Genomic Variation and Diversification in Begomovirus Genome in Implication to Host and Vector Adaptation
Abstract
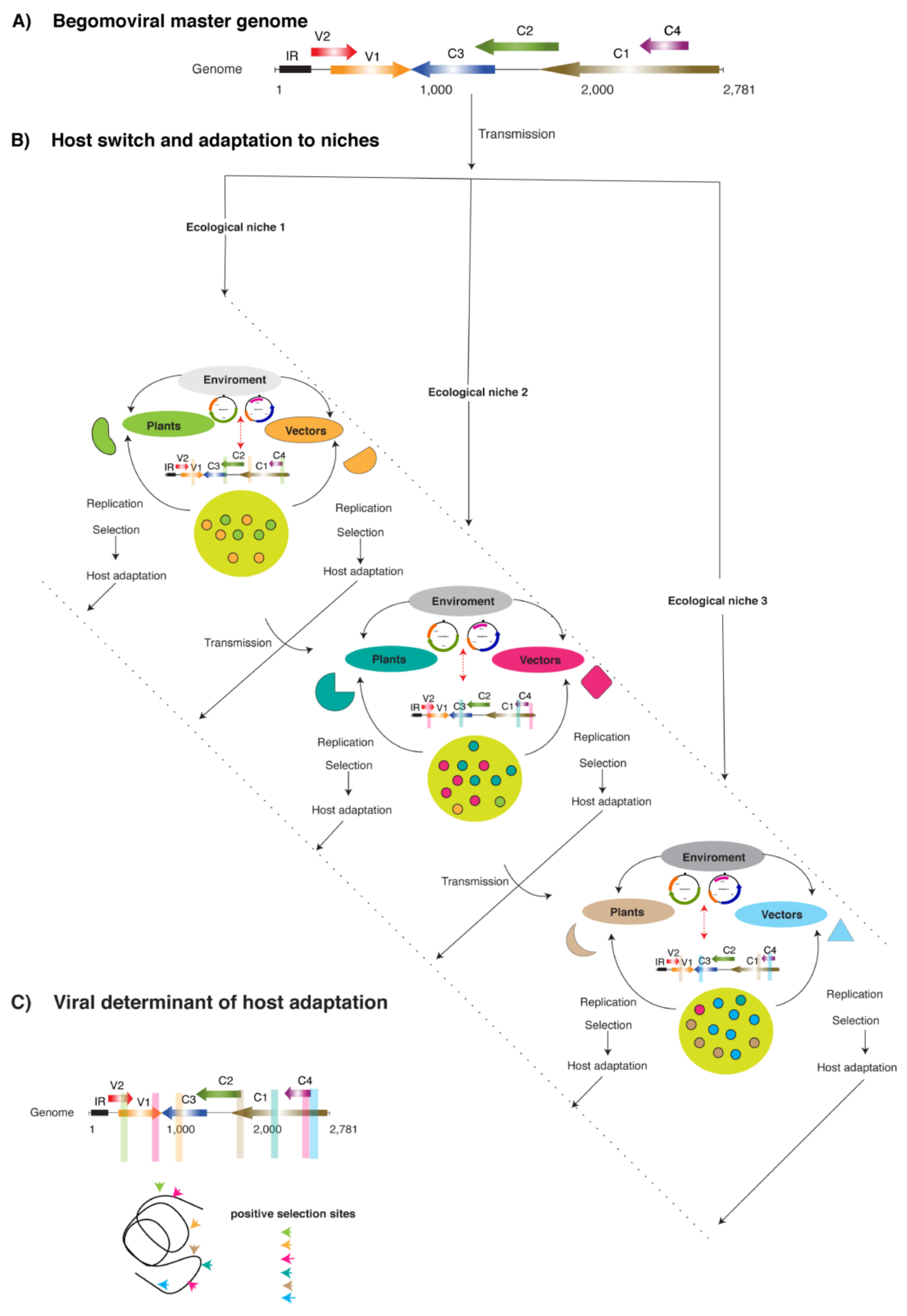
1. Introduction
2. Discussion
3. Conclusions
4. Prospects
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Krupovic, M.; Varsani, A.; Kazlauskas, D.; Breitbart, M.; Delwart, E.; Rosario, K.; Yutin, N.; Wolf, Y.I.; Harrach, B.; Zerbini, F.M.; et al. Cressdnaviricota: A Virus Phylum Unifying Seven Families of Rep-Encoding Viruses with Single-Stranded, Circular DNA Genomes. J. Virol. 2020, 94, e00582-20. [Google Scholar] [CrossRef]
- Dolja, V.V.; Krupovic, M.; Koonin, E.V. Deep roots and splendid boughs of the global plant virome. Annu. Rev. Phytopathol. 2020, 58, 23–53. [Google Scholar] [CrossRef]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A.; Consortium, I.R. ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Navas-Castillo, J. Molecular and biological characterization of a New World mono-/bipartite begomovirus/deltasatellite complex infecting Corchorus siliquosus. Front. Microbiol. 2020, 11, 1755. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Idriss, M.; Abdallah, N.; Aref, N.; Haridy, G.; Madkour, M. Biotypes of the castor bean whitefly Trialeurodes ricini (Misra)(Hom., Aleyrodidae) in Egypt: Biochemical characterization and efficiency of geminivirus transmission. J. Appl. Entomol. 1997, 121, 501–509. [Google Scholar] [CrossRef]
- Sangeetha, B.; Malathi, V.; Alice, D.; Suganthy, M.; Renukadevi, P. A distinct seed-transmissible strain of tomato leaf curl New Delhi virus infecting Chayote in India. Virus Res. 2018, 258, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Sohrab, S.S. Molecular diagnosis of begomovirus associated with Chilli leaf curl disease in Jeddah, Saudi Arabia. Saudi J. Biol. Sci. 2020, 27, 3060–3064. [Google Scholar] [CrossRef] [PubMed]
- Torres-Herrera, S.I.; Romero-Osorio, A.; Moreno-Valenzuela, O.; Pastor-Palacios, G.; Cardenas-Conejo, Y.; Ramírez-Prado, J.H.; Riego-Ruiz, L.; Minero-García, Y.; Ambriz-Granados, S.; Argüello-Astorga, G.R. A lineage of begomoviruses encode Rep and AC4 proteins of enigmatic ancestry: Hints on the evolution of geminiviruses in the New World. Viruses 2019, 11, 644. [Google Scholar] [CrossRef]
- Lazarowitz, S.G.; Wu, L.C.; Rogers, S.G.; Elmer, J.S. Sequence-specific interaction with the viral AL1 protein identifies a geminivirus DNA replication origin. Plant Cell 1992, 4, 799–809. [Google Scholar]
- Argüello-Astorga, G.; Guevara-Gonzalez, R.; Herrera-Estrella, L.; Rivera-Bustamante, R. Geminivirus replication origins have a group-specific organization of iterative elements: A model for replication. Virology 1994, 203, 90–100. [Google Scholar] [CrossRef] [PubMed]
