X.; Liu, M.; Ma, H.; Ge, X. OsERF101, an ERF family transcription factor, regulates drought stress response in reproductive tissues. Plant Mol. Biol. 2018, 98, 51–65. [Google Scholar] [CrossRef]
- Zhu, M.; Meng, X.; Cai, J.; Li, G.; Dong, T.; Li, Z. Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol. 2018, 18, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-S.; Yu, J.-G.; Lee, G.-H.; Park, Y.-D. Drought tolerance induction in transgenic tobacco through RNA interference of BrDST71, a drought responsive gene from Chinese cabbage. Hortic. Environ. Biotechnol. 2018, 59, 749–757. [Google Scholar] [CrossRef]
- Wang, W.; Wang, W.; Wu, Y.; Li, Q.; Zhang, G.; Shi, R.; Yang, J.; Wang, Y.; Wang, W. The involvement of wheat U box E3 ubiquitin ligase TaPUB1 in salt stress tolerance. J. Integr. Plant Biol. 2019, 62, 631–651. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wang, X.; Mu, X.; Zhang, L. RNAi mediated silencing of dehydrin gene WZY2 confers osmotic stress intolerance in transgenic wheat. Funct. Plant Biol. 2019, 46, 877–884. [Google Scholar] [CrossRef]
- Oerke, E.C.; Dehne, H.W.; Schonbeck, F.; Weber, A. Crop Production and Crop Protection: Estimated Losses in Major Food and Cash Crops; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Waterhouse, P.M.; Graham, M.W.; Wang, M.-B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 1998, 95, 13959–13964. [Google Scholar] [CrossRef] [Green Version]
- Missiou, A.; Kalantidis, K.; Boutla, A.; Tzortzakaki, S.; Tabler, M.; Tsagris, M. Generation of transgenic potato plants highly resistant to potato virus Y (PVY) through RNA silencing. Mol. Breed. 2004, 14, 185–197. [Google Scholar] [CrossRef] [Green Version]
- Andika, I.B.; Kondo, H.; Tamada, T. Evidence that RNA silencing-mediated resistance to beet necrotic yellow vein virus is less effective in roots than in leaves. Mol. Plant-Microbe Interact. 2005, 18, 194–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hily, J.-M.; Ravelonandro, M.; Damsteegt, V.; Bassett, C.; Petri, C.; Liu, Z.; Scorza, R. Plum pox virus coat protein gene Intron-hairpin-RNA (ihpRNA) constructs provide resistance to plum pox virus in Nicotiana benthamiana and Prunus domestica. J. Am. Soc. Hortic. Sci. 2007, 132, 850–858. [Google Scholar] [CrossRef] [Green Version]
- Bonfim, K.; Faria, J.C.; Nogueira, E.O.P.L.; Mendes, É.A.; Aragão, F.J.L. RNAi-mediated resistance to Bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Mol. Plant-Microbe Interact. 2007, 20, 717–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanitharani, R.; Chellappan, P.; Fauquet, C.M. Short interfering RNA-mediated interference of gene expression and viral DNA accumulation in cultured plant cells. Proc. Natl. Acad. Sci. USA 2003, 100, 9632–9636. [Google Scholar] [CrossRef] [Green Version]
- Vanderschuren, H.; Alder, A.; Zhang, P.; Gruissen, W. Dose-dependent RNAi-mediated germinivirus resistance in the tropical root crop cassava. Plant Mol. Biol. 2009, 70, 265–272. [Google Scholar] [CrossRef] [Green Version]
- Patil, B.; Ogwok, E.; Wagaba, H.; Mohammed, I.U.; Yadav, J.S.; Bagewadi, B.; Taylor, N.J.; Kreuze, J.; Maruthi, M.N.; Alicai, T.; et al. RNAi mediated resistance to diverse isolates belonging to two virus species involved in Cassava brown streak disease. Mol. Plant Pathol. 2010, 12, 31–41. [Google Scholar] [CrossRef]
- Pradeep, K.; Satya, V.K.; Selvapriya, M.; Vijayasamundeeswari, A.; Ladhalakshmi, D.; Paranidharan, V.; Rabindran, R.; Samiyappan, R.; Balasubramanian, P.; Velazhahan, R. Engineering resistance against tobacco streak virus (TSV) in sunflower and tobacco using RNA interference. Biol. Plant 2012, 56, 735–741. [Google Scholar] [CrossRef]
- Zhou, Y.; Yuan, Y.; Yuan, F.; Wang, M.; Zhong, H.; Gu, M.; Liang, G. RNAi-directed down-regulation of RSV results in increased resistance in rice (Oryza sativa L.). Biotechnol. Lett. 2012, 34, 965–972. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, M.-J.; Pak, J.H.; Im, H.H.; Lee, D.H.; Kim, K.-H.; Lee, J.-H.; Kim, D.-H.; Choi, H.K.; Jung, H.W.; et al. RNAi-mediated Soybean mosaic virus (SMV) resistance of a Korean Soybean cultivar. Plant Biotechnol. Rep. 2016, 10, 257–267. [Google Scholar] [CrossRef] [Green Version]
- Pooggin, M.M.; Shivaprasad, P.V.; Veluthambi, K.; Hohn, T. RNAi targeting of DNA virus in plants. Nat. Biotechnol. 2003, 21, 131–132. [Google Scholar] [CrossRef]
- Worrall, E.; Bravo-Cazar, A.; Nilon, A.T.; Fletcher, S.J.; Robinson, K.E.; Carr, J.; Mitter, N. Exogenous application of RNAi-inducing double-stranded RNA inhibits aphid-mediated transmission of a plant virus. Front. Plant Sci. 2019, 10, 265. [Google Scholar] [CrossRef] [Green Version]
