Arbuscular Mycorrhizal Fungi from Argentinean Highland Puna Soils Unveiled by Propagule Multiplication
Abstract
1. Introduction
2. Results and Discussion
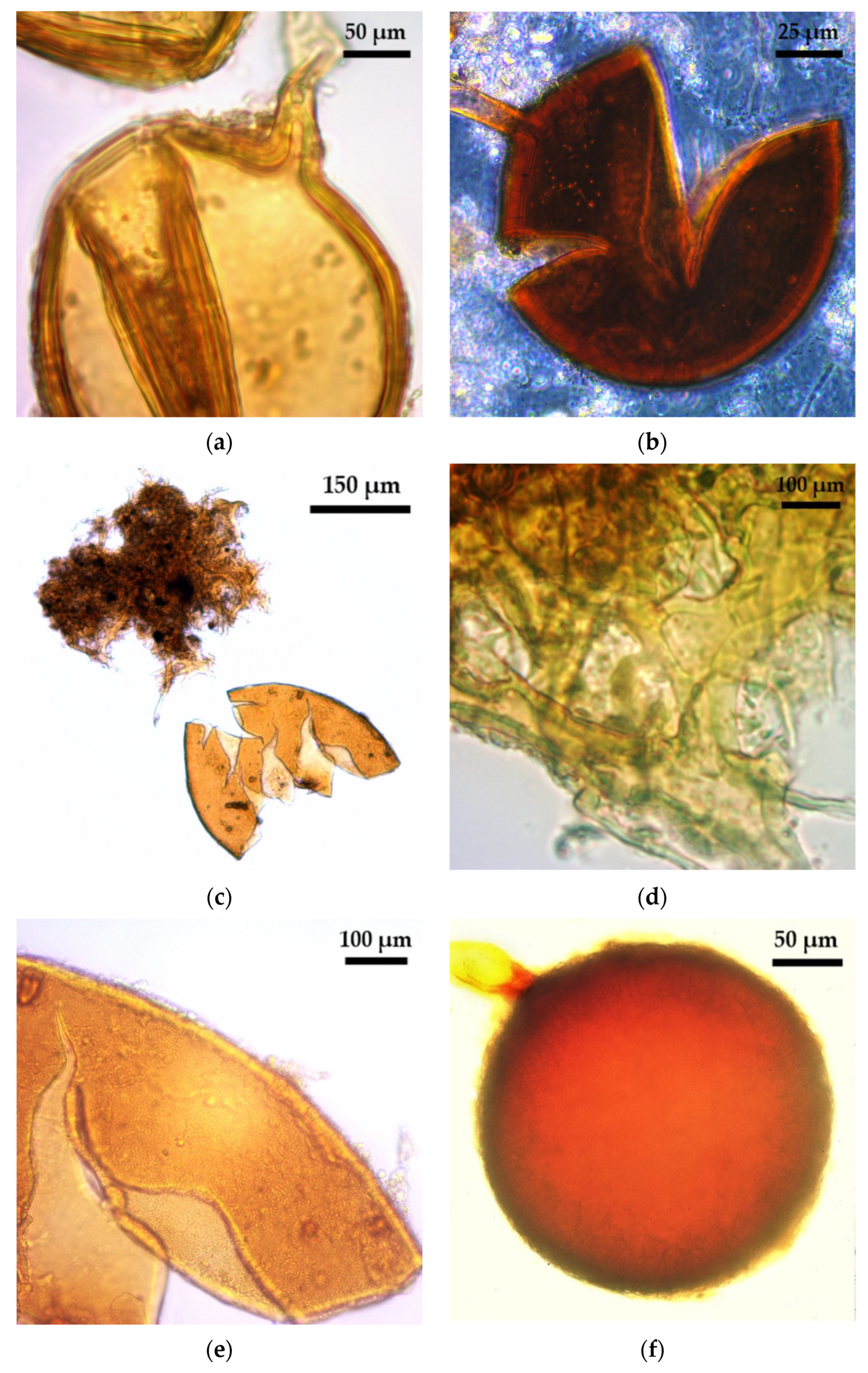
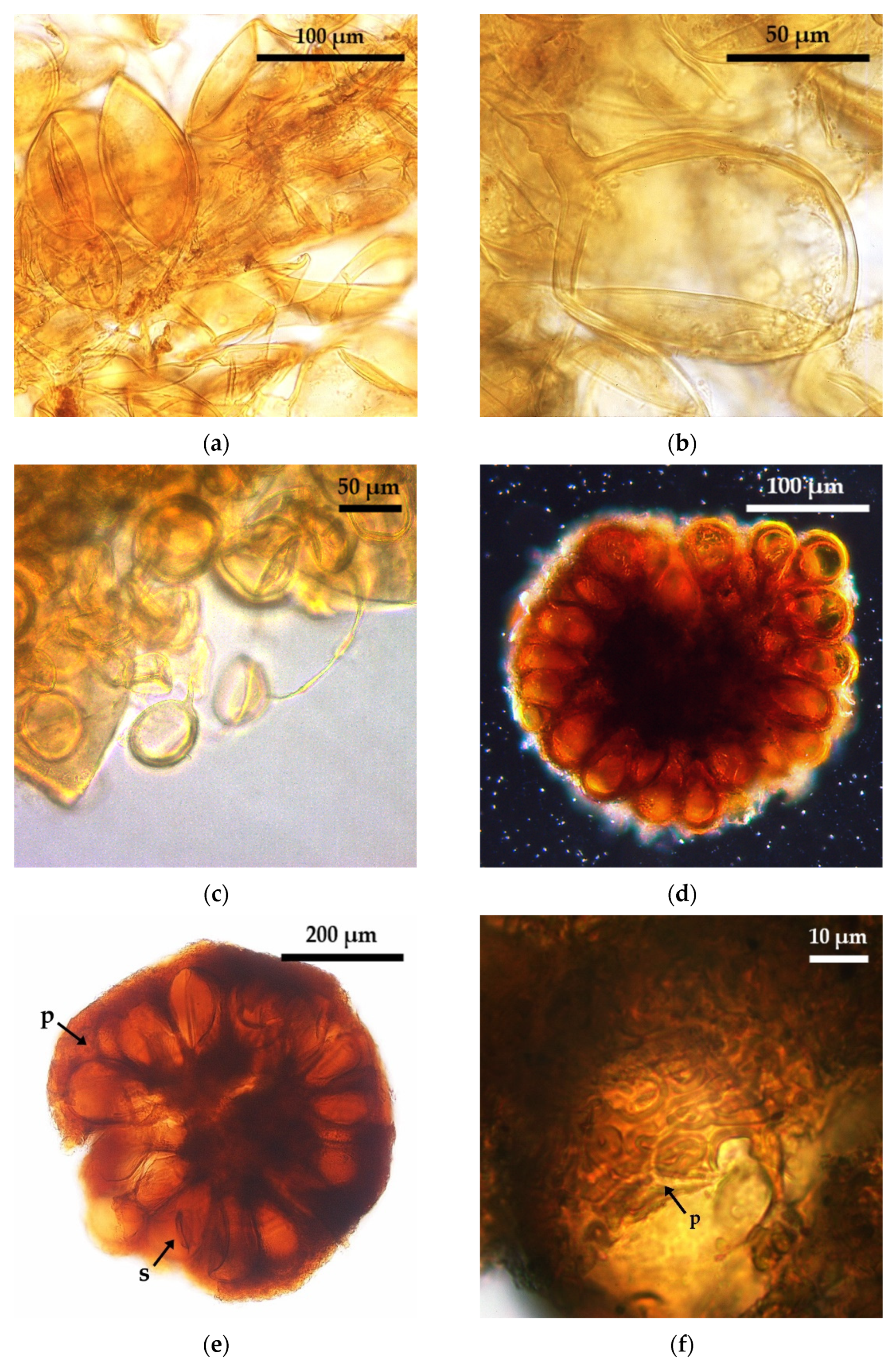
2.1. Glomoid AMF Diversity Assessed by Spore Morphology
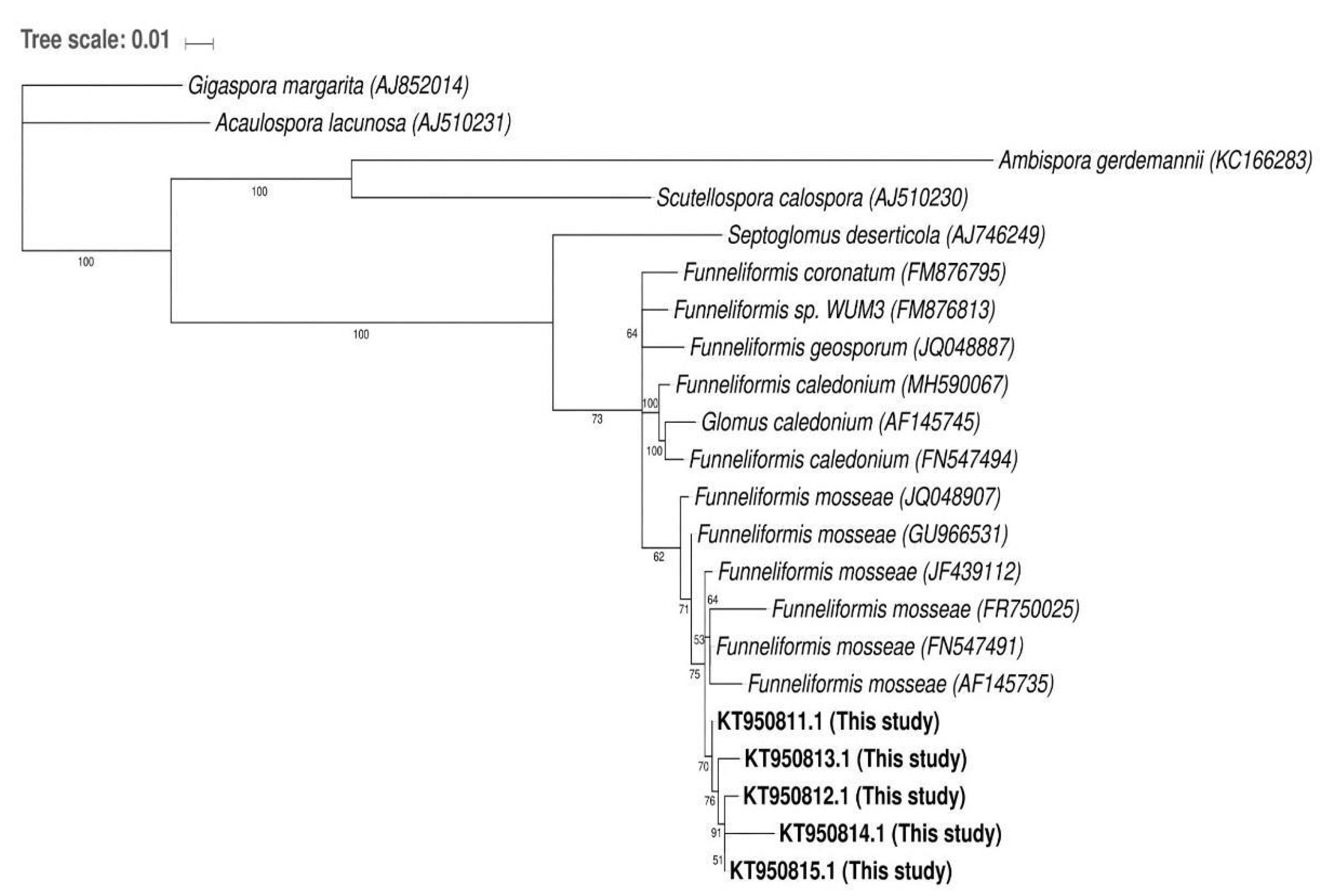
2.2. Molecular Analysis of Glomoid AMF Diversity
3. Materials and Methods
3.1. Study Area and Soil Sampling
3.2. AMF Propagule Multiplication
3.3. Glomoid AMF Diversity Assessed by Spore Morphological Approach
3.4. Molecular Analysis of Glomoid AMF Diversity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Powell, J.R.; Rillig, M.C. Biodiversity of arbuscular mycorrhizal fungi and ecosystem function. New Phytol. 2018, 220, 1059–1075. [Google Scholar] [CrossRef]
- Brundrett, M.; Bougher, N.; Dell, B.; Grove, T.; Malajczuk, N. Working with Mycorrhizas in Forestry and Agriculture; CSIRO: Canberra, Australia, 1996; p. 374. [Google Scholar]
- Stürmer, S.L.; Bever, J.D.; Morton, J.B. Biogeography of arbuscular mycorrhizal fungi (Glomeromycota): A phylogenetic perspective on species distribution patterns. Mycorrhiza 2018, 28, 587–603. [Google Scholar] [CrossRef] [PubMed]
- Trejo-Aguilar, D.; Lara-Capistrán, L.; Maldonado-Mendoza, I.E.; Zulueta-Rodríguez, R.; Sangabriel-Conde, W.; Mancera-López, M.E.; Negrete-Yankelevich, S.; Barois, I. Loss of arbuscular mycorrhizal fungal diversity in trap cultures during long-term subculturing. IMA Fungus 2013, 4, 161–167. [Google Scholar] [CrossRef]
- Davison, J.; Moora, M.; Öpik, M.; Adholeya, A.; Ainsaar, L.; Bâ, A.; Burla, S.; Diedhiou, A.G.; Hiiesalu, I.; Jairus, T.; et al. Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 2015, 349, 970–973. [Google Scholar] [CrossRef]