- García-Arriaza, J.; Manrubia, S.C.; Toja, M.; Domingo, E.; Escarmís, C. Evolutionary transition toward defective RNAs that are infectious by complementation. J. Virol. 2004, 78, 11678–11685. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Patil, B.L.; Bagewadi, B.; Nawaz-ul-Rehman, M.S.; Fauquet, C.M. Distinct evolutionary histories of the DNA-A and DNA-B components of bipartite begomoviruses. BMC Evol. Biol. 2010, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Romay, G.; Geraud-Pouey, F.; Chirinos, D.T.; Mahillon, M.; Gillis, A.; Mahillon, J.; Bragard, C. Tomato twisted leaf virus: A novel indigenous new world monopartite begomovirus infecting tomato in Venezuela. Viruses 2019, 11, 327. [Google Scholar] [CrossRef]
- Czosnek, H.; Hariton-Shalev, A.; Sobol, I.; Gorovits, R.; Ghanim, M. The incredible journey of begomoviruses in their whitefly vector. Viruses 2017, 9, 273. [Google Scholar] [CrossRef]
- De Barro, P.J.; Liu, S.-S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A statement of species status. Annu. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef]
- LIU, S.-S.; Colvin, J.; De Barro, P.J. Species concepts as applied to the whitefly Bemisia tabaci systematics: How many species are there? J. Integr. Agric. 2012, 11, 176–186. [Google Scholar] [CrossRef]
- Firdaus, S.; Vosman, B.; Hidayati, N.; Jaya Supena, E.D.; GF Visser, R.; van Heusden, A.W. The Bemisia tabaci species complex: Additions from different parts of the world. Insect Sci. 2013, 20, 723–733. [Google Scholar] [CrossRef]
- Manivannan, K.; Renukadevi, P.; Malathi, V.G.; Karthikeyan, G.; Balakrishnan, N. A new seed-transmissible begomovirus in bitter gourd (Momordica charantia L.). Microb. Pathog. 2019, 128, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Chowda-Reddy, R.; Kirankumar, M.; Seal, S.E.; Muniyappa, V.; Valand, G.B.; Govindappa, M.; Colvin, J. Bemisia tabaci phylogenetic groups in India and the relative transmission efficacy of Tomato leaf curl Bangalore virus by an indigenous and an exotic population. J. Integr. Agric. 2012, 11, 235–248. [Google Scholar] [CrossRef]
- Gong, S. Investigating Vector-Virus-Plant Interactions Influencing Transmission Efficiency of Tomato Yellow Leaf Curl Virus and Tomato Mottle Virus by Bemisia tabaci. 2018. Available online: https://etd.auburn.edu/handle/10415/6317 (accessed on 13 August 2021).
- Bucciarelli, G.; Golani, D.; Bernardi, G. Genetic cryptic species as biological invaders: The case of a Lessepsian fish migrant, the hardyhead silverside Atherinomorus lacunosus. J. Exp. Mar. Biol. Ecol. 2002, 273, 143–149. [Google Scholar] [CrossRef]
