Abstract
The root-knot nematode (Meloidogyne incognita) causes huge economic losses in the agricultural industry throughout the world. Control methods against these polyphagous plant endoparasites are sparse, the preferred one being the deployment of plant cultivars or rootstocks bearing resistance genes against Meloidogyne species. Our previous study has cloned one resistance gene, PsoRPM3, from Xinjiang wild myrobalan plum (Prunus sogdiana). However, the function of PsoRPM3 remains elusive. In the present study, we have investigated the regulatory mechanism of PsoRPM3 in plant defense responses to M. incognita. Our results indicate that fewer giant cells were detected in the roots of the PsoRPM3 transgenic tobacco than wild tobacco lines after incubation with M. incognita. Transient transformations of full-length and TN structural domains of PsoRPM3 have induced significant hypersensitive responses (HR), suggesting that TIR domain might be the one which caused HR. Further, yeast two-hybrid results revealed that the full-length and LRR domain of PsoRPM3 could interact with the transcription factor Pso9TF. The addition of Pso9TF increased the ROS levels and induced HR. Thus, our data revealed that the LRR structural domain of PsoRPM3 may be associated with signal transduction. Moreover, we did not find any relative inductions of defense-related genes PsoEDS1, PsoPAD4 and PsoSAG101 in P. sogdiana, which has been incubated with M. incognita. In summary, our work has shown the key functional domain of PsoRPM3 in the regulation of defense responses to M. incognita in P. sogdiana.
1. Introduction
Root-knot nematodes (Meloidogyne spp.) are widespread throughout the world and can parasitize more than 3000 species of plants. Each year, root-knot nematodes cause around 5% of all agricultural losses worldwide [1,2]. However, because root-knot nematodes have a short life cycle and high reproduction rate, they are particularly difficult to control, and the methods used to control root knot nematodes in production need to be further explored. In terms of chemical control, commonly used organic phosphate chemical nematicides, such as sebufos, arbofuran and fenamiphos [3], are non-specific and harmful to humans, birds and fish. In recent years, they have been removed from the pesticide market. The ozone-depleting methyl bromide fumigation method is also being eliminated, and the use of biological fumigation instead of harmful methyl bromide to control nematodes is becoming more and more popular [4]. Another potential method for controlling plant parasitic nematode is microbial preparations, such as bacteria [5], fungi [6] and actinomycetes [7], which have certain control effects on root-knot nematodes, among which Pseudomonas fluorescens and Streptomyces avermitilis are commercially used for the control of plant parasitic nematodes in many countries [8,9]. The current methods for controlling nematodes are either harmful to the environment or costly, and are not very friendly to agricultural production. With the gradual development of modern molecular biology technology, it is necessary to choose resistant rootstocks or cultivate resistant stocks. Resistant crops are an effective way to control root-knot nematode disease [10].
Most disease-resistance genes in plants have nucleotide-binding-leucine-rich repeat structures (NLRs). Each structural domain plays a different role in plant disease resistance. The N-terminal CC or TIR domain is associated with signaling transduction [11] and is involved in the protein–protein interactions [12,13], which usually self-associate or form complexes through interacting with other proteins [14,15,16]. The intermediate NB-ARC domain is a nucleotide-binding domain that activates or inhibits NLR protein structures by binding to or hydrolyzing ADP/ATP, and is a molecular switch that regulates the activity of NLRs and has a broad role in influencing the self-conjugation of the N-terminal CC or TIR structural domain [17,18,19,20,21]. The C-terminal LRR structural domain is usually responsible for recognizing effectors, preventing auto-activation or auto-inhibition of downstream signals, and is associated with the strength of the immune response [22]. Deletion of the LRR structural domain in some NLRs can lead to autoimmune responses [23,24,25]. The interaction of different structural domains of NLR determines the structure and activity of NLR, resulting in different functions of individual structural domains. However, there are also differences in the functions of the different structural domains for specific NLR proteins, suggesting that the NLR has extremely complex properties. Mi-1.2 [26], Mi-9 [27], CaMi [28] and Ma [29] are resistant to root-knot nematodes, while Hero-A [30], Gpa2 [31] and Gro1–4 [32] are resistant to cyst nematodes. Gro1–4 and Ma encode TNL, while others encode CNL, in which the Ma protein has a highly polymorphic LRR region, which is considered to be very important for the recognition of PPN (plant parasitic nematode) [33]. In addition, 185 genes with TNL domains have been found in the genome of peach that may be resistant to root-knot nematodes. They all have LRR domains, but there are no more in-depth research reports [34].
Transcription factors are the most common proteins that can interact with the NLR and regulate the NLR-mediated resistant and HR responses [35,36]. A WRKY domain was found at the C-terminus of the immune receptor TNL1 of Prunus plants conferring resistance against root-knot nematodes [33]. It has been reported that the transcription factor WRKY can regulate the expression of defense-related genes in the immunity triggered by the effector [29], and the presence of the WRKY domain in TNL1 indicates that the NLR protein’s ability to recognize nematode invasion is related to the presence of transcription factors [33,37]. Similarly, in Arabidopsis, TIR-NBS-LRR disease-resistant proteins DSC1 and WRKY19 typically regulate the resistance of Arabidopsis to M. incognita [38]. Therefore, NLR-associated signalings in plant immunity were conducted via recruiting different transcription factors. However, little is known about the downstream components of NLR immune signaling.
It has been shown that a key downstream component of TIR-NLR immune signaling requires EDS1 (enhanced disease susceptibility 1) [39,40], an adipose-like protein that interacts with PAD4 (phytoalexin deficient 4) and SAG101 (senescence-associated gene 101) adipose-like proteins, respectively. EDS1–SAG101 interaction occurs only in the nucleus, whereas EDS1–PAD4 interaction is present in the cytoplasm and nucleus [41,42]. EDS1 also facilitates the interaction between PAD4 and SAG101 and forms a larger complex [42]. Co-expression of EDS1 and PAD4 leads to autoimmunity [43]. EDS1 can affect basal immunity and ETI processes as well as HR responses in plants [44]. In contrast, EDS1, SAG101 and PAD4 are all highly conserved in higher plants [45]. However, the mechanisms of activation and signaling between them remain unknown.