- Hamada, W.; Spanu, P.D. Co-suppression of the hydrophobin gene HCf-1 is correlated with antisense RNA biosynthesis in Cladosporium fulvum. Mol. Genet. Genom. 1998, 259, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, A.; van Kan, J.; Plummer, K.M. Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet. Biol. 2004, 41, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Goldoni, M.; Azzalin, G.; Macino, G.; Cogoni, C. Efficient gene silencing by expression of double stranded RNA in Neurospora crassa. Fungal Genet. Biol. 2004, 41, 1016–1024. [Google Scholar] [CrossRef]
- Kadotani, N.; Nakayashiki, H.; Tosa, Y.; Mayama, S. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2003, 16, 769–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández, I.; Chacón, O.; Rodriguez, R.; Portieles, R.; López, Y.; Pujol, M.; Borrás-Hidalgo, O. Black shank resistant tobacco by silencing of glutathione S-transferase. Biochem. Biophys. Res. Commun. 2009, 387, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, Q.; Huang, M.; Liu, Y.; Liu, Z.; Liu, X.; Ma, Z. Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Sci. Rep. 2015, 5, srep12500. [Google Scholar] [CrossRef] [Green Version]
- Yogindran, S.; Rajam, M.V. RNAi for Crop Improvement. In Plant Biology and Biotechnology; Bahadur, B., Rajam, M.V., Sahijram, L., Krishnamurthy, K., Eds.; Springer: New Delhi, India, 2015; pp. 623–637. [Google Scholar] [CrossRef]
- Gheysen, G.; Vanholme, B. RNAi from plants to nematodes. Trends Biotechnol. 2007, 25, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA 2006, 103, 14302–14306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sindhu, A.S.; Maier, T.R.; Mitchum, M.G.; Hussey, R.S.; Davis, E.L.; Baum, T.J. Effective and specific in planta RNAi in cyst nematodes: Expression interference of four parasitism genes reduces parasitic success. J. Exp. Bot. 2008, 60, 315–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Kakrana, A.; Sirohi, A.; Subramaniam, K.; Srinivasan, R.; Abdin, M.Z.; Jain, P.K. Host-delivered RNAi-mediated root-knot nematode resistance in Arabidopsis by targeting splicing factor and integrase genes. J. Gen. Plant Pathol. 2017, 83, 91–97. [Google Scholar] [CrossRef]
- Tsygankova, V.A.; Andrusevich, Y.V.; Shysha, E.N.; Biliavska, L.O.; Galagan, T.O.; Galkin, A.P.; Yemets, A.; Iutynska, G.A.; Blume, Y.B. RNAi-mediated resistance against plant parasitic nematodes of wheat plants obtained in vitro using bioregulators of microbiological origin. Curr. Chem. Biol. 2019, 13, 73–89. [Google Scholar] [CrossRef]
- Mao, Y.-B.; Cai, W.-J.; Wang, J.-W.; Hong, G.-J.; Tao, X.; Wang, L.-J.; Huang, Y.-P.; Chen, X. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef]
- A Baum, J.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Thakur, N.; Upadhyay, S.K.; Verma, P.C.; Chandrashekar, K.; Tuli, R.; Singh, P.K. Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase A gene. PLoS ONE 2014, 9, e87235. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Dong, Y.; Desneux, N.; Niu, C. RNAi silencing of the HaHMG-CoA reductase gene inhibits oviposition in the Helicoverpa armigera cotton bollworm. PLoS ONE 2013, 8, e67732. [Google Scholar] [CrossRef] [PubMed]
- Pardo, J.M. Biotechnology of water and salinity stress tolerance. Curr. Opin. Biotechnol. 2010, 21, 185–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.-H.; Liu, H.; Yang, Y.-L.; Zhen, P.-P.; Liang, J.-S. Down-regulated expression of RACK1 gene by RNA interference enhances drought tolerance in rice. Rice Sci. 2009, 16, 14–20. [Google Scholar] [CrossRef]
- Manavalan, L.P.; Chen, X.; Clarke, J.; Salmeron, J.; Nguyen, H.T. RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice. J. Exp. Bot. 2012, 63, 163–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Liu, H.; Liwen, W.; Chen, H. Identification and characterization of maize microRNAs involved in the very early stage of seed germination. BMC Genom. 2011, 12, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Chen, L.; Zhao, M.; Tian, Q.; Zhang, W.-H. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genom. 2011, 12, 367. [Google Scholar] [CrossRef] [Green Version]
- Jan, A.; Maruyama, K.; Todaka, D.; Kidokoro, S.; Abo, M.; Yoshimura, E.; Shinozaki, K.; Nakashima, K.; Yamaguchi-Shinozaki, K. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013, 161, 1202–1216. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Wang, L.; Zhao, R.; Sheng, J.; Zhang, S.; Li, R.; Shen, L. Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol. 2019, 19, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alam, I.; Salimullah, M. Genetic engineering of eggplant (Solanum melongena L.): Progress, controversy and potential. Hoticulture 2021, 7, 78. [Google Scholar] [CrossRef]