- Johnson, R.N.; Starks, P.T. A surprising level of genetic diversity in an invasive wasp: Polistes dominulus in the northeastern United States. Ann. Entomol. Soc. Am. 2004, 97, 732–737. [Google Scholar] [CrossRef]
- Miura, O. Molecular genetic approaches to elucidate the ecological and evolutionary issues associated with biological invasions. Ecol. Res. 2007, 22, 876–883. [Google Scholar] [CrossRef]
- Rosario, K.; Marr, C.; Varsani, A.; Kraberger, S.; Stainton, D.; Moriones, E.; Polston, J.E.; Breitbart, M. Begomovirus-associated satellite DNA diversity captured through vector-enabled metagenomic (VEM) surveys using whiteflies (Aleyrodidae). Viruses 2016, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Tovar, R.; Navas-Castillo, J. Deciphering the biology of deltasatellites from the New World: Maintenance by New World begomoviruses and whitefly transmission. New Phytol. 2016, 212, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Idris, A.; Alteri, C.; Stenger, D.C. Emergence of a new cucurbit-infecting begomovirus species capable of forming viable reassortants with related viruses in the Squash leaf curl virus cluster. Phytopathology 2002, 92, 734–742. [Google Scholar] [CrossRef]
- Xie, Y.; Wu, P.; Liu, P.; Gong, H.; Zhou, X. Characterization of alphasatellites associated with monopartite begomovirus/betasatellite complexes in Yunnan, China. Virol. J. 2010, 7, 1–10. [Google Scholar] [CrossRef]
- Hameed, U.; Zia-Ur-Rehman, M.; Herrmann, H.-W.; Haider, M.; Brown, J.K. First report of Okra enation leaf curl virus and associated cotton leaf curl Multan betasatellite and cotton leaf curl Multan alphasatellite infecting cotton in Pakistan: A new member of the cotton leaf curl disease complex. Plant Dis. 2014, 98, 1447. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.M.; Shahid, M.S.; Briddon, R.W.; Khan, A.; Zhu, J.-K.; Brown, J.K. An unusual alphasatellite associated with monopartite begomoviruses attenuates symptoms and reduces betasatellite accumulation. J. Gen. Virol. 2011, 92, 706–717. [Google Scholar] [CrossRef]
- Rahman, M.-u.; Khan, A.Q.; Rahmat, Z.; Iqbal, M.A.; Zafar, Y. Genetics and genomics of cotton leaf curl disease, its viral causal agents and whitefly vector: A way forward to sustain cotton fiber security. Front. Plant Sci. 2017, 8, 1157. [Google Scholar] [CrossRef]
- Kumar, R.V.; Singh, D.; Singh, A.K.; Chakraborty, S. Molecular diversity, recombination and population structure of alphasatellites associated with begomovirus disease complexes. Infect. Genet. Evol. 2017, 49, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.; Brown, J.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X.; Fauquet, C. Recommendations for the classification and nomenclature of the DNA-β satellites of begomoviruses. Arch. Virol. 2008, 153, 763–781. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Usha, R. Bhendi yellow vein mosaic disease in India is caused by association of a DNA β satellite with a begomovirus. Virology 2003, 305, 310–317. [Google Scholar] [CrossRef]