In this study, we screened the transcription factor Pso9TF, a basic leucine zipper transcription factor A-like gene, which interacts with the PsoRPM3 protein, based on the PsoRPM3 gene with resistance to the root-knot nematode (M.incognita), obtained by cloning from a single resistant plant of Xinjiang wild myrobalan plum (Prunus sogdiana) and using CO-IP assay—Pso9TF was named because this gene is ranked ninth in the CO-IP screening results [46]. Further, we observed the formation of giant cells in the root system of tobacco trans-PsoRPM3 gene using paraffin sections. The resistance of PsoRPM3 protein to root-knot nematode (M. incognita) was further confirmed by observing the changes of reactive oxygen species (ROS) and HR response in leaves of N. benthamian through truncated PsoRPM3 assay. The interactions between different structural domains of PsoRPM3 and Pso9TF were determined by yeast two-hybrid assay, and it was found that Pso9TF could influence the disease resistance of PsoRPM3 protein. We further measured the expression of TNL-related genes EDS1, PAD4 and SAG101 in Xinjiang wild myrobalan plum (Prunus sogdiana) to M. incognita.
2. Results
2.1. Histological Observations of Tobacco Transgenic for PsoRPM3
Changes in tissue structure were observed after inoculation of wild-type and PsoRPM3-transformed tobacco with the M. incognita, as shown in Figure 1. The root cells of wild-type and PsoRPM3-transformed tobacco developed normally (Figure 1A,E) before inoculation with M. incognita. After 3 days post-infection (dpi), a large number of J2 nematodes could be observed in wild-type tobacco roots invading vascular tissues, and there was an expansion of the nematodes (Figure 1B). At 14 dpi, the feeding sites were large, occupying almost the entire longitudinal section of the root system, and each giant cell in the feeding site was cytoplasmically dense, by which time the nematodes had changed to third instar nematodes (Figure 1C). At 30 dpi, the nematodes were already laying eggs and discharging egg masses in large numbers outside the roots (Figure 1D). Invasion of nematodes was also observed in the vascular tissue of a small number of roots at 3 dpi in tobacco transgenic for PsoRPM3 (Figure 1E). Feeding sites were also formed in some roots at 14 dpi, but the development of giant cells was not full (Figure 1F). Only a few nematodes in the roots became J4 stage at 30 dpi, but no eggs were laid. At this time, the number of giant cells in the roots was low and the development of feeding sites was incomplete (Figure 1G).
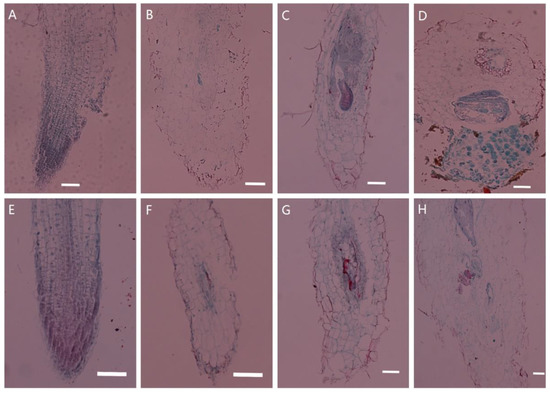
Figure 1.
Tissue section to observe the disease resistance response of PsoRPM3 transgenic tobacco after nematode inoculation. The paraffin section technique was used to observe the nematode invasion status of tobacco roots infected by M. incognita, and to compare the difference in disease resistance between the transgenic and wild-type tobacco W38. (A–D) The observation of wild-type tobacco W38 roots which were inoculated with M. incognita at 0 d, 3 d, 14 d and 30 d as a negative control; (E–H) The observation of transgenic PsoRPM3 tobacco roots which were inoculated with M. incognita at 0 d, 3 d, 14 d and 30 d; scale bar is 100 μm.
2.2. Disease Resistance Analysis of the PsoRPM3 Protein
Although the resistant role of PsoRPM3 protein to M. incognita has been identified, the mode of action of PsoRPM3 remains unclear. Firstly, we analyzed whether PsoRPM3 proteins could form homodimers by yeast two-hybrid system, and the results showed that PsoRPM3 proteins could not form homodimers (Figure 2A). We further divided the PsoRPM3 protein into full-length, TIR+NBS (TN) and NBS+LRR (NL) domains (Figure 2B) to analyze their respective roles in the plant defense responses, for instance, the ROS (reactive oxygen species) burst and HR. When tobacco leaves were injected with empty vectors, no ROS bursts were detected on the leaves. When the PsoRPM3 FL (full-length), TN and NL structural domains were injected separately, the full length of PsoRPM3 protein produced significant ROS production, while both the TN and NL structural domains also produced ROS, but the NL structural domain produced more ROS than the TN structural domain (Figure 2C). Further, we constructed overexpression vectors for transient injection into tobacco leaves to observe the HR response. We found that no HR response occurred in the no-load and a weak HR response occurred in the NL domain, while the TN domain and the full-length PsoRPM3 protein elicited a significant HR response (Figure 2D). Statistical analysis of the necrotic area of cells eliciting HR responses revealed that the TN structural domain and PsoRPM3 full-length (FL) mean necrotic area was larger, while the NL structural domain had a slightly larger necrotic area than the empty vector (Figure 2E).
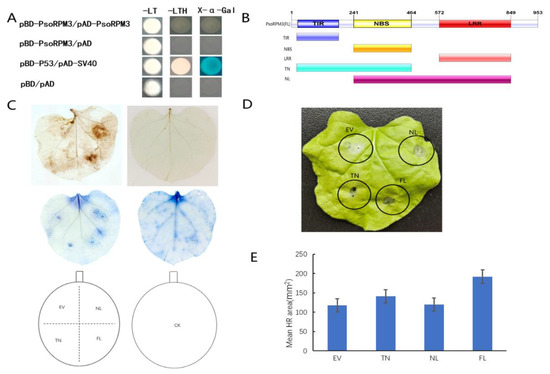
Figure 2.
ROS (Reactive oxygen species) burst detection and cell death staining in the PsoRPM3 disease resistance response. (A) Analysis of interactions between PsoRPM3 and PsoRPM3 using the yeast two-hybrid system. α-Galactosidase activity was detected using X-α-Gal as a substrate. (B) Segmentation pattern of the structural domain of PsoRPM3 protein. (C) The brown color is DAB for reactive oxygen species staining in HR reaction, and the blue color is trypan blue for cell necrosis staining. (D) HR responses observed in tobacco (N. benthamiana) leaves transiently overexpressing empty vector, TN, NL, and FL 1 d after inoculation with M. incognita. (E) Determination of cell necrosis area to (D).