- Pandolfini, T. Seedless fruit production by hormonal regulation of fruit set. Nutrients 2009, 1, 168–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varoquaux, F.; Blanvillain, R.; Delseny, M.; Gallois, P. Less is better: New approaches for seedless fruit production. Trends Biotechnol. 2000, 18, 233–242. [Google Scholar] [CrossRef]
- Denna, D.W. Effects of genetic parthenocarpy and gynoecious flowering habit on fruit production and growth of cucumber Cucumis sativus L. J. Am. Soc. Hortic. Sci. 1973, 98, 602–604. [Google Scholar]
- Tiedjens, V.A. Sex ratios in cucumber flowers as affected by different conditions of soil and light. J. Agric. Res. 1928, 36, 721–746. [Google Scholar]
- Falavigna, A.; Soressi, G.P. Influence of the pat-sha gene on plant and fruit traits in tomato (L. esculentum Mill.). In Proceedings of the 10th Meeting of the EUCARPIA Tomato Working Group, Salerno, Italy, 2–6 September 1987; p. 128. [Google Scholar]
- De Jong, M.; Wolters-Arts, M.; Feron, R.; Mariani, C.; Vriezen, W.H. The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 2009, 57, 160–170. [Google Scholar] [CrossRef]
- Schijlen, E.G.; de Vos, C.R.; Martens, S.; Jonker, H.H.; Rosin, F.M.; Molthoff, J.W.; Tikunov, Y.M.; Angenent, G.C.; van Tunen, A.J.; Bovy, A.G. RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol. 2007, 144, 1520–1530. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, M.; Ahuja, P.S.; Yadav, S.K. Post-transcriptional silencing of flavonol synthase mRNA in tobacco leads to fruits with arrested seed set. PLoS ONE 2011, 6, e28315. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Pandolfini, T.; Rotino, G.L.; Dani, V.; Spena, A. Aucsia gene silencing causes parthenocarpic fruit development in tomato. Plant Physiol. 2009, 149, 534–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takei, H.; Shinozaki, Y.; Yano, R.; Kashojiya, S.; Hernould, M.; Chevalier, C.; Ezura, H.; Ariizumi, T. Loss-of-function of a tomato receptor-like kinase impairs male fertility and induces parthenocarpic fruit set. Front. Plant Sci. 2019, 10, 403. [Google Scholar] [CrossRef]
- Xiong, A.-S.; Yao, Q.-H.; Peng, R.-H.; Li, X.; Han, P.-L.; Fan, H.-Q. Different effects on ACC oxidase gene silencing triggered by RNA interference in transgenic tomato. Plant Cell Rep. 2005, 23, 639–646. [Google Scholar] [CrossRef]
- Gupta, A.; Pal, R.K.; Rajam, M.V. Delayed ripening and improved fruit processing quality in tomato by RNAi-mediated silencing of three homologs of 1-aminopropane-1-carboxylate synthase gene. J. Plant Physiol. 2013, 170, 987–995. [Google Scholar] [CrossRef]
- Meli, V.S.; Ghosh, S.; Prabha, T.N.; Chakraborty, N.; Chakraborty, S.; Datta, A. Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 2413–2418. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Zhang, J.; Li, J.; Yang, C.; Wang, T.; Ouyang, B.; Li, H.; Giovannoni, J.; Ye, Z. A STAY GREEN protein SlSGR1 regulates lycopene and β-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol. 2013, 198, 442–452. [Google Scholar] [CrossRef]
- Elitzur, T.; Yakir, E.; Quansah, L.; Zhangjun, F.; Vrebalov, J.; Khayat, E.; Giovannoni, J.J.; Friedman, H. Banana MaMADS transcription factors are necessary for fruit ripening and molecular tools to promote shelf-life and food security. Plant Physiol. 2016, 171, 380–391. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Huang, W.; Xiong, F.; Xian, Z.; Su, D.; Ren, M.; Li, Z. Silencing of Sl PL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 2017, 15, 1544–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davuluri, G.R.; Van Tuinen, A.; Fraser, P.D.; Manfredonia, A.; Newman, R.; Burgess, D.; Brummell, D.; King, S.R.; Palys, J.; Uhlig, J.; et al. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 2005, 23, 890–895. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Hajirezaei, M.R.; Zanor, M.; Hornyik, C.; Debast, S.; Lacomme, C.; Fernie, A.R.; Sonnewald, U.; Börnke, F. RNA interference-mediated repression of sucrose phosphatase in transgenic potato tubers (Solanum tuberosum) strongly affects the hexose to sucrose ratio upon cold storage with only minor effects on total soluble carbohydrate accumulation. Plant Cell Environ. 2007, 31, 165–176. [Google Scholar] [CrossRef]
- Nawaz-Ul-Rehman, M.S.; Mansoor, S.; Khan, A.A.; Zafar, Y.; Briddon, R.W. RNAi-mediated male sterility of tobacco by silencing TA29. Mol. Biotechnol. 2007, 36, 159–165. [Google Scholar] [CrossRef]
- Tehseen, M.; Imran, M.; Hussain, M.; Irum, S.; Ali, S.; Mansoor, S.; Zafar, Y. Development of male sterility by silencing Bcp1 gene of Arabidopsis through RNA interference. Afr. J. Biotechnol. 2010, 9, 2736–2741. Available online: http://www.academicjournals.org/AJB (accessed on 27 August 2021).