- Saunders, K.; Briddon, R.W.; Stanley, J. Replication promiscuity of DNA-β satellites associated with monopartite begomoviruses; deletion mutagenesis of the Ageratum yellow vein virus DNA-β satellite localizes sequences involved in replication. J. Gen. Virol. 2008, 89, 3165–3172. [Google Scholar] [CrossRef]
- Fauquet, C.; Briddon, R.; Brown, J.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X. Geminivirus strain demarcation and nomenclature. Arch. Virol. 2008, 153, 783–821. [Google Scholar] [CrossRef]
- Simmonds, P.; Aiewsakun, P.; Katzourakis, A. Prisoners of war—Host adaptation and its constraints on virus evolution. Nat. Rev. Microbiol. 2019, 17, 321–328. [Google Scholar] [CrossRef]
- Morales, F.J.; Anderson, P.K. The emergence and dissemination of whitefly-transmitted geminiviruses in Latin America. Arch. Virol. 2001, 146, 415–441. [Google Scholar] [CrossRef]
- Duffy, S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018, 16, e3000003. [Google Scholar] [CrossRef]
- Nigam, D.; Garcia-Ruiz, H. Variation profile of the orthotospovirus genome. Pathogens 2020, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Lima, A.; Silva, J.C.; Silva, F.N.; Castillo-Urquiza, G.P.; Silva, F.F.; Seah, Y.M.; Mizubuti, E.S.; Duffy, S.; Zerbini, F.M. The diversification of begomovirus populations is predominantly driven by mutational dynamics. Virus Evol. 2017, 3, vex005. [Google Scholar] [CrossRef] [PubMed]
- Nigam, D.; LaTourrette, K.; Garcia-Ruiz, H. Mutations in virus-derived small RNAs. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nigam, D.; LaTourrette, K.; Souza, P.F.; Garcia-Ruiz, H. Genome-wide variation in potyviruses. Front. Plant Sci. 2019, 10, 1439. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.T.; Sobrinho, R.R.; Gonzalez-Aguilera, J.; Rocha, C.S.; Silva, S.J.; Xavier, C.A.; Silva, F.N.; Duffy, S.; Zerbini, F.M. Synonymous site variation due to recombination explains higher genetic variability in begomovirus populations infecting non-cultivated hosts. J. Gen. Virol. 2013, 94, 418–431. [Google Scholar] [CrossRef]
- Rocha, C.S.; Castillo-Urquiza, G.P.; Lima, A.T.; Silva, F.N.; Xavier, C.A.; Hora-Júnior, B.T.; Beserra-Júnior, J.E.; Malta, A.W.; Martin, D.P.; Varsani, A. Brazilian begomovirus populations are highly recombinant, rapidly evolving, and segregated based on geographical location. J. Virol. 2013, 87, 5784–5799. [Google Scholar] [CrossRef]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef]