2.3. Interaction of PsoRPM3 Protein with Downstream Transcription Factor Pso9TF
The above studies have shown that PsoRPM3 protein could activate the production of ROS and induce HR responses, for which the components downstream of the disease resistance signal were not clear. We screened a number of complexes bound to PsoRPM3 protein in Xinjiang wild myrobalan plum (P. sogdiana) by CO-IP based on its localization in the nucleus, including a transcription factor Pso9TF. We cloned the Pso9TF gene from P. sogdiana. The phylogenetic tree analysis revealed that Pso9TF was a basic leucine zipper transcription factor or kinase (Figure 3A), and its protein structure contained the PB1 structural domain (Figure 3B). The subcellular localization result showed that Pso9TF was localized in the nucleus (Figure 3C). The transcriptional self-activation assay revealed found Pso9TF has transcriptional self-activation activity (Figure 3D). Further, yeast two-hybrid assays showed that there was an interaction between Pso9TF and the full-length (FL) and LRR structural domains of the PsoRPM3 protein, but not with the TIR, NBS, TN and NL structural domains (Figure 3E). Their relationships were also verified by BiFC assays (Figure 3F).
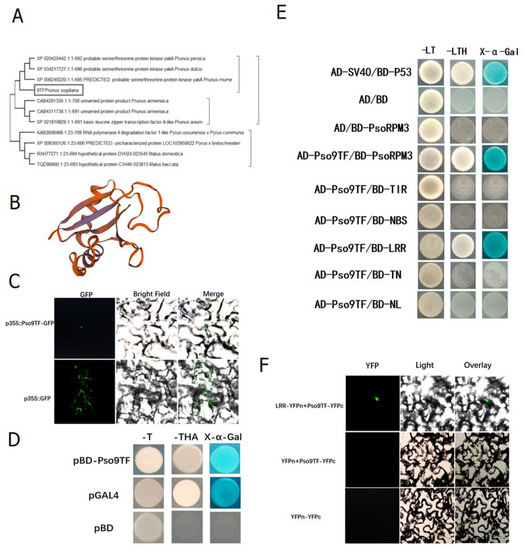
Figure 3.
Basic characteristics of Pso9TF and detection of interactions with PsoRPM3. (A) Phylogenetic tree containing Pso9TF (rectangle highlighted) and homologous proteins from other related plant species. (B) The predicted three-dimensional structures of the Pso9TF. (C) Pso9TF-GFP localized in the nucleus of N. benthamiana cells, GFP alone localized throughout the whole cells. Fluorescence (left), bright field (middle), and merged images (right) were obtained at 48 h post-agroinfiltration by using Leica confocal microscopy. (D) Transcriptional self-activation of Pso9TF using a yeast one-hybrid. (E) Interaction of Pso9TF with each structural domain of PsoRPM3 using yeast two-hybrid. (F) Interactions between PsoRPM3 and its three chaperonin proteins were determined by BiFC. YFP fluorescence in the upper epidermis cells of N. benthamiana leaves was detected by laser scanning confocal microscopy.
2.4. Pso9TF and PsoRPM3 Proteins together Cause Disease Resistance Responses
Due to the reciprocal relationship between PsoRPM3 protein and Pso9TF, a question is proposed: do they have a joint effect on disease resistance? The injection of PsoRPM3 protein full-length followed by Pso9TF on tobacco leaves has enhanced the production of ROS and HR response, and the same results were observed for the LRR structural domain of PsoRPM3 protein, but the TN structural domain did not change much (Figure 4).
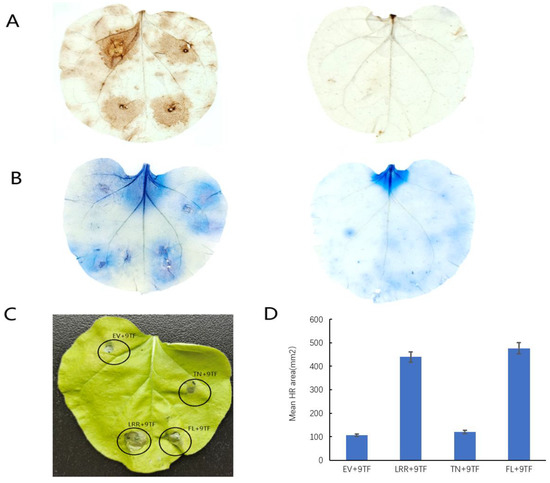
Figure 4.
Effect of Pso9TF on PsoRPM3 disease resistance. (A) Pso9TF and GFP empty vectors, Pso9TF and TN, Pso9TF and LRR, Pso9TF and FL were transiently co-expressed on tobacco leaves of N. benthamian, and then injected into nematodes after 1 d, and then stained with DAB for reactive oxygen species. (B) Samples stained with trypan blue for necrotic cells; the left was the test group, the right was the control group. (C) HR reaction observed directly without any dyeing treatment. (D) Measurement and counting of the area of the HR response.
2.5. Expression of Pso9TF and Related Disease Resistance Genes in Xinjiang Wild Myrobalan Plum
Expression of PsoRPM3, Pso9TF, PsoPAD4, PsoEDS1 and PsoSAG101 genes was analyzed using roots of Xinjiang wild myrobalan plum cuttings after inoculation with M. incognita. The results showed that the expression of the PsoRPM3 gene increased continuously from 1 to 5 days after inoculation, with the highest on the fifth day (Figure 5A); the Pso9TF gene was abundantly expressed in both disease-resistant and susceptible plants from 1 to 5 days after inoculation, but the expression was significantly higher in disease-resistant plants than in susceptible plants, while the expression of Pso9TF gene decreased significantly before and 7 days after inoculation (Figure 5B). Both PsoPAD4 and PsoEDS1 genes strongly expressed in susceptible plants in different time points (Figure 5C,D). The PsoSAG101 gene was only expressed in susceptible plants at 5 days after inoculation and was expressed at low levels in the rest of the period (Figure 5E). In all the tested time points, we found that the expression levels of Pso9TF and PsoRPM3 have shown a similar trend: both genes showed significantly more induction in resistant plants than susceptible plants.
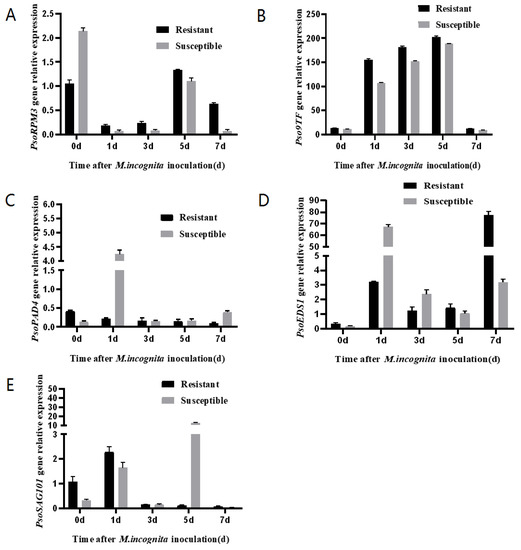
Figure 5.
