1. Introduction
Xylella fastidiosa subspecies
pauca (
Xfp)
, a Gram-negative bacterium in the family
Xanthomonadaceae (γ-proteobacteria), is one of the most dangerous plant pathogens worldwide [
1]. It colonizes the xylem of the host, and is transmitted by several xylem sap-feeding insect vectors (Homoptera, Auchenorrhyncha) [
2,
3]. Formerly restricted to the Americas, a very aggressive genotype,
Xfp ST53, has been reported in Apulia [
4,
5] as responsible for the Olive Quick Decline Syndrome (OQDS), a vascular disease causing the death of millions of young and centenarian olive trees [
6]. The disease appeared on the west coast of the Salento Peninsula (Apulia region, southeastern Italy) in 2013 and spread rapidly, causing devastating effects on olive trees cultivation, with serious impact on the economy, landscape, environment, and cultural heritage of the region [
7]. According to a recent estimation, around 4 million of olive trees have been killed or severely impaired in their productivity in the Salento areas, thus, causing an economic loss of approximately 390M€ [
8]. Subsequently,
Xfp and other subspecies (i.e.,
X. f. subspecies
multiplex, and
X. f. subspecies
fastidiosa) and genotypes, were also detected in insular and mainland France and Spain, as well as in Portugal, thus, spreading in the world’s largest olive-growing area (more than 2.5 million hectares) and threatening the whole Mediterranean agriculture (grapevine, almond, and citrus) [
9]. Based on these data, it could be foreseen that for the entire European territory, the risk and the economic losses arising from
Xylella fastidiosa might be much higher [
6].
The localization of the pathogen into the xylematic vessels determines an alteration of the raw sap movement, thus, impairing the water and nutrition supply in the affected twigs and leading to withering and desiccation of scattered shoots and small branches in the canopy. Bacterial cells attach to the vessel wall and, by multiplying, they aggregate in a biofilm matrix that includes nucleic acids, proteins, humic substances, and exopolysaccharide (EPS) protecting bacterial communities [
10]. This biofilm is the main factor responsible for the blockage of water movement in
X.
fastidiosa-infected plants, even if some studies have found that vessel occlusions are the active host plants responses as defense mechanism [
11,
12,
13,
14].
The diagnosis of
X. fastidiosa is difficult, because the pathogen infects a wide range of wild and cultivated plant species without causing any apparent symptoms for variable length of time. In the case of olive trees, it has been estimated that the asymptomatic period lasts approximately 1.2 years [
8,
9,
10,
11,
12,
13,
14,
15]. The initial symptom can be observed on the terminal leaf tip of the affected twigs, which turns dark yellow to brown, a condition that spreads inward and leads to complete desiccation. However, the appearance of these symptoms identifies an advanced stage of the infection that is no longer reversible and leads to the death of the plant [
16]. After the appearance of the initial symptoms, the desiccation extends to the rest of the canopy and the collapse of the entire tree follows within 3.5–4 years approximately [
8], although significant variations may occur, depending on the olive cultivar and the age of the plants [
5].
The variation of plant reflectance responses, measured by means of spectral indices in Visible and Near Infrared (NIR) spectral range, has been widely used as an indicator of disease development in the last years, being a feasible and not invasive monitoring means [
17,
18]. Since a typical symptom of
Xf infection is the leaf scorching and desiccation, it might be interesting to assess the water stress in the infected olive plants. Sun et al. (2008) [
19] studied the effect induced by a water stress on the reflectance spectrum response of olive trees. As a result, they found that 16 relevant wavelengths were responsive: (1) 524 nm, 555 nm, 572 nm, and 673 nm in visible region; 715 nm, 919 nm, 958 nm, and 1098 nm in NIR region; 1140 nm, 1202 nm, 1225 nm, 1272 nm, 1391 nm, 1455 nm, 1502 nm, and 1656 nm in the shortwave infrared region (SWIR). They developed an algorithm, the Relative Reflectance Increment (RRI), able to quantify and compare the change of reflectance spectra at different reference wavelengths in response to abiotic water stress.