- Ge, L.; Zhang, J.; Zhou, X.; Li, H. Genetic structure and population variability of tomato yellow leaf curl China virus. J. Virol. 2007, 81, 5902–5907. [Google Scholar] [CrossRef] [PubMed]
- Pita, J.; Fondong, V.; Sangare, A.; Otim-Nape, G.; Ogwal, S.; Fauquet, C. Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 2001, 82, 655–665. [Google Scholar] [CrossRef]
- Farooq, T.; Umar, M.; She, X.; Tang, Y.; He, Z. Molecular Phylogenetics and Evolutionary Analysis of a Highly Recombinant Begomovirus, Cotton Leaf Curl Multan Virus and Associated Satellites. Virus Evol. 2021, 1–15. [Google Scholar] [CrossRef]
- Ismayil, A.; Yang, M.; Haxim, Y.; Wang, Y.; Li, J.; Han, L.; Wang, Y.; Zheng, X.; Wei, X.; Nagalakshmi, U. Cotton leaf curl Multan virus βC1 protein induces autophagy by disrupting the interaction of autophagy-related protein 3 with glyceraldehyde-3-phosphate dehydrogenases. Plant Cell 2020, 32, 1124–1135. [Google Scholar] [CrossRef]
- Zhang, X.-P.; Liu, D.-S.; Yan, T.; Fang, X.-D.; Dong, K.; Xu, J.; Wang, Y.; Yu, J.-L.; Wang, X.-B. Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta. PLoS Pathog. 2017, 13, e1006522. [Google Scholar] [CrossRef] [PubMed]
- Watt, L.G.; Crawshaw, S.; Rhee, S.-J.; Murphy, A.M.; Canto, T.; Carr, J.P. The cucumber mosaic virus 1a protein regulates interactions between the 2b protein and ARGONAUTE 1 while maintaining the silencing suppressor activity of the 2b protein. PLoS Pathog. 2020, 16, e1009125. [Google Scholar] [CrossRef]
- Pasin, F.; Shan, H.; García, B.; Müller, M.; San León, D.; Ludman, M.; Fresno, D.H.; Fátyol, K.; Munné-Bosch, S.; Rodrigo, G. Abscisic acid connects phytohormone signaling with RNA metabolic pathways and promotes an antiviral response that is evaded by a self-controlled RNA virus. Plant Commun. 2020, 1, 100099. [Google Scholar] [CrossRef] [PubMed]
- Ismayil, A.; Haxim, Y.; Wang, Y.; Li, H.; Qian, L.; Han, T.; Chen, T.; Jia, Q.; Yihao Liu, A.; Zhu, S. Cotton Leaf Curl Multan virus C4 protein suppresses both transcriptional and post-transcriptional gene silencing by interacting with SAM synthetase. PLoS Pathog. 2018, 14, e1007282. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Pérez, M.; Aparicio, F.; López-Gresa, M.P.; Bellés, J.M.; Sánchez-Navarro, J.A.; Pallás, V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef]
- Devendran, R.; Kumar, M.; Ghosh, D.; Yogindran, S.; Karim, M.J.; Chakraborty, S. Capsicum-infecting begomoviruses as global pathogens: Host–virus interplay, pathogenesis, and management. Trends Microbiol. 2021. S0966-842X(21)00129-3. Available online: https://www.cell.com/trends/microbiology/fulltext/S0966-842X(21)00129-3 (accessed on 13 August 2021).