- Hijri, M.; Sanders, I.R. Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei. Nature 2005, 433, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, S.; Cusant, L.; Roux, C.; Corradi, N. Arbuscular mycorrhizal fungi: Intraspecific diversity and pangenomes. New Phytol. 2018, 220, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Alguacil, M.M.; Roldán, A.; Torres, M.P. Assessing the diversity of AM fungi in arid gypsophilous plant communities. Environ. Microbiol. 2009, 11, 2649–2659. [Google Scholar] [CrossRef]
- Thougnon Islas, A.J.; Hernandez Guijarro, K.; Eyherabide, M.; Sainz Rozas, H.R.; Echeverría, H.E.; Covacevich, F. Can soil properties and agricultural land use affect arbuscular mycorrhizal fungal communities indigenous from the Argentinean Pampas soils? Appl. Soil Ecol. 2016, 101, 47–56. [Google Scholar] [CrossRef]
- Commatteo, J.G.; Consolo, V.F.; Barbieri, P.A.; Covacevich, F. Indigenous arbuscular mycorrhiza and Trichoderma from systems with soybean predominance can improve tomato growth. Soil Environ. 2019, 38, 151–161. [Google Scholar] [CrossRef]
- Lugo, M.A.; Menoyo, E. Chapter 12: Southern Highlands: Fungal Endosymbiotic Associations. In Mycorrhizal Fungi in South America; Life Science Series, Fungal Biology; Pagano, M.C., Lugo, M.A., Eds.; Springer: Cham, Switzerland, 2019; pp. 217–255. ISBN 9783030152277. [Google Scholar]
- Lugo, M.A.; Ferrero, M.; Menoyo, E.; Estévez, M.C.; Siñeriz, F.; Anton, A.M. Arbuscular mycorrhizal fungi and rhizospheric bacteria diversity along an altitudinal gradient in South American Puna grassland. Microbiol. Ecol. 2008, 55, 705–713. [Google Scholar] [CrossRef]
- Oehl, F.; Sieverding, E.; Ineichen, K.; Mäder, P.; Boiler, T.; Wiemken, A. Impact of land use intensity on the species diversity of arbuscular mycorrhizal fungi in agroecosystems of Central Europe. Appl. Environ. Microbiol. 2003, 69, 2816–2824. [Google Scholar] [CrossRef]
- Songachan, L.S.; Kayang, H. Diversity of arbuscular mycorrhizal fungi associated with Flemingia vestita Benth. ex Baker. Mycology 2013, 4, 85–95. [Google Scholar] [CrossRef][Green Version]
- Liu, R.J.; Wang, F.Y. Selection of appropriate host plants used in trap culture of arbuscular mycorrhizal fungi. Mycorrhiza 2003, 13, 123–127. [Google Scholar] [CrossRef]