Zarco-Tejada et al. (2018) [
20] compared Support Vector Machine (SVM), neural networks, and Linear Discriminant Analysis (LDA) procedures for detecting infected plants. They classified olive trees: in the field, scoring visually the plants as symptomatic and asymptomatic, as well as in the laboratory, determining the DNA of
Xfp by quantitative Polymerase Chain Reaction (qPCR) and rating the plants as infected and uninfected. As a result, they found that the Normalized Phaeophytinization Index (NPQI) [
21,
22] measured by remote sensing on board of aircraft, together with Crop Water Stress Index (CWSI), and the thermal-based stress trait best distinguished
X. fastidiosa symptomatic from asymptomatic trees. The accuracy of disease detection was confirmed by qPCR and exceeded the 80% when high-resolution fluorescence and thermal stress indicators were coupled with photosynthetic traits sensitive to rapid pigment dynamics and degradation.
Therefore, considering the relevant socio-cultural and economic impact of Xfp on olive cultivation, as well as the infectiousness and the destructiveness of the disease, the monitoring of the infected area must be fast and reliable, thus, allowing an early diagnosis of the disease also when the symptoms are not yet visible. In fact, any delay would preclude the effectiveness of the mandatory phytosanitary measures to slow down the epidemic progression (i.e., felling of the infected trees), thus, increasing the infection risk for the surrounding plants. Considering the recent developments of hyperspectral spectrometry for the detection of Xfp-infected plants, the objectives of this work were to select specific wavelengths, to assess the potential of hyperspectral data analyzed with a proper statistical approach and to discriminate, with a sufficient degree of accuracy, between uninfected and infected but asymptomatic olive leaves. Therefore, olive leaves collected from twigs tested for the presence/absence of the pathogen were analyzed with a hyperspectral apparatus, and then submitted to qPCR analysis. An integrated approach was used, including different statistical techniques: Variable Importance in the Projection (VIP) statistic determination with Partial Least Square (PLS) regression (PLS-VIP), Canonical Discriminant Analysis (CDA), stepwise selection, and unsupervised classification, each one adding an extra piece in the process of extracting information from hyperspectral data related to the infection by Xfp.
2. Materials and Methods
2.1. Site Description
Three demarcated areas were identified in Apulia by the Phytosanitary Authority, in order to deal with the diffusion of
Xfp disease in southern Apulia, and according to the EU Decision 2015/789 and subsequent modifications: (1) the infected zone, the southernmost area of the Salento peninsula where the disease had been diagnosed initially; (2) the containment zone, comprising the last 20 km toward North of the infected zone, where the eradication of the pathogen was mandatory; (3) the buffer zone, 10 Km wide strip after the containment zone, not yet contaminated by
Xfp (
Figure 1). Two experimental fields were selected in the Municipality of Oria (Brindisi), located on the edge between the infected and the containment zones at the time of survey (
Figure 1).
2.2. Diagnostic Assessment of Plants and Evaluation of Their Phytosanitary Status
The survey was carried out in March 2019. The experimental fields were commercial olive orchards (cv. Cellina di Nardò, approximately 50 years old) located at Oria (province of Brindisi, southeastern Italy). Twenty-five plants were selected in the same proportions of infected (30%) and uninfected (70%) trees present in the experimental fields in a previous survey of October 2018. Before the survey of March 2019, the phytosanitary status of the selected plants was carefully tested, to exclude the occurrence of pathogens causing vascular disease or leaf desiccation other than Xfp. The occurrence of vascular fungal pathogens was ascertained by isolation on Potato Dextrose Agar (PDA) amended with antibiotics (500 mgL−1, streptomycin sulphate, Sigma-Aldrich). Woody samples (twigs and branchlets) from the selected trees were superficially disinfected with ethanol (80%), then peeled to expose the xylem tissue and examined for the presence of wood discoloration. Subsequently, 10 small wood chips (2 × 2 mm) per sample were seeded onto PDA plates. The developing colonies were checked after 7–14 days incubation at 23 ± 1 °C in dark and transferred in purity to obtain monoconidial isolates. Subsequently, colonies were identified based on their morphological and molecular characteristics.