- Gaur, R.K.; Khurana, S.P.; Sharma, P.; Hohn, T. Plant Virus-Host Interaction: Molecular Approaches and Viral Evolution; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Zhao, P.; Yao, X.; Cai, C.; Li, R.; Du, J.; Sun, Y.; Wang, M.; Zou, Z.; Wang, Q.; Kliebenstein, D.J. Viruses mobilize plant immunity to deter nonvector insect herbivores. Sci. Adv. 2019, 5, eaav9801. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Kumar Kushwaha, N.; Kumar Singh, A.; Pankaj Sahu, P.; Vinoth Kumar, R.; Chakraborty, S. Dynamics of a geminivirus-encoded pre-coat protein and host RNA-dependent RNA polymerase 1 in regulating symptom recovery in tobacco. J. Exp. Bot. 2018, 69, 2085–2102. [Google Scholar] [CrossRef]
- Mei, Y.; Zhang, F.; Wang, M.; Li, F.; Wang, Y.; Zhou, X. Divergent symptoms caused by geminivirus-encoded C4 proteins correlate with their ability to bind NbSKη. J. Virol. 2020, 94, e01307-20. [Google Scholar] [CrossRef]
- Martins, L.G.; Raimundo, G.A.; Ribeiro, N.G.; Silva, J.C.F.; Euclydes, N.C.; Loriato, V.A.; Duarte, C.E.; Fontes, E.P. A begomovirus nuclear shuttle protein-interacting immune hub: Hijacking host transport activities and suppressing incompatible functions. Front. Plant Sci. 2020, 11, 398. [Google Scholar] [CrossRef]
- Wang, X.-R.; Wang, C.; Ban, F.-X.; Ghanim, M.; Pan, L.-L.; Qian, L.-X.; Liu, Y.-Q.; Wang, X.-W.; Liu, S.-S. Apoptosis in a whitefly vector activated by a begomovirus enhances viral transmission. Msystems 2020, 5, e00433-20. [Google Scholar] [CrossRef]
- Ohnesorge, S.; Bejarano, E. Begomovirus coat protein interacts with a small heat-shock protein of its transmission vector (Bemisia tabaci). Insect Mol. Biol. 2009, 18, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.; Pinner, M.; Stanley, J.; Markham, P. Geminivirus coat protein gene replacement alters insect specificity. Virology 1990, 177, 85–94. [Google Scholar] [CrossRef]
- Roberts, I.; Robinson, D.; Harrison, B. Serological relationships and genome homologies among geminiviruses. J. Gen. Virol. 1984, 65, 1723–1730. [Google Scholar] [CrossRef]
- Mondal, D.; Mandal, S.; Shil, S.; Sahana, N.; Pandit, G.K.; Choudhury, A. Genome wide molecular evolution analysis of begomoviruses reveals unique diversification pattern in coat protein gene of Old World and New World viruses. Virusdisease 2019, 30, 74. [Google Scholar] [CrossRef]
- Pan, L.-L.; Chi, Y.; Liu, C.; Fan, Y.-Y.; Liu, S.-S. Mutations in the coat protein of a begomovirus result in altered transmission by different species of whitefly vectors. Virus Evol. 2020, 6, veaa014. [Google Scholar] [CrossRef]
- Xie, K.; Yang, Y. RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.-J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 1–10. [Google Scholar] [CrossRef]
- El-Mounadi, K.; Morales-Floriano, M.L.; Garcia-Ruiz, H. Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front. Plant Sci. 2020, 11, 56. [Google Scholar] [CrossRef]
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.-e.-A.; Mahfouz, M.M. CRISPR/Cas9-mediated immunity to geminiviruses: Differential interference and evasion. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.-e.-A.; Mahas, A.; Vanderschuren, H.; Mahfouz, M.M. Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 2020, 21, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Stürchler, A.; Anjanappa, R.B.; Zaidi, S.S.-e.-A.; Hirsch-Hoffmann, M.; Gruissem, W.; Vanderschuren, H. Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 2019, 20, 1–10. [Google Scholar] [CrossRef]
- Gupta, N.; Reddy, K.; Bhattacharyya, D.; Chakraborty, S. Plant responses to geminivirus infection: Guardians of the plant immunity. Virol. J. 2021, 18, 1–25. [Google Scholar] [CrossRef]
