Morphological Analysis, Protein Profiling and Expression Analysis of Auxin Homeostasis Genes of Roots of Two Contrasting Cultivars of Rice Provide Inputs on Mechanisms Involved in Rice Adaptation towards Salinity Stress
Abstract
:1. Introduction
2. Results
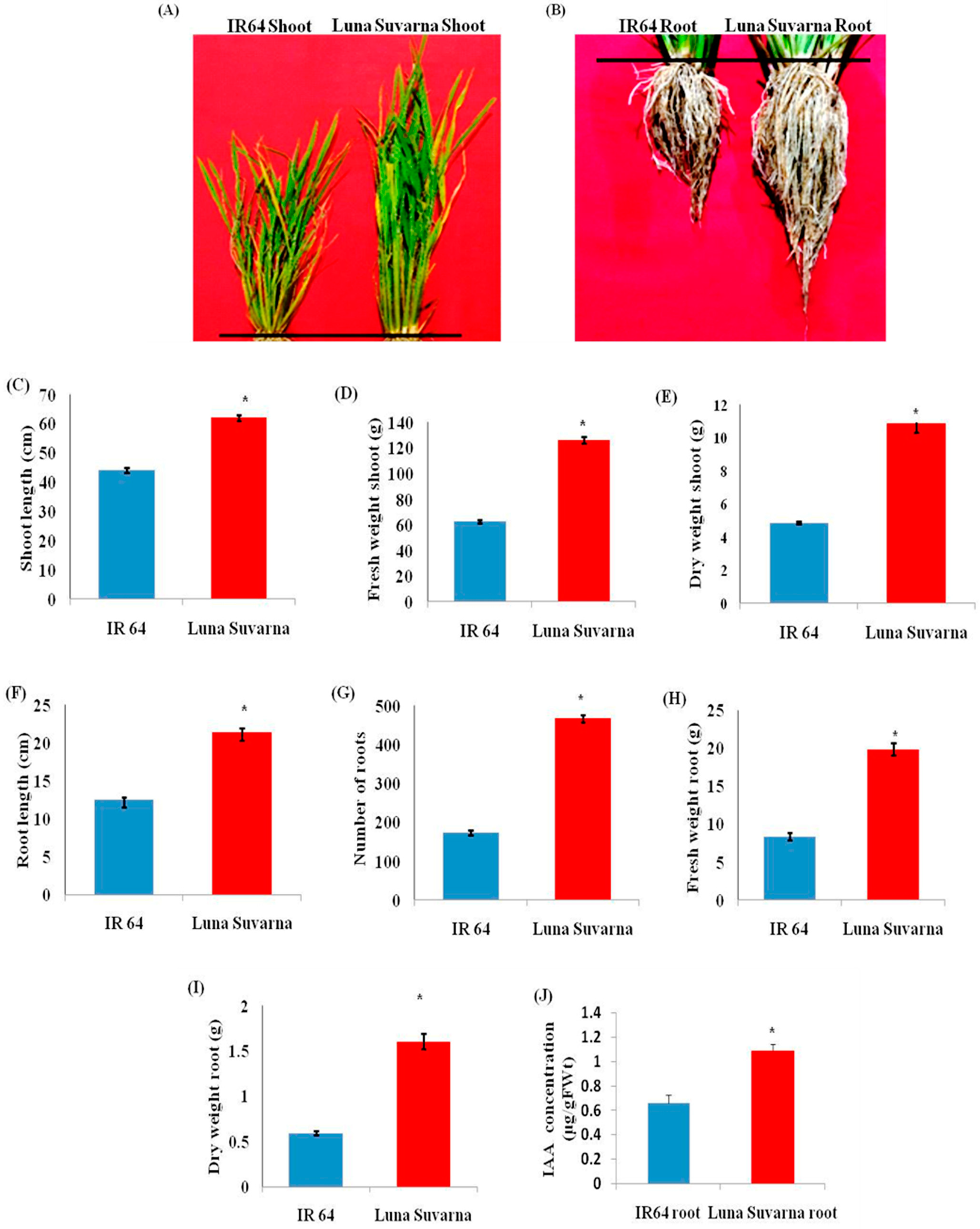
2.1. Analysis of Morphological Parameters and IAA Quantification in IR64 and LS
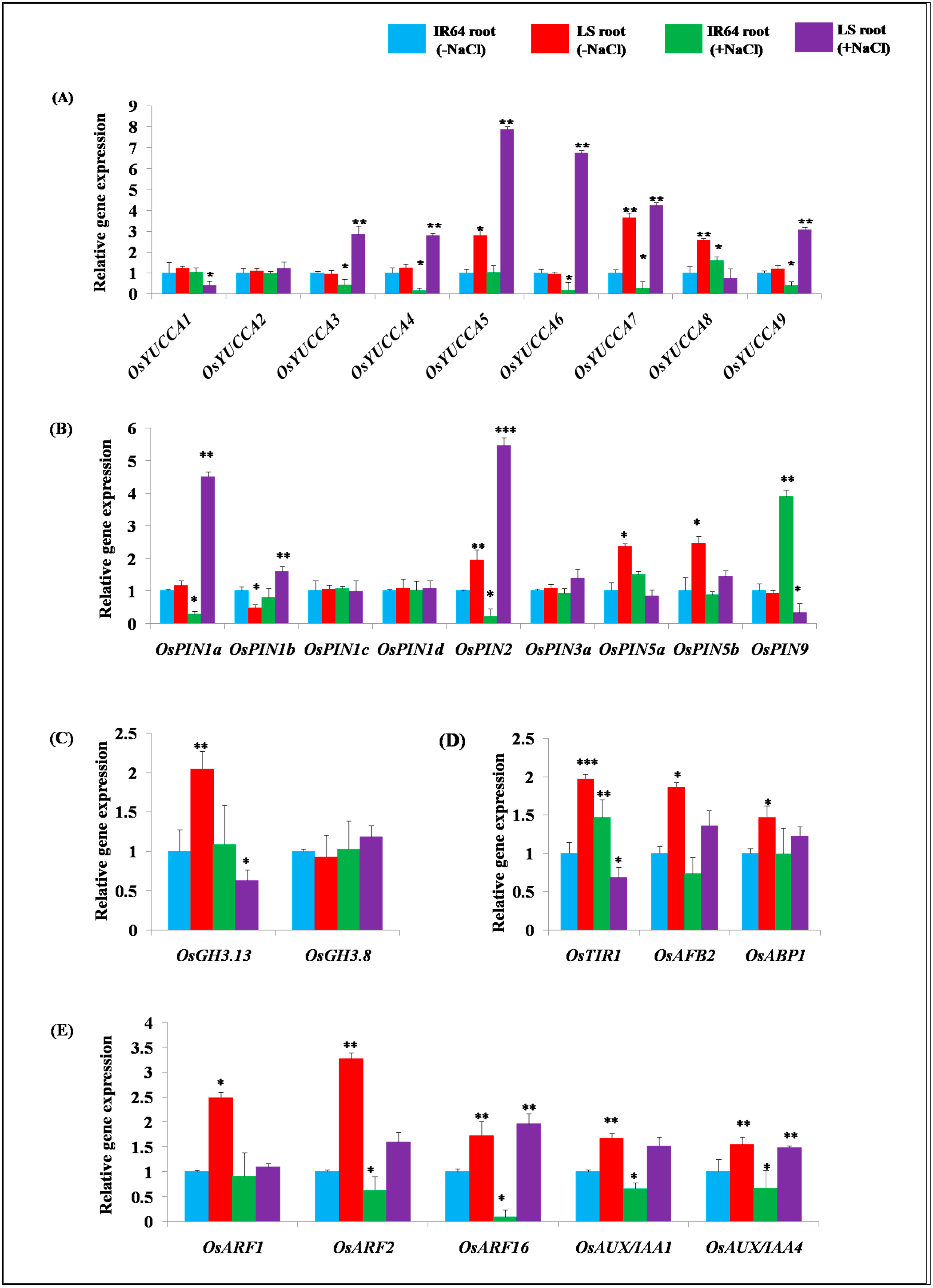
2.2. Expression Analysis of Genes Involved in Auxin Homeostasis in IR64 and LS Roots
2.3. 2-DE Analysis of Root Proteins in IR64 and LS
3. Discussion
| Protein | Functions | Reference |
|---|---|---|
| Calcineurin B-like interacting protein kinases | (1) Enhances shoot to root auxin transport. (2) Mediates root development and lateral root formation.(3) Promotes salt stress tolerance. | [52,53,54] |
| 4-Coumarate CoA ligase | (1) Enhances salt stress tolerance. (2) Increases lignification of salt stress-tolerant varieties. | [57,58] |
| β-glucosidases | (1) In vacuoles, it converts ABA glucopyranosides into free ABA. (2) It leads to the adaptation of plants to salt stress conditions. | [67,68] |
| Phytosulfokines | (1) Emerged as a novel kind of plant hormone recently and involved in immunity and homeostasis. (2) Involved in root growth and development; and inhibits ethylene production. | [62,63,64] |
| Plant intracellular Ras-group-related LRR protein 2 | (1) Promotes root development. (2) Promotes salt stress tolerance by encoding PGIPs. | [68,70] |
| Minichromosome maintenance 6 (MCM6) | (1) It confers salinity stress tolerance in pea by additional uptake of Na+ | [65,69] |
| B3 Domain containing protein | (1) RAV (Related to ABI3 and VP1) has AP2 and B3 domain. (2) Promotes salinity tolerance, enhances stress marker genes. | [71] |
| Cyanate hydratase | (1) Involved in detoxification of cyanate. (2) Provide alternative sources of nitrogen and carbon for enhancing salt stress tolerance. | [60,61] |
| Delta pyrroline-5-carboxylate synthase | (1) Maintenance of cell turgor, osmotic balance and lipid synthesis. (2) Salt stress resistance. | [55,56] |
| DEAD-box ATP-dependent RNA helicase 53 (DEAD) | (1) ReduceS oxidative stress through activation of ROS scavenging system. (2) Improves plant’s photosynthesis machinery, enhancing plant growth and development and mitigates salt stress. | [65,66] |