- Sinha, R.; Rajam, M.V. RNAi silencing of three homologues of S-adenosylmethionine decarboxylase gene in tapetal tissue of tomato results in male sterility. Plant Mol. Biol. 2013, 82, 169–180. [Google Scholar] [CrossRef]
- Sandhu, A.P.S.; Abdelnoor, R.V.; Mackenzie, S.A. Transgenic induction of mitochondrial rearrangements for cytoplasmic male sterility in crop plants. Proc. Natl. Acad. Sci. USA 2007, 104, 1766–1770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishihara, M.; Nakatsuka, T.; Yamamura, S. Flavonoid components and flower color change in transgenic tobacco plants by suppression of chalcone isomerase gene. FEBS Lett. 2005, 579, 6074–6078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamiishi, Y.; Otani, M.; Takagi, H.; Han, D.-S.; Mori, S.; Tatsuzawa, F.; Okuhara, H.; Kobayashi, H.; Nakano, M. Flower color alteration in the liliaceous ornamental Tricyrtis sp. by RNA interference-mediated suppression of the chalcone synthase gene. Mol. Breed. 2012, 30, 671–680. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Mishiba, K.-I.; Abe, Y.; Kubota, A.; Kakizaki, Y.; Yamamura, S.; Nishihara, M. Flower color modification of gentian plants by RNAi-mediated gene silencing. Plant Biotechnol. 2008, 25, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Nakatsuka, T.; Mishiba, K.-I.; Kubota, A.; Abe, Y.; Yamamura, S.; Nakamura, N.; Tanaka, Y.; Nishihara, M. Genetic engineering of novel flower colour by suppression of anthocyanin odification genes in gentian. J. Plant Physiol. 2010, 167, 231–237. [Google Scholar] [CrossRef]
- He, H.; Ke, H.; Keting, H.; Qiaoyan, X.; Silan, D. Flower colour modification of chrysanthemum by suppression of F3′H and overexpression of the exogenous Senecio cruentus F3′5′H gene. PLoS ONE 2013, 8, e74395. [Google Scholar] [CrossRef] [Green Version]
- White, P.J.; Broadley, M. Biofortification of crops with seven mineral elements often lacking in human diets–iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 2009, 182, 49–84. [Google Scholar] [CrossRef]
- Flores, T.; Karpova, O.; Su, X.; Zeng, P.; Bilyeu, K.; Sleper, D.A.; Nguyen, H.T.; Zhang, Z.J. Silencing of GmFAD3 gene by siRNA leads to low α-linolenic acids (18:3) of fad3-mutant phenotype in soybean [Glycine max (Merr.)]. Transgenic Res. 2008, 17, 839–850. [Google Scholar] [CrossRef]
- Ozseyhan, M.E.; Li, P.; Na, G.; Li, Z.; Wang, C.; Lu, C. Improved fatty acid profiles in seeds of Camelina sativa by artificial microRNA mediated FATB gene suppression. Biochem. Biophys. Res. Commun. 2018, 503, 621–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusaba, M.; Miyahara, K.; Iida, S.; Fukuoka, H.; Takano, T.; Sassa, H.; Nishimura, M.; Nishio, T. Low glutelin content1: A dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 2003, 15, 1455–1467. [Google Scholar] [CrossRef] [Green Version]
- Regina, A.; Bird, A.; Topping, D.; Bowden, S.; Freeman, J.; Barsby, T.; Kosar-Hashemi, B.; Li, Z.; Rahman, S.; Morell, M. High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc. Natl. Acad. Sci. USA 2006, 103, 3546–3551. [Google Scholar] [CrossRef] [Green Version]
- Weise, S.E.; Aung, K.; Jarou, Z.J.; Mehrshahi, P.; Li, Z.; Hardy, A.C.; Carr, D.J.; Sharkey, T.D. Engineering starch accumulation by manipulation of phosphate metabolism of starch. Plant Biotechnol. J. 2012, 10, 545–554. [Google Scholar] [CrossRef]
- Yu, B.; Lydiate, D.J.; Young, L.W.; Schäfer, U.A.; Hannoufa, A. Enhancing the carotenoid content of Brassica napus seeds by downregulating lycopene epsilon cyclase. Transgenic Res. 2008, 17, 573–585. [Google Scholar] [CrossRef]
- Aggarwal, S.; Kumar, A.; Bhati, K.K.; Kaur, G.; Shukla, V.; Tiwari, S.; Pandey, A.K. RNAi-mediated downregulation of inositol pentakisphosphate kinase (IPK1) in wheat grains decreases phytic acid levels and increases Fe and Zn accumulation. Front. Plant Sci. 2018, 9, 259. [Google Scholar] [CrossRef]
- Borgio, J.F. RNA interference (RNAi) technology: A promising tool for medicinal plant research. J. Med. Plant Res. 2009, 3, 1176–1183. Available online: http://www.academicjournals.org/JMPR (accessed on 27 August 2021).