Three semi-lignified twigs containing at least six mature and asymptomatic leaves, were selected and identified on each monitored plant, to verify without any reasonable doubt, the occurrence of
Xfp infection. The day before the spectral analysis, two leaves per twig were collected and used for a quantitative measurement of the bacterial DNA in the tissues by qPCR [
23,
24]. The leaves were collected with scissors mounted on a telescopic rod, closed in a double plastic bag, and transported to the laboratory where they were stored at 5 °C, to be analyzed within 24 hours from the collection. Leaves were disinfected by dipping for 1 min in an aqueous ethanol solution (70%), and then rinsed in sterile distilled water. For each couple of disinfected leaves, midribs portion (approximately 0.1 g) was used to extract Total Nucleic Acid (TNA), following a standard CTAB-based procedure [
25]. Aliquots of 2.0 μL of TNA were tested in triplicate with quantitative real-time PCR (qPCR) in 25.0 μL of final reaction volume, according to the protocol of Harper et al. (2010) [
26] and using a CFX96 Touch Real-Time PCR apparatus (Biorad, Hercules, CA, USA). As positive control, DNA from a pure culture of
X. fastidiosa subspecies
pauca ST53, growing on buffered charcoal yeast-extract agarized medium (BCYE), was used [
15].
Xfp level occurring in the tested leaves, expressed as the concentration of the bacterial DNA (ng µL
−1), was determined from the standard curve of a serial dilution (10−10
−7 ng µL
−1) of pure bacterial DNA. The genomic DNA quality and quantity were determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA) and a Qubit 2.0 fluorimeter (Life Technologies Ltd., Paisley, UK). For values of cycles threshold (Ct) ≥ 38, twigs were considered negative.
The day of the spectral analysis, one leaf was randomly chosen on each selected twig and the spectral analysis was performed in field. After the test, the three leaves per tree (one per twig) were pooled and used in laboratory for the detection of
Xfp by qPCR as specified above [
25,
26].
Data from the qPCR measurements were subjected to different elaborations, depending on whether they were considered as continuous or binary numerical (0 negative/1 positive) value. In the first case, data were expressed as ng µL−1 of bacterial DNA; in the latter, the value 0 was associated with a DNA amount ≤ the detection limit, so declaring the leaves as negative or uninfected; conversely, 1 was attributed to DNA amount > the detection limit, the leaves were considered as positive or infected.
Each olive tree used in the test was defined as “asymptomatic” when it resulted negative or positive for the Xfp, but neither desiccated leaves nor fungal pathogens (i.e., Verticillium dahliae, Phaeoacremonium spp., Pseudophaeomoniella spp., Pleurostomophora richardsiae, Neofusicoccum spp.) were found following both the visual inspection and the isolation procedure.
2.3. Spectral Analysis
Leaf spectral measurements were performed in the field with Field Spec IV spectroradiometer (Analytical Spectral Devices Inc., Boulder, CO, USA) using artificial light able to detect a spectral signature in a range of 350–2500 nm. The instrument was equipped with three spectrometers: one for the 350–1000 nm region characterized by a sampling interval of 1.4 nm, the second for 1000–1800 nm region and the third for 1800–2500 nm, these last two with a sampling interval of 2 nm.
Field Spec IV provided spectra with 2151 bands having a resolution of 1 nm. The spectral reflectance data were then averaged over 10-nm intervals, to reduce the number of wavelengths from 2151 to 215, to smooth the spectra and to keep down the risk of over-fitting [
27].
A specifically developed leaf clip covering a spot of 10 mm diameter was related to the fiber-optic of spectroradiometer to a halogen lamp 6.5 W, as a light source. In this way, the influence of the atmosphere with the measurements was avoided.
The values of radiance were transformed into spectral reflectance as the ratio between the radiance reflected by the plant and the one from a standard white reference 10 × 10 mm2 disk (Spectralon panel, Lab-sphere, Inc., North Sutton, NH, USA). The calibration was repeated for each tree, thus, increasing the comparability of measurements. One measurement on a spot of 10-mm diameter was replicated three times on the leaf selected for each twig: nine individual spectra were collected for each tree and classified with qPCR performed on the pooled sample of the three leaves (as described before). Then 75 leaves were analyzed with the spectroradiometer (three for each of the 25 trees) for a total of 225 spectra collected. The spectra were analyzed individually rather than averaged at leaf level, because the objective of this work was to assess if the methodology provided by hyperspectral analysis can be useful to monitor the presence of Xylella infection.