- Kjøller, R.; Rosendahl, S. Detection of arbuscular mycorrhizal fungi (Glomales) in roots by nested PCR and SSCP (Single Stranded Conformation Polymorphism). Plant Soil 2000, 226, 189–196. [Google Scholar] [CrossRef]
- Rosendahl, S.; Stukenbrock, E.H. Community structure of arbuscular mycorrhizal fungi in undisturbed vegetation revealed by analyses of LSU rDNA sequences. Mol. Ecol. 2004, 13, 3179–3186. [Google Scholar] [CrossRef]
- Wetzel, K.; Silva, G.; Matczinski, U.; Oehl, F.; Fester, T. Superior differentiation of arbuscular mycorrhizal fungal communities from till and no-till plots by morphological spore identification when compared to T-RFLP. Soil Biol. Biochem. 2014, 72, 88–96. [Google Scholar] [CrossRef]
- Säle, V.; Aguilera, P.; Laczko, E.; Mäder, P.; Berner, A.; Zihlmann, U.; van der Heijden, M.G.A.; Oehl, F. Impact of conservation tillage and organic farming on the diversity of arbuscular mycorrhizal fungi. Soil Biol. Biochem. 2015, 84, 38–52. [Google Scholar] [CrossRef]
- Leal, P.L.; De Carvalho, T.S.; Siqueira, J.O.; Moreira, F.M.S. Assessment of the occurrence and richness of arbuscular mycorrhizal fungal spores by direct analysis of field samples and trap culture—A comparative study. Anais da Academia Brasileira de Ciências 2018, 90, 2359–2373. [Google Scholar] [CrossRef]
- Sun, X.; Hu, W.; Tang, M.; Chen, H. Characterizing and handling different kinds of AM fungal spores in the rhizosphere. World J. Microbiol. Biotechnol. 2016, 32, 97. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Gao, J.L.; Zhu, H.H.; Long, L.K.; Xing, Q.X.; Chen, J.Z. Evaluation of the potential of trap plants to detect arbuscular mycorrhizal fungi using polymerase chain reaction-denaturing gradient gel electrophoresis analysis. Soil Sci. Plant Nutr. 2010, 56, 205–211. [Google Scholar] [CrossRef]