4. Materials and Methods
4.1. Plant Material
4.2. Study of Morphological Parameters
4.3. IAA Estimation
4.4. RNA Extraction and cDNA Synthesis
4.5. Quantitative Real-Time (qRT) PCR Analysis
4.6. Protein Extraction
4.7. Protein Solubilization and Quantification
4.8. Two-Dimensional Gel Electrophoretic (2-DE) Analysis
4.9. Gel Staining, Imaging, and Analysis
4.10. Protein in-Gel Digestion and Mass Spectrometry (MS) Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jackson, M. Hormones from roots as signals for the shoots of stressed plants. Trends Plant Sci. 1997, 2, 22–28. [Google Scholar] [CrossRef]
- Saini, S.; Kaur, N.; Pati, P.K. Reactive oxygen species dynamics in roots of salt sensitive and salt tolerant cultivars of rice. Anal. Biochem. 2018, 550, 99–108. [Google Scholar] [CrossRef]
- Gerona, M.E.B.; Deocampo, M.P.; Egdane, J.A.; Ismail, A.M.; Dionisio-Sese, M.L. Physiological Responses of Contrasting Rice Genotypes to Salt Stress at Reproductive Stage. Rice Sci. 2019, 26, 207–219. [Google Scholar] [CrossRef]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum. 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi. J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Naveen, K.; Arora, N.K. Salt-Tolerant Plant Growth Promoting Rhizobacteria for Enhancing Crop Productivity of Saline Soils. Front. Microbiol. 2019, 10, 2791. [Google Scholar] [CrossRef] [Green Version]
- Solis, C.A.; Yong, M.T.; Vinarao, R.; Jena, K.; Holford, P.; Shabala, L.; Zhou, M.; Shabala, S.; Chen, Z. Back to the Wild: On a Quest for Donors toward Salinity Tolerant Rice. Front. Plant Sci. 2020, 11, 323. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Zinta, G.; Hegab, M.M.; Pandey, R.; Asard, H.; Abuelsoud, W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 2016, 7, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Zhang, H.; Wang, T.; Chen, S.; Dai, S. Proteomics-based investigation of salt-responsive mechanisms in plant roots. J. Proteomics 2013, 2, 230–253. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Xu, J. Abiotic stress responses in plant roots: A proteomics perspective. Front. Plant Sci. 2014, 5, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, H.; Cho, Y. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar] [CrossRef]