- Prasad, A.; Sharma, N.; Hari-Gowthem, G.; Muthamilarasan, M.; Prasad, M. Tomato yellow leaf curl virus: Impact, challenges, and management. Trends Plant Sci. 2020, 25, 897–911. [Google Scholar] [CrossRef]
- Prabhandakavi, P.; Pogiri, R.; Kumar, R.; Acharya, S.; Esakky, R.; Chakraborty, M.; Pinnamaneni, R.; Palicherla, S.R. Pyramiding Ty-1/Ty-3, Ty-2, ty-5 and ty-6 genes into tomato hybrid to develop resistance against tomato leaf curl viruses and recurrent parent genome recovery by ddRAD sequencing method. J. Plant Biochem. Biotechnol. 2021, 30, 462–476. [Google Scholar] [CrossRef]
- Prabhandakavi, P.; Kumar, R.; Acharya, S.; Chakraborty, M.; Rambabu, P.; Palicherla, S.R.; Pinnamaneni, R. Evaluation of Tomato Inbred Lines Harboring Ty Gene (s) for Resistance Against Monopartite and Bipartite Begomoviruses. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2021, 91, 45–52. [Google Scholar] [CrossRef]
- Gill, U.; Scott, J.W.; Shekasteband, R.; Ogundiwin, E.; Schuit, C.; Francis, D.M.; Sim, S.-C.; Smith, H.; Hutton, S.F. Ty-6, a major begomovirus resistance gene on chromosome 10, is effective against Tomato yellow leaf curl virus and Tomato mottle virus. Theor. Appl. Genet. 2019, 132, 1543–1554. [Google Scholar] [CrossRef]
- Colvin, J.; Omongo, C.; Govindappa, M.; Stevenson, P.C.; Maruthi, M.; Gibson, G.; Seal, S.; Muniyappa, V. Host-plant viral infection effects on arthropod-vector population growth, development and behaviour: Management and epidemiological implications. Adv. Virus Res. 2006, 67, 419–452. [Google Scholar]
- Legg, J.; French, R.; Rogan, D.; Okao-Okuja, G.; Brown, J. A distinct Bemisia tabaci (Gennadius)(Hemiptera: Sternorrhyncha: Aleyrodidae) genotype cluster is associated with the epidemic of severe cassava mosaic virus disease in Uganda. Mol. Ecol. 2002, 11, 1219–1229. [Google Scholar] [CrossRef]
- Colvin, J.; Omongo, C.; Maruthi, M.; Otim-Nape, G.; Thresh, J. Dual begomovirus infections and high Bemisia tabaci populations: Two factors driving the spread of a cassava mosaic disease pandemic. Plant Pathol. 2004, 53, 577–584. [Google Scholar] [CrossRef]
- Mansoor, S.; Amin, I.; Iram, S.; Hussain, M.; Zafar, Y.; Malik, K.; Briddon, R. Breakdown of resistance in cotton to cotton leaf curl disease in Pakistan. Plant Pathol. 2003, 52, 784. [Google Scholar] [CrossRef]
- Longdon, B.; Brockhurst, M.A.; Russell, C.A.; Welch, J.J.; Jiggins, F.M. The evolution and genetics of virus host shifts. PLoS Pathog. 2014, 10, e1004395. [Google Scholar] [CrossRef]
- Bakhshandeh, B.; Sorboni, S.G.; Javanmard, A.-R.; Mottaghi, S.S.; Mehrabi, M.-R.; Sorouri, F.; Abbasi, A.; Jahanafrooz, Z. Variants in ACE2; potential influences on virus infection and COVID-19 severity. Infect. Genet. Evol. 2021, 90, 104773. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Fares, M.A.; Elena, S.F. Molecular evolution of viral multifunctional proteins: The case of potyvirus HC-Pro. J. Mol. Evol. 2014, 78, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Priya, P.; Shanker, A. Coevolutionary forces shaping the fitness of SARS-CoV-2 spike glycoprotein against human receptor ACE2. Infect. Genet. Evol. 2021, 87, 104646. [Google Scholar] [CrossRef] [PubMed]
- Brito, A.F.; Pinney, J.W. Protein-protein interactions in virus-host systems. Front. Microbiol. 2017, 8, 1557. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, S.; Torbett, B.E. Interactions of HIV-1 Capsid with Host Factors and Their Implications for Developing Novel Therapeutics. Viruses 2021, 13, 417. [Google Scholar] [CrossRef] [PubMed]