Expression of Pso9TF on downstream disease-resistance related genes. (A–E) Expression of PsoRPM3, Pso9TF, PsoPAD4, PsoEDS1 and PsoSAG101 in the roots of resistant and susceptible Xinjiang wild myrobalan plum (Prunus sogdiana) individuals at 0, 1, 3, 5, and 7 d post-infection. Real-time PCR data were calculated based on three biological and three technical replicates. Error bars = SD.
3. Discussion
The formation of giant cells and the establishment of feeding sites are necessary steps for the successful invasion of root-knot nematodes (M. incognita) on plant roots. The giant cells can provide nematodes with various nutrients and support their growth and development by intranuclear replication [47]. The giant cells are surrounded by bast and xylem and cortical cells which gradually produce swollen root knots in the roots [48]. Our observations of the PsoRPM3-transformed tobacco roots showed fewer giant cells in the transgenic roots than the control, and giant cell development was delayed, ultimately leading to a failure to form eggs. These findings indicated that the PsoRPM3-transformed tobacco significantly improved resistance to M. incognita, consistent with previous results of functional validation of the PsoRPM3 gene [46].
The PsoRPM3 gene belongs to the TIR-NBS-LRR class of resistance proteins, and there are functional differences between their different structural domains. Studies in Arabidopsis have shown that the TIR of RPS4 and RRS1 can form homodimers and heterodimers for signaling transduction [49]. The TIR domain of the ZAR1 disease resistance protein forms homodimers and then transduces signals through enzymatic cleavage functions [50]. The TIR domain is thought to function primarily as an articulatory sub-structural domain that mediates protein interactions [51]. The PsoRPM3 protein did not self-associate in this study, suggesting that PsoRPM3 may not be involved in the disease resistance process through homodimerization. The full-length, TIR+NBS (TN) and NBS+LRR (NL) structural domains of PsoRPM3 all induced the production of reactive oxygen species, but only the full-length and TN structural domains of PsoRPM3 induced a strong HR response, while the NL structural domain caused an inadequate HR response. Our data suggest that the structural domains that induce the HR response are mainly TIR domains.
Resistance proteins generally work together and form complexes in the process of disease resistance in plants. Transcriptional regulators are the most commonly observed interacting proteins with NLRs [52]. Most of these interactions occur in the nucleus, contributing to the accumulation of NLR or transcription factors in the nucleus, regulating the expression of defense genes and thus influencing defense signaling [53,54]. Previous studies have shown that the combination of transcription factors with NLR protein can regulate disease resistance. For example, the combination of rice NLR protein Pb1 and transcription factor WRKY45 in the nucleus will stimulate disease resistance [54].Rice BPH14 is compounded with WRKY46/WRKY76, and the combination of the WRKY46/WRKY76 will enhance the expression of the defense gene RLCK281 and the immune response [55].
The NLR modulates immune signaling by recruiting different transcription factors [53]. The Pso9TF transcription factor that interacted with PsoRPM3 in this study contained the PB1 structural domain, the PsoRPM3 full-length and LRR structural domains interacted with Pso9TF, and both PsoRPM3 and Pso9TF were localized in the nucleus [46], indicating that this interaction occurred in the nucleus. In a transient tobacco injection assay, the full-length (FL) and LRR domains of PsoRPM3 induced an increase in ROS and an HR response after the addition of Pso9TF, suggesting that the LRR domain of PsoRPM3 may be involved in signal transduction and the HR response.
For most plants, the EDS1 gene is required downstream of immune signaling by TNLs [39,40,56]. In the Arabidopsis, EDS1 and PAD4 genes work closely to stimulate the production of the defense hormone salicylic acid (SA) [57] to limit the growth of pathogens, which is very important for the ETI immune response [58]. Further analysis of Arabidopsis mutants showed that EDS1 and PAD4 are also involved in disease resistance controlled by other types of intracellular receptors or pathogen proteins [59]. This indicates that EDS1 protein usually works with PAD4 as a regulatory ‘node’ to coordinate the plant’s immune response to various environmental stimuli [57]. It has been reported that the antagonism of the EDS1/PAD4 complex on the transcription factor MYC2 enhances the Arabidopsis effector to trigger the salicylic acid defense in immunity [60], although PAD4 can bind to EDS1 to trigger defense in all aspects of the response, EDS1 can also interact with SAG101 in the absence of PAD4 [61]. The EDS1–SAG101 complex is also necessary for TNL-mediated immunity in N. benthamian [62]. In addition, studies have shown that chickpea CaRGA protein interacts with WRKY64 in the nucleus to positively regulate EDS1 transcription and cell death signaling [63]; furthermore, in studies of resistance to soybean cyst nematodes, the downstream signaling of PAD4 is involved in regulating the salicylate signaling pathway and, thus, positively regulates resistance to cyst nematodes [64]. In P. sogdiana, PsoEDS1 was abundantly expressed in disease-resistant plants 7 days after inoculation, whereas the expression of PsoPAD4 and PsoSAG101 did not change significantly, suggesting that the disease resistance response generated by PsoRPM3 through interactions with Pso9TF did not directly affect the changes in the PsoEDS1, PsoPAD4 and PsoSAG101 genes. In Arabidopsis, PAD4 and SAG101 generally interact with each other to form a complex, thereby affecting the transmission of disease resistance signals. In this study, we found that the resistance of these two genes and PsoRPM3 to M. incognita was not clear. The synergistic relationship indicates that the disease resistance signal pathway of Xinjiang wild myrobalan plum may be different from that of Arabidopsis. After the PsoRPM3 gene initiates disease resistance, the signal pathway needs to be further studied. In the follow-up research, we will further explore other related genes.
Taken together, the current study investigated the structure and function of the screened nematode M. incognita-resistant PsoRPM3 gene and its reciprocal transcription factor Pso9TF. Moreover, the molecular mechanism of nematode resistance in Xinjiang wild myrobalan plum (P. sogdiana) mediated by PsoRPM3 and Pso9TF was clarified in the study, providing an adequate theoretical basis for the selection of nematode-resistant rootstocks. Moreover, this study uses wild rare nematode-resistant rootstocks as test materials and combines transgenic, yeast double hybrid and other molecular biology techniques to further elucidate the molecular mechanism of the nematode-resistant gene PsoRPM3, which benefits the selection of disease-resistant rootstocks and the development of the fruit industry.
4. Materials and Methods
4.1. Plant Materials and Inoculation with M. incognita
Xinjiang wild myrobalan plum (Prunus sogdiana) seedlings were planted at the Shangzhuang test station at the China Agricultural University. Resistant and susceptible Xinjiang wild myrobalan plum individuals were identified after inoculation with root-knot nematode (M. incognita) over eight consecutive years (2007–2015). Robust shoots were harvested yearly in early May and sliced into 15–20 cm cuttings, with an upper horizontal cut and a lower diagonal cut, with each cutting bearing only 1–2 leaves. The cuttings were rooted in a 1:1 mixture of perlite and vermiculite.