- Hong, H.; Pruden, A.; Reardon, K.F. Comparison of CE-SSCP and DGGE for monitoring a complex microbial community remediating mine drainage. J. Microbiol. Methods 2007, 69, 52–64. [Google Scholar] [CrossRef]
- Ontivero, R.E.; Voyron, S.; Risio Allione, L.V.; Bianco, P.; Bianciotto, V.; Iriarte, H.J.; Lugo, M.A.; Lumini, E. Impact of land use history on the arbuscular mycorrhizal fungal diversity in arid soils of Argentinean farming fields. FEMS Microbiol. Lett. 2020, 367, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Martínez Carretero, E. La Puna Argentina: Delimitación general y división en distritos florísticos. Boletín de la Sociedad Argentina de Botánica 1995, 31, 27–40. [Google Scholar]
- Panigatti, J.L. Argentina: 200 Años, 200 Suelos; INTA: Buenos Aires, Argentina, 2010; p. 345. Available online: https://inta.gob.ar/sites/default/files/script-tmp-inta-200-suelos.pdf (accessed on 7 July 2021).
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani LK, T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Deng, C. Outline of fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Benbouza, H.; Jacquemin, J.M.; Baudoin, J.P.; Mergeai, G. Optimization of a reliable, fast, cheap and sensitive silver staining method to detect SSR markers in polyacrylamide gels. Biotechnol. Agron. Soc. Environ. 2006, 10, 77–81. [Google Scholar]
- Höfle, M.G.; Kirchman, D.; Christen, R.; Brettar, I. Molecular diversity of bacterioplankton: Link to a predictive biogeochemistry of pelagic ecosystems. Aquat. Microb. Ecol. 2008, 53, 39–58. [Google Scholar] [CrossRef]
- Cui, J.; Bai, L.; Liu, X.; Jie, W.; Cai, B. Arbuscular mycorrhizal fungal communities in the rhizosphere of a continuous cropping soybean system at the seedling stage. Environ. Microbiol. 2018, 49, 240–247. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Bray, R.H.; Kurtz, L.T. Determination of total, organic and available forms of phosphorus in soils. Soil Sci. 1945, 59, 39–45. [Google Scholar] [CrossRef]

| ABRA DEL CÓNDOR | |||
|---|---|---|---|
| Trap Culture Strategy | AC Field | ACM1 | ACM2 |