- Korver, R.A.; Koevoets, I.T.; Testerink, C. Out of Shape during Stress: A Key Role for Auxin. Trends Plant Sci. 2018, 23, 783–793. [Google Scholar] [CrossRef]
- Quint, M.; Gray, W.M. Auxin signaling. Curr. Opin. Plant Biol. 2006, 9, 448–453. [Google Scholar] [CrossRef]
- Davies, P.J. Plant hormones: Their nature, occurrence, and functions. In Plant Hormones; Davies, P.J., Ed.; Springer: Dordrecht, The Netherlands, 2010; pp. 1–15. [Google Scholar]
- Shi, H.; Chen, L.; Ye, T.; Liu, X.; Ding, K.; Chan, Z. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol. Biochem. 2014, 82, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Jadamba, C.; Kang, K.; Paek, N.; Lee, S.I.; Yoo, S. Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice. Int. J. Mol. Sci. 2020, 21, 454. [Google Scholar] [CrossRef] [Green Version]
- Fukaki, H.; Okushima, Y.; Tasaka, M. Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 2007, 256, 111–137. [Google Scholar]
- Di Mambro, R.; De Ruvo, M.; Pacifici, E.; Salvi, E.; Sozzani, R.; Benfey, P.N.; Busch, W.; Novak, O.; Ljung, K.; Di Paola, L.; et al. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2017, 114, 641–649. [Google Scholar] [CrossRef] [Green Version]
- Rosquete, M.R.; Barbez, E.; Kleine-Vehn, J. Cellular auxin homeostasis: Gatekeeping is housekeeping. Mol. Plant 2012, 5, 772–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saini, S.; Sharma, I.; Pati, P.K. Integrating the Knowledge of Auxin Homeostasis with Stress Tolerance in Plants. In Mechanism of Plant Hormone Signaling under Stress; Pandey, G.K., Ed.; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 53–70. [Google Scholar]
- Ribba, T.; Garrido-Vargas, F.; O’Brien, J.A. Auxin-mediated responses under salt stress: From developmental regulation to biotechnological applications. J. Exp. Bot. 2020, 71, 3843–3853. [Google Scholar] [CrossRef] [PubMed]
- Vaseva, I.I.; Mishev, K.; Depaepe, T.; Vassileva, V.; Van Der Straeten, D. The Diverse Salt-Stress Response of Arabidopsis ctr1-1 and ein2-1 Ethylene Signaling Mutants is Linked to Altered Root Auxin Homeostasis. Plants 2021, 10, 452. [Google Scholar] [CrossRef]
- Zhang, H.; Han, B.; Wang, T.; Chen, S.; Li, H.; Zhang, Y.; Dai, S. Mechanisms of plant salt response: Insights from proteomics. J. Proteome Res. 2012, 11, 49–67. [Google Scholar] [CrossRef]