Root-knot nematodes (M. incognita) were sourced from the laboratory of Jian Heng from the Institute of Plant Protection of the China Agricultural University. Nematode cultures were maintained according to a previously published method [65] with slight modifications: eggs were collected from susceptible tobacco W38 roots, placed on nylon netting floating in water, and maintained in darkness at 30 °C for 5 days, at which point juvenile nematodes (J2) were collected for analysis.
Xinjiang wild myrobalan plum seedlings were inoculated with root-knot nematode (M. incognita). Seedlings at similar stages of growth were inoculated with nematodes, using 2000 J2 M. incognita per seedling. Five resistant and five susceptible seedlings were irrigated with identical amounts of water to serve as experimental controls. All of the root tips were collected at 0, 1, 3, 5 and 7 days post-infection (dpi) for gene cloning and expression assays.
Seedlings of tobacco W38 (Nicotiana tabacum cv. W38) and Nicotiana benthamiana were cultured in a light incubator at 24–25 °C. The PsoRPM3-transformed tobacco seedlings were grown in sterile glass flasks to the height of about 25 cm and then transplanted into soil substrate in a light incubator at 24–25 °C.
4.2. Gene Cloning and RT-qPCR Assays
Total RNA was isolated from Xinjiang wild Myrobalan plum roots using an EASY Spin Kit (Beijing Biomed Biotechnology Co., Ltd., Beijing, China) and used to prepare cDNA using oligo-dT18 primers (Takara Biomedical Technology Co., Ltd., Beijing, China). Using the data of protein XP_021810829.1, which was screened and compared in the gene bank of plum genus using CO-IP results, primers were designed in the 5′UTR and 3′UTR regions to clone Pso9TF using cDNAs from the root of disease-resistant Xinjiang wild myrobalan plum as templates. We performed quantitative real-time PCR (qRT-PCR) with the cDNAs on an ABIPRISM 7500 (Thermo Fisher Scientific, Waltham, MA, USA) based on the manufacturer’s instructions, with 40 cycles of 95 °C for 10 s and 60 °C for 30 s, using SuperReal PreMix Plus (Tiangen Biotech Co., Ltd., Beijing, China). The relative RNA abundance was calculated using the 2△△CT method [66], using PsoRPII as the reference gene [67]. All primers were designed using Primer 5.0 software. The primers of PsoRPM3, Pso9TF, PsoEDS1, PsoPAD4, and PsoSAG101 in this study were listed in Supplementary Table S1.
4.3. Paraffin Sectioning and Histological Observation
The roots of wild tobacco W38 and transgenic PsoRPM3 tobacco were collected after inoculation with 2000 J2 M. incognita for 0 d, 3 d, 14 d, 30 d; then, the roots were removed from the soil and cleaned with water. The samples were fixed in paraformaldehyde (FAA), dehydrated, embedded, sectioned, and stained with Safranin Fast Green dye prior to microscopy. Refer to reference [68] for the specific method of the test.
4.4. Protein Location Analysis of Pso9TF
Using the 35S promoter-driven Pso9TF-GFP, Agrobacterium-mediated transient expression was performed in N. benthamian leaves, with 35S: GFP serving as a negative control. The Agrobacterium strain transformed with the vector was cultured in YEP medium, harvested, and diluted to OD600 = 0.6–0.8, and then infiltrated into N. benthamian leaves using a syringe without a needle. At 48 h post-injection, infected leaves’ epidermis were analyzed under an Olympus BX61 fluorescence microscope (Olympus Corporation, Tokyo, Japan).
4.5. The Hypersensitive Response in Tobacco Leaves
A 35S-driven PsoRPM3 overexpression vector was constructed for Agrobacterium-mediated transient expression. Tobacco leaves were infiltrated with the Agrobacterium of p35S: GFP or full-length p35S: PsoRPM3-GFP or several structural domains of p35S: PsoRPM3-GFP or with Pso9TF after 24 h inoculation. Approximately 300 nematodes were injected into each treatment on the leaves of tobacco (N. benthamian), and their respective necrotic areas were analyzed in ImageJ software (https://imagej.nih.gov/ij/ accessed on 9 January 2021). The number of replicates per treatment was at least 30 leaves.
Reactive oxygen species (ROS) staining referred to the method in this literature [69]; the necrotic cells were stained with trypan blue, and the test method referred to in [70].
4.6. Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was conducted using the yeast strain AH109. The bait vector used was pGBKT7, and the prey vector was pGADT7. Competent yeast cells were transformed with specific vector combinations using the freeze–thaw method. The AH109 yeast strain was grown on SD/-Leu-Trp medium for 2–3 days at 30 °C, and 9 independent clones were picked and cultured on SD/-His-Leu-Trp medium at 30 °C for 2–3 days. Then, they were analyzed for α-galactosidase activity via color development using X-α-Gal (4 mg/mL, 5 µL per colony).
4.7. Bimolecular Fluorescence Complementation (BiFC)
The coding sequences of PsoRPM3-LRR and Pso9TF were amplified and individually inserted into the vector pUC-SPYNE-YFPn and pUC-SPYNE-YFPc. Each vector was transformed into Agrobacterium tumefaciens GV3101 and cultured for 2–4 days on YEP plates containing kanamycin and rifampin. Each positively transformed colony was cultured in YEP liquid medium overnight, harvested by centrifugation, and resuspended in inducing buffer (10 mM MgCl2, 0.2 mM acetosyringone, and 10 mM MES, pH 5.6) to a final concentration of OD600 = 1.0. After four hours, bacteria harboring YFPn-PsoRPM3-LRR and YFPc-Pso9TF were mixed at a ratio of 1:1 and infiltrated into the N. benthamiana leaves for transient expression. At 48 h post-infection (hpi), the fluorescence of the N. benthamiana epidermal cells was imaged using a confocal laser fluorescence microscope (Olympus BX61).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants10081561/s1, Table S1: Primers used in this study.
Author Contributions
Conceptualization, J.H.; methodology, J.H., K.X., Z.L., Y.W.; validation, H.Z.; analysis, H.Z., W.P., C.G., S.L., Y.X.; writing—original draft preparation, H.Z. and J.H.; writing—review and editing, J.H. and P.G.; project administration, J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the National Natural Science Foundation of China (31972360).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We thank Heng Jian from China Agricultural University for providing the root-knot nematode (Meloidogyne incognita).
Conflicts of Interest
We declare that we have no conflict of interact.