- Allen, R.S.; Millgate, A.G.; A Chitty, J.; Thisleton, J.; Miller, J.A.C.; Fist, A.J.; Gerlach, W.L.; Larkin, P.J. RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat. Biotechnol. 2004, 22, 1559–1566. [Google Scholar] [CrossRef]
- Ogita, S.; Uefuji, H.; Yamaguchi, Y.; Koizumi, N.; Sano, H. Producing decaffeinated coffee plants. Nature 2003, 423, 823. [Google Scholar] [CrossRef]
- Mohanpuria, P.; Kumar, V.; Ahuja, P.S.; Yadav, S.K. Producing low-caffeine tea through post-transcriptional silencing of caffeine synthase mRNA. Plant Mol. Biol. 2011, 76, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, H.; Liang, X.; Yan, Y.; Xia, P.; Jia, Y.; Liang, Z. Enhanced production of phenolic acids in Salvia miltiorrhiza hairy root cultures by combing the RNAi-mediated silencing of chalcone synthase gene with salicylic acid treatment. Biochem. Eng. J. 2015, 103, 185–192. [Google Scholar] [CrossRef]
- Wang, Q.; Reddy, V.; Panicker, D.; Mao, H.-Z.; Kumar, N.; Rajan, C.; Venkatesh, P.N.; Chua, N.-H.; Sarojam, R. Metabolic engineering of terpene biosynthesis in plants using a trichome specific transcription factor Ms YABBY 5 from spearmint (Mentha spicata). Plant Biotechnol. J. 2016, 14, 1619–1632. [Google Scholar] [CrossRef] [Green Version]
- Jamaluddin, N.D.; Rohani, E.R.; Noor, N.M.; Goh, H.-H. Transcriptome-wide effect of DE-ETIOLATED1 (DET1) suppression in embryogenic callus of Carica papaya. J. Plant Res. 2019, 132, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Du, H.; Wang, J.; Pu, Y.; Yang, C.; Yan, R.; Yang, H.; Cheng, H.; Yu, D. Multiplex CRISPR/Cas9 mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 2019, 18, 1384–1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puchta, H.; Fauser, F. Synthetic nucleases for genome engineering in plants: Prospects for a bright future. Plant J. 2013, 78, 727–741. [Google Scholar] [CrossRef]
- Li, Q.; Sapkota, M.; Van Der Knaap, E. Perspectives of CRISPR/Cas-mediated cis-engineering in horticulture: Unlocking the neglected potential for crop improvement. Hortic. Res. 2020, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [Green Version]
- Jansen, R.; Van Embden, J.D.A.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mojica, F.J.; Diez-Villasenor, C.; Garcia-Martinez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef] [Green Version]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Kunin, V.; Sorek, R.; Hugenholtz, P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007, 8, R61–R67. [Google Scholar] [CrossRef] [Green Version]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef] [Green Version]
- Brouns, S.J.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.H.; Snijders, A.P.L.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; Van Der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garneau, J.E.; Dupuis, M.-È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. Genet. 2011, 9, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Brant, E.; Budak, H.; Zhang, B. CRISPR/Cas: A Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J. Zhejiang Univ. Sci. B 2021, 22, 253–284. [Google Scholar] [CrossRef] [PubMed]
- Sinkunas, T.; Gasiunas, G.; Fremaux, C.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011, 30, 1335–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Terns, R.M.; Terns, M.P. Cas9 function and host genome sampling in Type II-A CRISPR–Cas adaptation. Genes Dev. 2015, 29, 356–361. Available online: http://www.genesdev.org/cgi/doi/10.1101/gad.257550.114 (accessed on 27 August 2021). [CrossRef] [Green Version]
- Chylinski, K.; Le Rhun, A.; Charpentier, E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013, 10, 726–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonin, E.V.; Makarova, K.S. CRISPR-Cas: Evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013, 10, 679–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science 2008, 322, 1843–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marraffini, L.A.; Sontheimer, E.J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nat. Cell Biol. 2010, 463, 568–571. [Google Scholar] [CrossRef] [Green Version]