2.4. Pre-Processing Methods
Some spectral pre-processing methods were required to reduce the impact of multiplicative and additive effects of eventual backscattering inside the instrument and to achieve a sufficient degree of accuracy for the prediction models [
28].
In this regard, two pretreatments were carried out on reflectance spectra: (1) multiple scatter correction (MSC); and (2) smoothing/denoising with Savitzky–Golay polynomials [
29].
Multiplicative scatter correction (MSC) works primarily when the scatter effect is the dominant source of variability and removes additive and multiplicative components. MSC simplifies the calibration model reducing the number of components needed and may improve the linearity between prediction and predictors [
30].
Savitzky–Golay (SG) first-order polynomial algorithm reduces the random noise of the measurements. The algorithm uses a moving polynomial fit of any order and the size of the filter consists of (2
n + 1) points, where n is the half-width of the smoothing window (
w). The polynomial fit interpolates the points between the 2
n’s. A window size (
w) of 11 (
w = 2
n + 1) and the second polynomial order were applied here [
31].
Spectral data pre-processing was performed with ParLeS software [
32].
When the response variable (DNA content) departed from normal distribution, it was transformed in Gaussian ranks by SAS/RANK procedure: the ranks divided by the total number of observations form values in the range 0 to 1, which were used in subsequent processing. Predictors (spectral data) and response variable (Gaussian rank transformed DNA data) were centered and scaled to have the mean at zero and the variance at 1, to place both on the same relative position to their variation in the data in the process of prediction model estimation.
2.5. Prediction Model
Among the available multivariate statistical methods to estimate DNA content, Partial Least Squares Regression (PLSR) [
28,
33] was chosen, due to the high number of predictors. PLSR is based on decomposition of two sets of variables: the matrix
X of predictors (matrix
n x
NX, where
n is the number of observations, 225, and
NX is the number of reflectance data at different wavelengths, 215) and the vector
Y of the response variable (vector
n x
1 of DNA data). PLSR selects successive orthogonal factors (or latent variables) that maximize the covariance between predictors (
X reflectance) and response variable (
Y) and explains most of the variation in both. PLSR decomposes
X (1) and y (2) into factor scores (
T) and factor loadings (
P′ and
q) according to:
A good PLSR model has few PLSR factors (T) that explain most of the variation in both predictors and responses. The residuals E and f are negligible. Therefore, by combining the two previous equations, the variable y (DNA content) can be estimated as a linear combination of a smaller number of spectra (x), through the estimated loadings [
33]. For more details on PLSR method, see, e.g., Martens and Næs (1989) [
28].
Cross-validation was performed to determine the number of latent variables that minimized the predicted residual sum of squares, followed by Van der Voet test (1994) [
34] used to identify the simplest model with a not significantly larger error than the absolute minimum.
In validation, the same data set was randomly split into two datasets: two-third of the total data set formed the calibration set and the remaining one-third of the samples, the validation set. This procedure was repeated one hundred times, to verify robustness and stability of the models and then the calculated statistics at each iter were averaged. The performance of PLSR prediction model was evaluated by means of three statistics: (1) the coefficient of determination
R2 of regression: observation vs. prediction; (2) root mean square error (RMSE); and 3) the ratio of performance to interquartile distance (RPIQ) [
35] defined as follows:
is the observed values, is their average, the predicted values and n is the number of samples used for validation with i = 1, 2, ..., n.
IQ is the difference between the third and first quartiles of y:
and measures the spread of the data. RMSE and
IQ have the same unit of DNA measurement; therefore, RPIQ is unitless and measures the accuracy of the prediction model [
36]. R
2 measures the proportion of variance of observations explained by the model.