References
- Abd-Elgawad, M.M.; Askary, T.H. Impact of phytonematodes on agriculture economy. In Biocontrol agents of phytonematodes; CAB International: Wallingford, UK, 2015; pp. 3–49. [Google Scholar] [CrossRef]
- McCarter, J. Nematology: Terra incognita no more. Nat. Biotechnol. 2008, 26, 882–884. [Google Scholar] [CrossRef]
- Patel, B.; Patel, H.J.T.R. Effect of physical, cultural and chemical methods of management on population dynamics of phytonematodes in bidi tobacco nursery. Tob. Res. 1999, 25, 51–60. [Google Scholar]
- Brennan, R.J.B.; Glaze-Corcoran, S.; Wick, R.; Hashemi, M. Biofumigation: An alternative strategy for the control of plant parasitic nematodes. J. Integr. Agric. 2020, 19, 1680–1690. [Google Scholar] [CrossRef]
- Stirling, G.R.; Wong, E.; Bhuiyan, S. Pasteuria, a bacterial parasite of plant-parasitic nematodes: Its occurrence in Australian sugarcane soils and its role as a biological control agent in naturally-infested soil. Australas. Plant. Pathol. 2017, 46, 563–569. [Google Scholar] [CrossRef]
- Hussain, M.; Zouhar, M.; Ryšánek, P. Suppression of Meloidogyne incognita by the Entomopathogenic Fungus Lecanicillium muscarium. Plant. Dis. 2018, 102, 977–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nimnoi, P.; Pongsilp, N.; Ruanpanun, P. Monitoring the efficiency of Streptomyces galilaeus strain KPS-C004 against root knot disease and the promotion of plant growth in the plant-parasitic nematode infested soils. Biol. Control. 2017, 114, 158–166. [Google Scholar] [CrossRef]
- Li, J.; Zou, C.; Xu, J.; Jianping, X.; Niu, X.; Yang, J.; Huang, X.; Zhang, K.-Q. Molecular Mechanisms of Nematode-Nematophagous Microbe Interactions: Basis for Biological Control of Plant-Parasitic Nematodes. Annu. Rev. Phytopathol. 2015, 53, 67–95. [Google Scholar] [CrossRef]
- Dong, L.Q.; Zhang, K.Q. Microbial control of plant-parasitic nematodes: A five-party interaction. Plant. Soil 2006, 288, 31–45. [Google Scholar] [CrossRef]
- Vieira, C.C.; Chen, P.; Usovsky, M.; Vuong, T.; Howland, A.D.; Nguyen, H.T.; Li, Z.; Mitchum, M.G.; Shannon, G. A major quantitative trait locus resistant to southern root-knot nematode sustains soybean yield under nematode pressure. Crop. Sci. 2021, 61, 1773–1782. [Google Scholar] [CrossRef]
- Zhang, X.; Dodds, P.; Bernoux, M. What Do We Know About NOD-Like Receptors in Plant Immunity? Annu. Rev. Phytopathol. 2017, 55, 205–229. [Google Scholar] [CrossRef]
- Cesari, S.; Moore, J.; Chen, C.; Webb, D.; Periyannan, S.; Mago, R.; Bernoux, M.; Lagudah, E.; Dodds, P.N. Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC–NLR proteins. Proc. Natl. Acad. Sci. USA 2016, 113, 10204–10209. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, T.; Cheng, W.; Spiridon, L.; Töller, A.; Lukasik, E.; Saijo, Y.; Liu, P.; Shen, Q.-H.; Micluta, M.A.; Somssich, I.; et al. Coiled-Coil Domain-Dependent Homodimerization of Intracellular Barley Immune Receptors Defines a Minimal Functional Module for Triggering Cell Death. Cell Host Microbe 2011, 9, 187–199. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Bernoux, M.; Bentham, A.; Newman, T.; Ve, T.; Casey, L.; Raaymakers, T.M.; Hu, J.; Croll, T.; Schreiber, K.J.; et al. Multiple functional self-association interfaces in plant TIR domains. Proc. Natl. Acad. Sci. USA 2017, 114, E2046–E2052. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.-F.; Balint-Kurti, P. Cytoplasmic and Nuclear Localizations Are Important for the Hypersensitive Response Conferred by Maize Autoactive Rp1-D21 Protein. Mol. Plant. Microbe Interact. 2015, 28, 1023–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Jeong, R.-D.; Lim, G.-H.; Yu, K.; Wang, C.; Chandra-Shekara, A.; Navarre, D.; Klessig, D.F.; Kachroo, A.; Kachroo, P. Double-Stranded RNA-Binding Protein 4 Is Required for Resistance Signaling against Viral and Bacterial Pathogens. Cell Rep. 2013, 4, 1168–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chen, T.; Han, M.; Qian, L.; Li, J.; Wu, M.; Han, T.; Cao, J.; Nagalakshmi, U.; Rathjen, J.P.; et al. Plant NLR immune receptor Tm-22 activation requires NB-ARC domain-mediated self-association of CC domain. PLoS Pathog. 2020, 16, e1008475. [Google Scholar] [CrossRef] [PubMed]
- El Kasmi, F.; Chung, E.-H.; Anderson, R.G.; Li, J.; Wan, L.; Eitas, T.K.; Gao, Z.; Dangl, J.L. Signaling from the plasma-membrane localized plant immune receptor RPM1 requires self-association of the full-length protein. Proc. Natl. Acad. Sci. USA 2017, 114, E7385–E7394. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.; Sornaraj, P.; de Courcy-Ireland, E.; Menz, R.I.; Kobe, B.; Ellis, J.G.; Dodds, P.; Anderson, P. An Autoactive Mutant of the M Flax Rust Resistance Protein Has a Preference for Binding ATP, Whereas Wild-Type M Protein Binds ADP. Mol. Plant. Microbe Interact. 2011, 24, 897–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tameling, W.I.; Vossen, J.H.; Albrecht, M.; Lengauer, T.; Berden, J.A.; Haring, M.A.; Cornelissen, B.J.; Takken, F.L. Mutations in the NB-ARC Domain of I-2 That Impair ATP Hydrolysis Cause Autoactivation. Plant. Physiol. 2006, 140, 1233–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedl, S.J.; Li, W.; Chao, Y.; Schwarzenbacher, R.; Shi, Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nat. Cell Biol. 2005, 434, 926–933. [Google Scholar] [CrossRef]