- Hale, C.R.; Zhao, P.; Olson, S.; Duff, M.O.; Graveley, B.R.; Wells, L.; Terns, R.M.; Terns, M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009, 139, 945–956. [Google Scholar] [CrossRef] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Ahmar, S.; Saeed, S.; Khan, M.; Khan, S.U.; Mora-Poblete, F.; Kamran, M.; Faheem, A.; Maqsood, A.; Rauf, M.; Saleem, S.; et al. A Revolution toward Gene-Editing Technology and Its Application to Crop Improvement. Int. J. Mol. Sci. 2020, 21, 5665. [Google Scholar] [CrossRef]
- Song, G.; Jia, M.; Chen, K.; Kong, X.; Khattak, B.; Xie, C.; Li, A.; Mao, L. CRISPR/Cas9: A powerful tool for crop genome editing. Crop J. 2016, 4, 75–82. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS 8 variants generated by CRISPR Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Pyott, D.E.; Sheehan, E.; Molnar, A. Engineering of CRISPR/Cas9 mediated potyvirus resistance in transgene free Arabidopsis plants. Mol. Plant Pathol. 2016, 17, 1276–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef]
- Shen, C.; Que, Z.; Xia, Y.; Tang, N.; Li, D.; He, R.; Cao, M. Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J. Plant Biol. 2017, 60, 539–547. [Google Scholar] [CrossRef]
- Ueta, R.; Abe, C.; Watanabe, T.; Sugano, S.S.; Ishihara, R.; Ezura, H.; Osakabe, Y.; Osakabe, K. Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Zhang, L.; Wang, L.; Chen, L.; Zhao, R.; Sheng, J.; Shen, L. Biotechnology and biological transformations CRISPR/Cas9-Mediated SlCBF1 mutagenesis reduces tomato plant chilling tolerance. J. Agric. Food Chem. 2018, 66, 9042–9051. [Google Scholar] [CrossRef]
- Tashkandi, M.; Ali, Z.; Aljedaani, F.; Shami, A.; Mahfouz, M.M. Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 2018, 13, e1525996. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Soyars, C.L.; Li, J.; Fei, Q.; He, G.; Peterson, B.A.; Meyers, B.C.; Nimchuk, Z.L.; Wang, X. CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant Direct 2018, 2, e00047. [Google Scholar] [CrossRef]
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Čermák, T.; Voytas, D.F.; Choi, I.; Chadha-Mohanty, P. Novel alleles of rice eIF4G generated by CRISPR/Cas9 targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 2018, 16, 1918–1927. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR / Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Okada, A.; Arndell, T.; Borisjuk, N.; Sharma, N.; Watson-Haigh, N.; Tucker, E.J.; Baumann, U.; Langridge, P.; Whitford, R. CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 2019, 17, 1905–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhang, N.; Zhang, Q.; Zhou, G.; Tian, H.; Hussain, S.; Ahmed, S.; Wang, T.; Wang, S. Genome editing to integrate seed size and abiotic stress tolerance traits in Arabidopsis reveals a role for DPA4 and SOD7 in the regulation of inflorescence architecture. Int. J. Mol. Sci. 2019, 20, 2695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paixão, J.F.R.; Gillet, F.-X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; De Melo, B.P.; De Almeida-Engler, J.; Grossi-De-Sa, M.F. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a Histone AcetylTransferase. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Kis, A.; Hamar, É.; Tholt, G.; Bán, R.; Havelda, Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 2019, 17, 1004–1006. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Zhang, L.; Tang, M.; Liu, J.; Liu, H.; Yang, H.; Fan, S.; Terzaghi, W.; Wang, H.; Hua, W. Knockout of two Bna MAX 1 homologs by CRISPR/Cas9 targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 2020, 18, 644–654. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR–Cas9 system. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. Generation of High Yielding and Fragrant Rice (Oryza sativa L.) Lines by CRISPR/Cas9 targeted mutagenesis of three homoeologs of cytochrome P450 gene family and OsBADH2 and transcriptome and proteome profiling of revealed changes triggered by mutations. Plants 2020, 9, 788. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.V.S.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Faal, P.G.; Farsi, M.; Seifi, A.; Kakhki, A.M. Virus-induced CRISPR-Cas9 system improved resistance against tomato yellow leaf curl virus. Mol. Biol. Rep. 2020, 47, 3369–3376. [Google Scholar] [CrossRef]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef]