- Sengupta, S.; Majumder, A.L. Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: A physiological and proteomic approach. Planta 2009, 29, 911–929. [Google Scholar] [CrossRef]
- Comas, L.H.; Becker, S.R.; Cruz, V.M.V.; Byrne, P.F.; Dierig, D.A. Root traits contributing to plant productivity under drought. Front. Plant Sci. 2013, 4, 442. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Chung, Y.S.; Lee, E.; Tripathi, P.; Heo, S.; Kim, K.H. Root response to drought stress in rice (Oryza sativa L.). Int. J. Mol. Sci. 2020, 21, 1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arif, M.R.; Islam, M.T.; Robin, A.H.K. Salinity stress alters root morphology and root hair traits in Brassica napus. Plants 2019, 8, 192. [Google Scholar] [CrossRef] [Green Version]
- Polania, J.; Poschenrieder, C.; Rao, I.; Beebe, S. Root traits and their potential links to plant ideotypes to improve drought resistance in common bean. Theor. Exp. Plant Physiol. 2017, 29, 143–154. [Google Scholar] [CrossRef] [Green Version]
- Ku, Y.; Sintaha, M.; Cheung, M.; Lam, H. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Yang, Y.; Chen, S.; Ning, N.; Hu, H. Arabidopsis IAR4 modulates primary root growth under salt stress through ROS-mediated modulation of auxin distribution. Front. Plant Sci. 2019, 10, 522. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zörb, C.; Geilfus, C.M.; Mühling, K.H.; Ludwig-Müller, J. The influence of salt stress on ABA and auxin concentrations in two maize cultivars differing in salt resistance. J. Plant Physiol. 2013, 170, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.M.; Testerink, C. Roots Withstanding their Environment: Exploiting Root System Architecture Responses to Abiotic Stress to Improve Crop Tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef] [Green Version]
- Saini, S.; Sharma, I.; Kaur, N.; Pati, P.K. Auxin: A master regulator in plant root development. Plant. Cell Rep. 2013, 32, 741–757. [Google Scholar] [CrossRef]
- Chen, Q.; Dai, X.; De-Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant. Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef] [Green Version]
- Kao, C. Mechanisms of Salt Tolerance in Rice Plants: Cell Wall-Related Genes and Expansins. J. Taiwan Agric. Res. 2017, 66, 87–93. [Google Scholar]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alarcón, M.; Salguero, J.; Lloret, P.G. Auxin modulated initiation of lateral roots is linked to pericycle cell length in maize. Front. Plant Sci. 2019, 10, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [Green Version]