After a preliminary cross-validation test using raw DNA data, the residuals were tested for normality with Shapiro–Wilk and Kolmogorov–Smirnov tests. As such, in case of distribution deviated from the normal one, the DNA data were submitted to normalizing rank transformation [
37,
38,
39]. From now on, all elaborations refer to the normal ranks of DNA data.
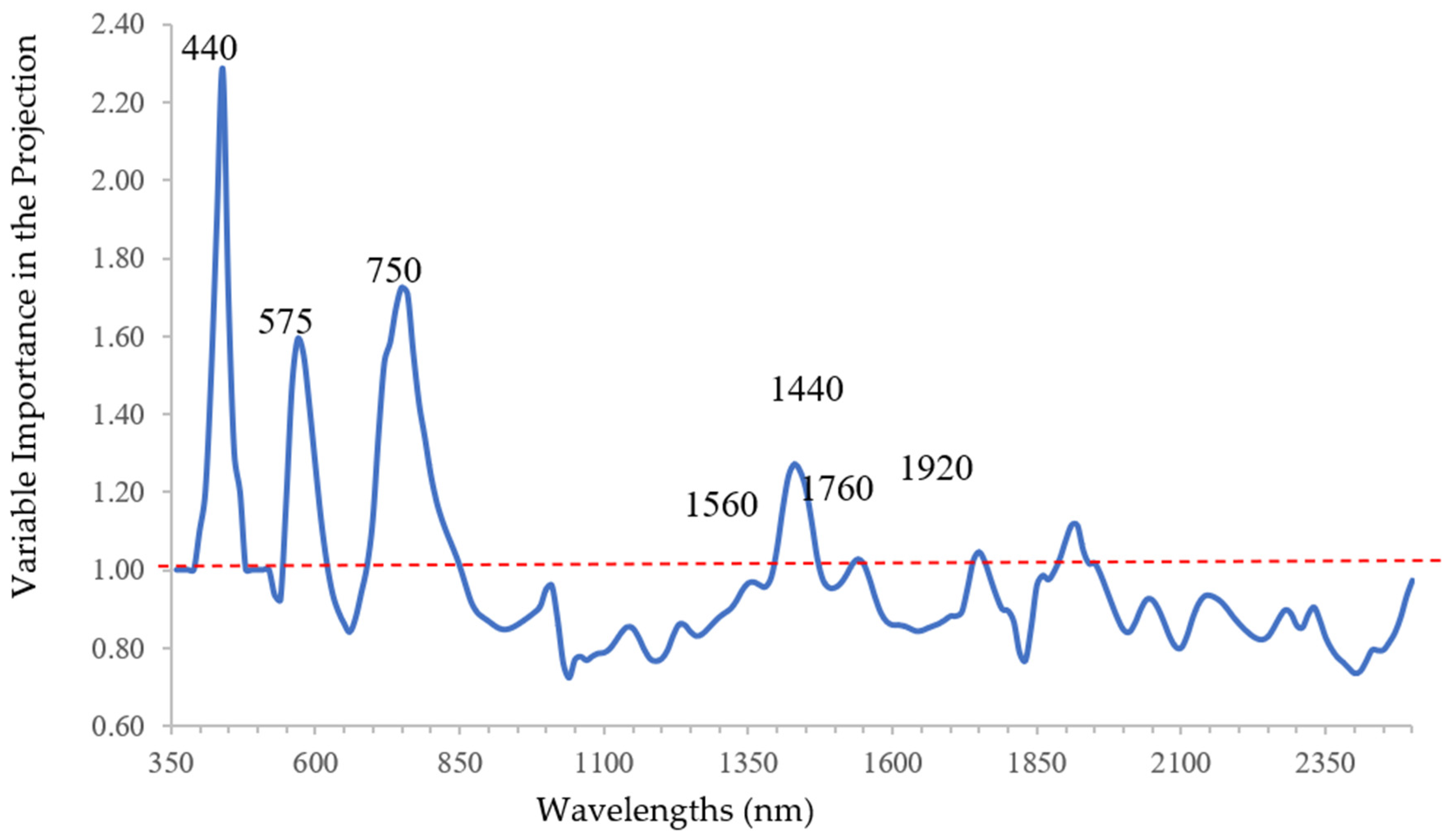
Variable Importance in the Projection (VIP) statistic was used for the selection of the relevant wavelengths since it gives some insights on the relative importance of wavelengths to predict DNA content by qPCR analysis [
40]. In fact, VIP statistics consider both predictors and response variables, because it is a weighted sum of squares of PLSR X-score coefficients (loadings) for the retained components (latent variables), with the weights calculated from the amount of dependent variable (y) variance explained by each retained component [
41]. Since the average of squared VIP scores equals 1 [
42,
43], a threshold value of 1 was chosen and the wavelengths with a VIP score greater than 1 were considered significant.