| Sampling site of Puna | Abra del Cóndor | Abra del Cóndor | Abra del Cóndor |
| Site elevation | 3870 m | 3870 m | 3870 m |
| Data source | [12] | This study | This study |
| Initial total spore density/100 g dry soil weight | - | 424 | 424 |
| 1st cycle time | - | 5 months | 5 months |
| 1st cycle TPS | - | Sorghum bicolor + Melilotus albus | S. bicolor |
| 2nd cycle time | - | - | 2 months |
| 2nd cycle TPS | - | - |
S. bicolor +M. albus +
Allium ampeloprasum var. porrum |
| Total time of trap culture | - | 5 months | 7 months |
| Glomoid AMF taxa |
Glomus
sp. G. ambisporum Rhizoglomus aggregatus (=Glomus aggregatum) Sclerocystis sinuosa (=Glomus sinuosum) |
Funneliformis
sp. F. geosporus Sclerocystis sp. S. sinuosa Septoglomus constrictum |
Funneliformis
sp. F. geosporus Septoglomus constrictum |
|
Richness of glomoid morphospecies with funnel pore | 0 | 3 | 3 |
| Richness of total glomoid morphospecies | 4 | 5 | 4 |
| H’ Diversity Index (SSCP analysis) | - | 1.44 | 1.16 |
| Sequence of an excised band (SSCP analysis) | - | KT950811 | KT950815 |
| ITURBE | ||||
|---|---|---|---|---|
| Trap Culture Strategy | It Field | ItM3-a1 | ItM3-a2 | ItM3-a3 |
| Sampling site of Puna | Iturbe | Iturbe | Iturbe | Iturbe |
| Site elevation | 3370 m | 3370 m | 3370 m | 3370 m |
| Data source | [12] | This study | This study | This study |
| Initial total spore density/100 g dry soil weight | - | 171 | 171 | 171 |
| 1st cycle time | - | 5 months | 5 months | 5 months |
| 1st cycle TPS | - |
Sorghum bicolor Melilotus albus Zea mays Pennisetum glaucum Avena sativa Secale cereale S. bicolor+ S. cereale P. glaucum+ S. cereale |
S. bicolor M. albus Z. mays P. glaucum A. sativa S. cereale S. bicolor+ S. cereale P. glaucum+ S. cereale |
S. bicolor M. albus Z. mays P. glaucum A. sativa S. cereale S. bicolor+ S. cereale P. glaucum+ S. cereale |
| 2nd cycle time | - | 6 months | 6 months | 6 months |
| 2nd cycle TPS | - | S. bicolor | S. bicolor | S. bicolor |