- Lindner, S.; Keller, B.; Singh, S.P.; Hasenkamp, Z.; Jung, E.; Müller, M.C.; Bourras, S.; Keller, B. Single residues in the LRR domain of the wheat PM3A immune receptor can control the strength and the spectrum of the immune response. Plant. J. 2020, 104. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yan, C.; Liu, P.; Huang, Z.; Ma, R.; Zhang, C.; Wang, R.; Zhang, Y.; Martinon, F.; Miao, D.; et al. Crystal Structure of NLRC4 Reveals Its Autoinhibition Mechanism. Science 2013, 341, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Tameling, W.I.; Nooijen, C.; Ludwig, N.; Boter, M.; Slootweg, E.J.; Goverse, A.; Shirasu, K.; Joosten, M.H. RanGAP2 Mediates Nucleocytoplasmic Partitioning of the NB-LRR Immune Receptor Rx in the Solanaceae, Thereby Dictating Rx Function. Plant. Cell 2010, 22, 4176–4194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ade, J.; DeYoung, B.J.; Golstein, C.; Innes, R.W. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA 2007, 104, 2531–2536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milligan, S.B.; Bodeau, J.; Yaghoobi, J.; Kaloshian, I.; Zabel, P.; Williamson, V.M.J.T.P.C. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant. Cell 1998, 10, 1307–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jablonska, B.; Ammiraju, J.S.; Bhattarai, K.K.; Mantelin, S.; de Ilarduya, O.M.; Roberts, P.A.; Kaloshian, I. The Mi-9 Gene from Solanum arcanum Conferring Heat-Stable Resistance to Root-Knot Nematodes Is a Homolog of Mi-1. Plant. Physiol. 2006, 143, 1044–1054. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Li, H.; Zhang, L.; Zhang, J.; Xiao, J.; Ye, Z. CaMi, a root-knot nematode resistance gene from hot pepper (Capsium annuum L.) confers nematode resistance in tomato. Plant. Cell Rep. 2007, 26, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant. Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [Green Version]
- Ernst, K.; Kumar, A.; Kriseleit, D.; Kloos, D.-U.; Phillips, M.S.; Ganal, M.W. The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. Plant. J. 2002, 31, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Van Der Vossen, E.A.G.; Van Der Voort, J.N.A.M.R.; Kanyuka, K.; Bendahmane, A.; Sandbrink, H.; Baulcombe, D.C.; Bakker, J.; Stiekema, W.J.; Klein-Lankhorst, R.M. Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: A virus and a nematode. Plant. J. 2000, 23, 567–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paal, J.; Henselewski, H.; Muth, J.; Meksem, K.; Menéndez, C.M.; Salamini, F.; Ballvora, A.; Gebhardt, C. Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant. J. 2004, 38, 285–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claverie, M.; Dirlewanger, E.; Bosselut, N.; Van Ghelder, C.; Voisin, R.; Kleinhentz, M.; Lafargue, B.; Abad, P.; Rosso, M.-N.; Chalhoub, B.; et al. The Ma Gene for Complete-Spectrum Resistance to Meloidogyne Species in Prunus Is a TNL with a Huge Repeated C-Terminal Post-LRR Region. Plant. Physiol. 2011, 156, 779–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Ghelder, C.; Esmenjaud, D. TNL genes in peach: Insights into the post-LRR domain. BMC Genom. 2016, 17, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, R.; Fang, H.; Zhang, C.; Zhang, F.; Hao, Z.; You, X.; Shi, X.; Park, C.H.; Hua, K.; et al. Two VOZ transcription factors link an E3 ligase and an NLR immune receptor to modulate immunity in rice. Mol. Plant. 2021, 14, 253–266. [Google Scholar] [CrossRef]
- Zang, Z.; Lv, Y.; Liu, S.; Yang, W.; Ci, J.; Ren, X.; Wang, Z.; Wu, H.; Ma, W.; Jiang, L.; et al. A Novel ERF Transcription Factor, ZmERF105, Positively Regulates Maize Resistance to Exserohilum turcicum. Front. Plant. Sci. 2020, 11, 850. [Google Scholar] [CrossRef]
- Van Der Hoorn, R.A.; Kamoun, S. From Guard to Decoy: A New Model for Perception of Plant Pathogen Effectors. Plant. Cell 2008, 20, 2009–2017. [Google Scholar] [CrossRef] [Green Version]
- Warmerdam, S.; Sterken, M.G.; Sukarta, O.C.A.; Van Schaik, C.C.; Oortwijn, M.E.P.; Lozano-Torres, J.L.; Bakker, J.; Smant, G.; Goverse, A. The TIR-NB-LRR pair DSC1 and WRKY19 contributes to basal immunity of Arabidopsis to the root-knot nematode Meloidogyne incognita. BMC Plant. Biol. 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Xiao, S.; Calis, O.; Patrick, E.; Zhang, G.; Charoenwattana, P.; Muskett, P.; Parker, J.E.; Turner, J.G. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant. J. 2005, 42, 95–110. [Google Scholar] [CrossRef]
- Chandra-Shekara, A.; Navarre, D.; Kachroo, A.; Kang, H.-G.; Klessig, D.; Kachroo, P. Signaling requirements and role of salicylic acid in HRT- and rrt-mediated resistance to turnip crinkle virus in Arabidopsis. Plant. J. 2004, 40, 647–659. [Google Scholar] [CrossRef]
- Wagner, S.; Stuttmann, J.; Rietz, S.; Guerois, R.; Brunstein, E.; Bautor, J.; Niefind, K.; Parker, J.E. Structural Basis for Signaling by Exclusive EDS1 Heteromeric Complexes with SAG101 or PAD4 in Plant Innate Immunity. Cell Host Microbe 2013, 14, 619–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Jeong, R.-D.; Venugopal, S.C.; Lapchyk, L.; Navarre, D.; Kachroo, A.; Kachroo, P. SAG101 Forms a Ternary Complex with EDS1 and PAD4 and Is Required for Resistance Signaling against Turnip Crinkle Virus. PLoS Pathog. 2011, 7, e1002318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Gobbato, E.; Kracher, B.; Qiu, J.; Bautor, J.; Parker, J.E. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2016, 213, 1802–1817. [Google Scholar] [CrossRef] [Green Version]
- García, A.V.; Blanvillain-Baufumé, S.; Huibers, R.P.; Wiermer, M.; Li, G.; Gobbato, E.; Rietz, S.; Parker, J.E. Balanced Nuclear and Cytoplasmic Activities of EDS1 Are Required for a Complete Plant Innate Immune Response. PLoS Pathog. 2010, 6, e1000970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiermer, M.; Feys, B.J.; Parker, J.E. Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant. Biol. 2005, 8, 383–389. [Google Scholar] [CrossRef] [Green Version]