- Li, C.; Liu, C.; Qi, X.; Wu, Y.; Fei, X.; Mao, L.; Cheng, B.; Li, X.; Xie, C. RNA guided Cas9 as an in vivo desired target mutator in maize. Plant Biotechnol. J. 2017, 15, 1566–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; He, M.; Li, J.; Chen, L.; Huang, Z.; Zheng, S.; Zhu, L.; Ni, E.; Jiang, D.; Zhao, B. Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBF s in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [Green Version]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [Green Version]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 1–4. [Google Scholar] [CrossRef]
- Zhan, X.; Zhang, F.; Zhong, Z.; Chen, R.; Wang, Y.; Chang, L.; Bock, R.; Nie, B.; Zhang, J. Generation of virus resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 2019, 17, 1814–1822. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of Ta EDR 1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef] [Green Version]
- Nadakuduti, S.S.; Enciso-Rodríguez, F. Advances in genome editing with CRISPR systems and transformation technologies for plant DNA manipulation. Front. Plant Sci. 2021, 11, 2267. [Google Scholar] [CrossRef]
- Montecillo, J.A.V.; Chu, L.L.; Bae, H. CRISPR-Cas9 system for plant genome editing: Current approaches and emerging developments. Agronomy 2020, 10, 1033. [Google Scholar] [CrossRef]
- Ren, Q.; Zhong, Z.; Wang, Y.; You, Q.; Li, Q.; Yuan, M.; He, Y.; Qi, C.; Tang, X.; Zheng, X.; et al. Bidirectional promoter-based CRISPR-Cas9 systems for plant genome editing. Front. Plant Sci. 2019, 10, 1173. [Google Scholar] [CrossRef]
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Kantor, A.; McClements, M.E.; MacLaren, R.E. CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci. 2020, 21, 6240. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
| Trait(s) | Crop Improved | Resistance Against | Targeted Gene(s) | References |
|---|---|---|---|---|
| Virus resistance | Nicotiana bethamiana | Chilli-infecting begomoviruses | AC1 AC2 βC1 | [43] |
| Triticum spp. | Triticum mosaic virus (TMV) | Coat protein (CP) | [44] | |
| Oryza sativa | Rice black streak dwarf virus (RBSDV) | S7-2 S8 | [45] | |
| Solanum tuberosum | Potato virus X (PVX), Potato virus Y (PVY) Potato virus S (PVS) | CP | [46] | |
| Glycine max | Soybean mosaic virus (SMV) | SMV P3 cistron | [47] | |
| Mungbean yellow mosaic virus (MYMIV) | CP | [48] | ||
| Arachis hypogaea | Tobacco streak virus (TSV) | CP | [49] | |
| O. sativa | Rice tungroo bacilliform virus (RTBV) Rice tungroo spherical virus (RTSV) | Coat protein 3 CP3 | [50] | |
| Glycine max | Soybean mosaic virus (SMV) | eIF4E1 | [51] | |
| N. bethamiana | Tomato yellow leaf curl Thailand virus (TYLCTV) | GSA | [52] | |
| Bacterial resistance | A. thaliana | Agrobacterium tumefaciens | iaaM ipt | [53] |
| Pseudomonas syringae | PPRL | [54] | ||
| Citrus limon | Xanthomonas citri | CalS1 | [55] | |
| Fungal resistance | S. tuberosum | Phytophthora infestans | Avr3a | [56] |
| T. aestivum | Fusarium graminearum | Chs 3b | [57] | |
| Musa spp. | F. oxysporum f. sp. cubense (Foc) | Foc velvet protein | [58] | |
| N. tabacum | Sclerotinia sclerotiorum | Chs | [59] | |
| S. lycopersicum | F. oxysporum | Fow2 chs V | [60] | |
| O. sativa | Magnaporthe oryzae | MoABC1 MoMAC1 MoPMK1 | [61] | |
| Rhizoctonia solani | RPMK1-1 RPMK1-2 | [62] | ||
| Zea mays | Aspergillus flavus | ZmPRms | [63] | |
| S. lycopersicum | F. oxysporum | Fmk1 Hog1 Pbs2 | [64] | |
| Z. mays | A. flavus | Amy1 | [65] | |
| S. tuberosum | Phytophthora infestans Alternaria solani | PVS1 PVS2 PVS3 PVS4 | [66] | |
| Glycine max | Phytophthora sojae | GmSnRK1.1 | [67] | |
| S. lycopersicum | F. oxysporum | ODC | [68] | |