| 3rd cycle time | - | 2 months | 2 months | 2 months |
| 3rd cycle TPS | - | Allium ampeloprasum var. porrum | S. bicolor | M. albus |
| Total time of trap culture | 13 months | 13 months | 13 months | |
| Glomoid AMF taxa |
Glomus
sp. G. ambisporum Rhizoglomus aggregatus (= Glomus aggregatum) S. sinuosa (= Glomus sinuosum) |
Funneliformis
sp. F. geosporus F. monosporus R. aggregatus R. microaggregatum |
Funneliformis
sp. F. geosporus R. aggregatus S. sinuosa |
F. geosporus R. microaggregatum R. aggregatus S. sinuosa |
|
Richness of glomoid morphospecies with funnel pore | 0 | 2 | 2 | 1 |
| Richness of total glomoid morphospecies | 4 | 5 | 4 | 4 |
| H’ Diversity Index (SSCP analysis) | - | 1.46 | 1.52 | 1.54 |
| Sequence of an excised band (SSCP analysis) | - | KT950814 | KT950813 | KT950812 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Covacevich, F.; Hernández Guijarro, K.; Crespo, E.M.; Lumini, E.; Rivero Mega, M.S.; Lugo, M.A. Arbuscular Mycorrhizal Fungi from Argentinean Highland Puna Soils Unveiled by Propagule Multiplication. Plants 2021, 10, 1803. https://doi.org/10.3390/plants10091803
Covacevich F, Hernández Guijarro K, Crespo EM, Lumini E, Rivero Mega MS, Lugo MA. Arbuscular Mycorrhizal Fungi from Argentinean Highland Puna Soils Unveiled by Propagule Multiplication. Plants. 2021; 10(9):1803. https://doi.org/10.3390/plants10091803
Chicago/Turabian StyleCovacevich, Fernanda, Keren Hernández Guijarro, Esteban M. Crespo, Erica Lumini, María Soledad Rivero Mega, and Mónica A. Lugo. 2021. "Arbuscular Mycorrhizal Fungi from Argentinean Highland Puna Soils Unveiled by Propagule Multiplication" Plants 10, no. 9: 1803. https://doi.org/10.3390/plants10091803
APA StyleCovacevich, F., Hernández Guijarro, K., Crespo, E. M., Lumini, E., Rivero Mega, M. S., & Lugo, M. A. (2021). Arbuscular Mycorrhizal Fungi from Argentinean Highland Puna Soils Unveiled by Propagule Multiplication. Plants, 10(9), 1803. https://doi.org/10.3390/plants10091803