- Donlin, M.J.; Szeto, B.; Gohara, D.W.; Aurora, R.; Tavis, J.E. Genome-wide networks of amino acid covariances are common among viruses. J. Virol. 2012, 86, 3050–3063. [Google Scholar] [CrossRef]
- Liang, X.-Z.; Lee, B.T.; Wong, S.-M. Covariation in the capsid protein of Hibiscus chlorotic ringspot virus induced by serial passaging in a host that restricts movement leads to avirulence in its systemic host. J. Virol. 2002, 76, 12320–12324. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sruthi, C.; Prakash, M.K. Statistical characteristics of amino acid covariance as possible descriptors of viral genomic complexity. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Martínez, E.S.; Bosque-Pérez, N.A.; Berger, P.H.; Zemetra, R.S.; Ding, H.; Eigenbrode, S.D. Volatile cues influence the response of Rhopalosiphum padi (Homoptera: Aphididae) to Barley yellow dwarf virus–infected transgenic and untransformed wheat. Environ. Entomol. 2004, 33, 1207–1216. [Google Scholar] [CrossRef]
- Bosque-Pérez, N.A. Eight decades of maize streak virus research. Virus Res. 2000, 71, 107–121. [Google Scholar] [CrossRef]
- Jeger, M.J. The epidemiology of plant virus disease: Towards a new synthesis. Plants 2020, 9, 1768. [Google Scholar] [CrossRef] [PubMed]
- Jeger, M.; Van Den Bosch, F.; Madden, L.; Holt, J. A model for analysing plant-virus transmission characteristics and epidemic development. Math. Med. Biol. A J. IMA 1998, 15, 1–18. [Google Scholar] [CrossRef]
- Jeger, M.; Holt, J.; Van Den Bosch, F.; Madden, L. Epidemiology of insect-transmitted plant viruses: Modelling disease dynamics and control interventions. Physiol. Entomol. 2004, 29, 291–304. [Google Scholar] [CrossRef]
- Madden, L.; Jeger, M.; Van den Bosch, F. A theoretical assessment of the effects of vector-virus transmission mechanism on plant virus disease epidemics. Phytopathology 2000, 90, 576–594. [Google Scholar] [CrossRef]
- Domingo, E. Molecular basis of genetic variation of viruses: Error-prone replication. Virus Popul. 2020, 35–71. [Google Scholar]
- Xu, X.; Qian, Y.; Wang, Y.; Li, Z.; Zhou, X. Iterons homologous to helper geminiviruses are essential for efficient replication of betasatellites. J. Virol. 2019, 93, e01532-18. [Google Scholar] [CrossRef]
- Briddon, R.W. Cotton leaf curl disease, a multicomponent begomovirus complex. Mol. Plant Pathol. 2003, 4, 427–434. [Google Scholar] [CrossRef]
- García-Arenal, F.; McDonald, B.A. An analysis of the durability of resistance to plant viruses. Phytopathology 2003, 93, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 1997, 35, 191–209. [Google Scholar] [CrossRef] [PubMed]
- Pasin, F. Oligonucleotide abundance biases aid design of a type IIS synthetic genomics framework with plant virome capacity. Biotechnol. J. 2021, 16, 2000354. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nigam, D. Genomic Variation and Diversification in Begomovirus Genome in Implication to Host and Vector Adaptation. Plants 2021, 10, 1706. https://doi.org/10.3390/plants10081706
Nigam D. Genomic Variation and Diversification in Begomovirus Genome in Implication to Host and Vector Adaptation. Plants. 2021; 10(8):1706. https://doi.org/10.3390/plants10081706
Chicago/Turabian StyleNigam, Deepti. 2021. "Genomic Variation and Diversification in Begomovirus Genome in Implication to Host and Vector Adaptation" Plants 10, no. 8: 1706. https://doi.org/10.3390/plants10081706
APA StyleNigam, D. (2021). Genomic Variation and Diversification in Begomovirus Genome in Implication to Host and Vector Adaptation. Plants, 10(8), 1706. https://doi.org/10.3390/plants10081706