- Xiao, K. Functional Verification and Mechanism Analysis of PsoRPM3 from Prunus sogdiana in Reponse to Meloidogyne Incognita. Ph.D. Thesis, China Agricultural University, Beijing, China, 2020. [Google Scholar]
- Sun, Y.; Zhu, Y.-X.; Balint-Kurti, P.J.; Wang, G.-F. Fine-Tuning Immunity: Players and Regulators for Plant NLRs. Trends Plant. Sci. 2020, 25, 695–713. [Google Scholar] [CrossRef]
- Hamada, N.; Yimer, H.; Williamson, V.M.; Siddique, S. Chemical Hide and Seek: Nematode’s Journey to Its Plant Host. Mol. Plant. 2020, 13, 541–543. [Google Scholar] [CrossRef]
- Mejias, J.; Truong, N.M.; Abad, P.; Favery, B.; Quentin, M. Plant Proteins and Processes Targeted by Parasitic Nematode Effectors. Front. Plant. Sci. 2019, 10, 970. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.J.; Sohn, K.H.; Wan, L.; Bernoux, M.; Sarris, P.F.; Segonzac, C.; Ve, T.; Ma, Y.; Saucet, S.B.; Ericsson, D.J.; et al. Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor. Science 2014, 344, 299–303. [Google Scholar] [CrossRef]
- Wan, L.; Essuman, K.; Anderson, R.G.; Sasaki, Y.; Monteiro, F.; Chung, E.-H.; Nishimura, E.O.; DiAntonio, A.; Milbrandt, J.; Dangl, J.L.; et al. TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 2019, 365, 799–803. [Google Scholar] [CrossRef]
- Nimma, S.; Ve, T.; Williams, S.; Kobe, B. Towards the structure of the TIR-domain signalosome. Curr. Opin. Struct. Biol. 2017, 43, 122–130. [Google Scholar] [CrossRef] [Green Version]
- Ng, D.W.-K.; Abeysinghe, J.K.; Kamali, M. Regulating the Regulators: The Control of Transcription Factors in Plant Defense Signaling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef] [Green Version]
- Inoue, H.; Hayashi, N.; Matsushita, A.; Xinqiong, L.; Nakayama, A.; Sugano, S.; Jiang, C.-J.; Takatsuji, H. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein–protein interaction. Proc. Natl. Acad. Sci. USA 2013, 110, 9577–9582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Wu, Y.; Wu, D.; Rao, W.; Guo, J.; Ma, Y.; Wang, Z.; Shangguan, X.; Wang, H.; Xu, C.; et al. The Coiled-Coil and Nucleotide Binding Domains of BROWN PLANTHOPPER RESISTANCE14 Function in Signaling and Resistance against Planthopper in Rice. Plant. Cell 2017, 29, 3157–3185. [Google Scholar] [CrossRef] [Green Version]
- Lapin, D.; Kovacova, V.; Sun, X.; Dongus, J.A.; Bhandari, D.; Von Born, P.; Bautor, J.; Guarneri, N.; Rzemieniewski, J.; Stuttmann, J.; et al. A Coevolved EDS1-SAG101-NRG1 Module Mediates Cell Death Signaling by TIR-Domain Immune Receptors. Plant. Cell 2019, 31, 2430–2455. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, K.; Sato, M.; Stoddard, T.; Glazebrook, J.; Katagiri, F. Network Properties of Robust Immunity in Plants. PLoS Genet. 2009, 5, e1000772. [Google Scholar] [CrossRef] [Green Version]
- Bartsch, M.; Gobbato, E.; Bednarek, P.; Debey, S.; Schultze, J.; Bautor, J.; Parker, J.E. Salicylic Acid–Independent ENHANCED DISEASE SUSCEPTIBILITY1 Signaling in Arabidopsis Immunity and Cell Death Is Regulated by the Monooxygenase FMO1 and the Nudix Hydrolase NUDT7. Plant. Cell 2006, 18, 1038–1051. [Google Scholar] [CrossRef] [Green Version]
- Venugopal, S.C.; Jeong, R.-D.; Mandal, M.K.; Zhu, S.; Chandra-Shekara, A.C.; Xia, Y.; Hersh, M.; Stromberg, A.J.; Navarre, D.; Kachroo, A.; et al. Enhanced Disease Susceptibility 1 and Salicylic Acid Act Redundantly to Regulate Resistance Gene-Mediated Signaling. PLoS Genet. 2009, 5, e1000545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Qiu, J.; Zhou, Y.; Bhandari, D.D.; Zhao, C.; Bautor, J.; Parker, J.E. Antagonism of Transcription Factor MYC2 by EDS1/PAD4 Complexes Bolsters Salicylic Acid Defense in Arabidopsis Effector-Triggered Immunity. Mol. Plant. 2018, 11, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Rietz, S.; Stamm, A.; Malonek, S.; Wagner, S.; Becker, D.; Medina-Escobar, N.; Vlot, A.C.; Feys, B.J.; Niefind, K.; Parker, J.E. Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 2011, 191, 107–119. [Google Scholar] [CrossRef]
- Gantner, J.; Ordon, J.; Kretschmer, C.; Guerois, R.; Stuttmann, J. An EDS1-SAG101 Complex Is Essential for TNL-Mediated Immunity in Nicotiana benthamiana. Plant. Cell 2019, 31, 2456–2474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, J.; Priya, P.; Dastidar, S.G.; Das, S. Physical interaction between nuclear accumulated CC-NB-ARC-LRR protein and WRKY64 promotes EDS1 dependent Fusarium wilt resistance in chickpea. Plant. Sci. 2018, 276, 111–133. [Google Scholar] [CrossRef]
- Youssef, R.M.; Macdonald, M.H.; Brewer, E.P.; Bauchan, G.R.; Kim, K.-H.; Matthews, B.F. Ectopic expression of AtPAD4 broadens resistance of soybean to soybean cyst and root-knot nematodes. BMC Plant. Biol. 2013, 13, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priya, D.B.; Somasekhar, N.; Prasad, J.; Kirti, P. Transgenic tobacco plants constitutively expressing Arabidopsis NPR1 show enhanced resistance to root-knot nematode, Meloidogyne incognita. BMC Res. Notes 2011, 4, 231. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef] [Green Version]
- Xiao, K.; Chen, W.; Chen, X.; Zhu, X.; Guan, P.; Hu, J. CCS52 and DEL1 function in root-knot nematode giant cell development in Xinjiang wild myrobalan plum (Prunus sogdiana Vassilcz). Protoplasma 2020, 257, 1333–1344. [Google Scholar] [CrossRef]
- Tran, B.Q.; Jung, S. Modulation of chloroplast components and defense responses during programmed cell death in tobacco infected with Pseudomonas syringae. Biochem. Biophys. Res. Commun. 2020, 528, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chang, C.; Gu, C.; Tang, S.Y.; Xie, Q.; Shen, Q.-H. An E3 Ligase Affects the NLR Receptor Stability and Immunity to Powdery Mildew. Plant. Physiol. 2016, 172, 2504–2515. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).