| Insect resistance | S. lycopersicum | Helicoverpa armigera | HaCHI | [69] |
| N. tabacum | Bemisia tabaci | AChE EcR | [70] | |
| Lettuce | B. tabaci | V-ATPase | [71] | |
| A. thaliana | Myzus persicae | MyCP | [72] | |
| Brassica rapa | Tetranychus urticae | COPB2 | [73] | |
| Nematodes Resistance | S. lycopersicum | Meloidogyne incognita | Mi-cpl1 | [74] |
| N. benthamiana | Radopholus similis | Rs-cps | [75] | |
| S. lycopersicum | M. incognita | PolA1 | [76] | |
| Glycine max | Heterodera glycines | Hg16B09 | [77] | |
| HgY25 HgPrp17 | [78] | |||
| A. thaliana | M. incognita | Mi-msp3 Mi-msp 5 Mi-msp18 Mi-msp24 | [79] | |
| Abiotic stress tolerance | N. tabacum | Salt tolerance | Nt ε-LCY | [80] |
| O. sativa | Salt tolerance | OsPEX11 | [81] | |
| B. rapa | Salt tolerance | GIGANTEA (GI) | [82] | |
| A. thaliana | Drought tolerance | PAD4 LSD1 EDS1 | [83] | |
| O. sativa | Drought tolerance | OsGRXS17 | [84] | |
| O. sativa | Drought tolerance | OsDSR-1 | [85] | |
| O. sativa | Drought tolerance | OsERF101 | [86] | |
| S. lycopersicum | Drought and salt tolerance | SlbZIP1 | [87] | |
| N. tabacum | Drought tolerance | BrDST71 | [88] | |
| T. aestivum | Salt tolerance | TaPUB-1 | [89] | |
| A. thaliana | Osmotic tolerance | WZY2 | [90] |
| Trait(s) | Crop Used | Targeted Gene(s) | References |
|---|---|---|---|
| Drought tolerance | Z. mays (Maize) | ARGOS8 | [201] |
| Turnip mosaic virus (TMV) resistance | A. thaliana | eIF(iso)4E | [202] |
| Cucumber vein yellowing virus (CMYV) resistance | Cucumis sativus | eIF4E | [203] |
| Drought tolerance | S. lycopersicum | SlMAPK3 | [204] |
| Cold tolerance | O. sativa | OsAnn3 | [205] |
| Parthenocarpic fruit development | S. lycopersicum | SlIAA9 | [206] |
| Chilling stress tolerance | S. lycopersicum | SlCBF1 | [207] |
| Tomato yellow leaf curl virus (TYLCV) resistance | S. lycopersicum, N. benthamiana | Coat protein (CP) Replicase (Rep) | [208] |
| Cauliflower mosaic virus (CMV) resistance | A. thaliana | CaMV CP | [209] |
| Rice tungro spherical virus (RTSV) resistance | O. sativa | eIF4G | [210] |
| Salt tolerance | OsRR22 | [211] | |
| Male-sterile development | T. aestivum | Ms1 | [212] |
| Heat stress tolerance | S. lycopersicum | SlMAPK3 | [127] |
| Drought and salt stress tolerance | A. thaliana | DAP4 SOD7 | [213] |
| Drought tolerance | AREB1 | [214] | |
| Wheat dwarf virus (WDV) resistance | Hordeum vulgare | CP Rep/Rep4 | [215] |
| Yield improvement | B. napus | BnaMAX1 | [216] |
| Yield improvement Stress tolerance | O. sativa (Nippobare) | OsPIN5b GS3 OsMYB30 | [217] |
| Yield improvement | O. sativa | Cyt P450 homeologs OsBADH2 | [218] |
| Drought and stress tolerance | OsDST | [219] | |
| Tomato yellow leaf curl virus (TYLCV) resistance | S. lycopersicum | rgsCaM | [220] |
| Soyabean mosaic virus (SMV) resistance | Glycine max | GmF3H1 GmF3H2 GmFNSII-1 | [171] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajput, M.; Choudhary, K.; Kumar, M.; Vivekanand, V.; Chawade, A.; Ortiz, R.; Pareek, N. RNA Interference and CRISPR/Cas Gene Editing for Crop Improvement: Paradigm Shift towards Sustainable Agriculture. Plants 2021, 10, 1914. https://doi.org/10.3390/plants10091914
Rajput M, Choudhary K, Kumar M, Vivekanand V, Chawade A, Ortiz R, Pareek N. RNA Interference and CRISPR/Cas Gene Editing for Crop Improvement: Paradigm Shift towards Sustainable Agriculture. Plants. 2021; 10(9):1914. https://doi.org/10.3390/plants10091914
Chicago/Turabian StyleRajput, Meenakshi, Khushboo Choudhary, Manish Kumar, V. Vivekanand, Aakash Chawade, Rodomiro Ortiz, and Nidhi Pareek. 2021. "RNA Interference and CRISPR/Cas Gene Editing for Crop Improvement: Paradigm Shift towards Sustainable Agriculture" Plants 10, no. 9: 1914. https://doi.org/10.3390/plants10091914
APA StyleRajput, M., Choudhary, K., Kumar, M., Vivekanand, V., Chawade, A., Ortiz, R., & Pareek, N. (2021). RNA Interference and CRISPR/Cas Gene Editing for Crop Improvement: Paradigm Shift towards Sustainable Agriculture. Plants, 10(9), 1914. https://doi.org/10.3390/plants10091914