Discriminant analysis is a multivariate statistical technique that is commonly used as a powerful classification approach when response variable is categorical: it uses multiple quantitative attributes to discriminate single classification variable. Each observation is placed in the class from which it has the smallest generalized squared distance (
D) [
44], calculated for each group (
j) according to the following formula:
where
X is the vector of multivariate observations for a given pixel of the map,
is the vector of the means of variates for the class
j and
COV is the covariance matrix [
45,
46,
47].
In the present study, discriminant analysis was used to investigate the radiometric differences between the two categories indicated with the value 0, when Xfp was absent, and the value 1 when it was present.
The two procedures, PLSR and discriminant analysis, should not be considered in an alternative but integrated way. The first allowed us to identify those bands that contribute most to the prediction of DNA content, considered as a continuous numerical variable. The discriminant analysis instead allowed us to identify those wavelengths that most contribute to the separation of the two classes previously defined and based on the fixed threshold value of DNA. It, therefore, depended strongly on the criterion used for this classification as well as, similarly to PLSR, on the quality of DNA measurements.
The discriminant analysis consisted of the following six steps:
4. Conclusions
Hyperspectral data were analyzed with a set of integrative statistical techniques, to evaluate their potential in detecting olive plants infected by Xfp. The different methods produced consistent and integrable results for a more comprehensive understanding of the phenomenon under investigation.
When spectral data were analyzed as predictors of bacterial DNA content, using PLSR, the bands related to chlorophyll function were the most relevant. As regards to the discrimination between infected and uninfected plants, previously classified, CDA and stepwise added other bands related to water content and lignin, since desiccation is a clear symptom of Xfp infection. These results emphasize the usefulness of using multiple statistical techniques, not in an alternative but in a complementary way.
The confusion matrix for CDA showed that the model had a good overall accuracy of 0.67, but with a better capability to discriminate infected plants as compared to the uninfected ones. This not excellent result was probably due to the use of a composite sample for real time-PCR analysis. In this regard, it should be advisable to run qPCR measurements on a single leaf, thus, improving the matching between radiometric and laboratory measurement supports.
Unsupervised classification, using the spectral data only, was able to discriminate the infected plants at a very early stage of infection or even asymptomatic, on which sampling/monitoring should be preferably directed, thus, saving time and financial resources. However, the analyses also highlighted the limitations of the hyperspectral radiometer sensor, which cannot be used as the unique diagnostic tool, even after accurate calibration, due to the plethora of factors that might produce similar symptoms. With the statistical analyses used, the selected wavelengths were associated to biological functions, thus, confirming the results from other works.
An interesting prospect for future research would be to verify if the same results could be obtained by analyzing both full hyperspectral spectra and discrete spectra, including only the wavelengths identified by discriminant analysis. This would make it possible to design lightweight and efficient hyperspectral radiometers to be used on board drones for a preventive survey of olive groves. Unfortunately, it cannot be established a priori, given the multitude of factors that can affect spectral signatures and are not uniquely linked to Xfp infection. Nevertheless, one of the most significant results of this research is marking out paths for future research.
Finally, an integrated system of statistical techniques for the processing of radiometric data has also been proposed, that could be used as decision support for planning an efficient and cost-effective sampling/monitoring scheme of the quarantine pathogens. Given the complexity of the phenomenon under study, an interdisciplinary approach including in the research team statisticians, experts in imaging analysis, plant pathologists, epidemiologist, and agronomists, should be strongly encouraged.