- Fukui, K.; Hayashi, K.I. Manipulation and sensing of auxin metabolism, transport and signaling. Plant Cell Physiol. 2018, 59, 1500–1510. [Google Scholar] [CrossRef]
- Hentrich, M.; Böttcher, C.; Düchting, P.; Cheng, Y.; Zhao, Y.; Berkowitz, O.; Masle, J.; Medina, J.; Pollmann, S. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 2013, 74, 626–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwiewka, M.; Bilanovičová, V.; Seifu, Y.W.; Nodzyński, T. The Nuts and Bolts of PIN Auxin Efflux Carriers. Front. Plant Sci. 2019, 10, 985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielach, A.; Hrtyan, M.; Tognetti, V.B. Plants under stress: Involvement of auxin and cytokinin. Int. J. Mol. Sci. 2017, 18, 1427. [Google Scholar] [CrossRef] [Green Version]
- Potters, G.; Pasternak, T.P.; Guisez, Y.; Jansen, M.A. Different stresses, similar morphogenic responses: Integrating a plethora of pathways. Plant Cell Environ. 2009, 32, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Mohanta, T.K.; Sinha, A.K. Unraveling the Intricate Nexus of Molecular Mechanisms Governing Rice Root Development: OsMPK3/6 and Auxin-Cytokinin Interplay. PLoS ONE 2015, 10, e0123620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balzan, S.; Johal, G.S.; Carraro, N. The role of auxin transporters in monocots development. Front. Plant Sci. 2014, 5, 393. [Google Scholar] [CrossRef] [Green Version]
- Kong, W.; Zhong, H.; Deng, X.; Gautam, M.; Gong, Z.; Zhang, Y.; Zhao, G.; Liu, C.; Li, Y. Evolutionary Analysis of GH3 Genes in Six Oryza Species/Subspecies and Their Expression Under Salinity Stress in Oryza sativa ssp. Japonica. Plants 2019, 8, 30. [Google Scholar] [CrossRef] [Green Version]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and lesstolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar]
- Xu, D.; Miao, J.; Yumoto, E.; Yokota, T.; Asahina, M.; Watahiki, M. YUCCA9 Mediated Auxin Biosynthesis and Polar Auxin Transport Synergistically Regulate Regeneration of Root Systems Following Root Cutting. Plant Cell Physiol. 2017, 58, 1710–1723. [Google Scholar] [CrossRef] [Green Version]
- Guseman, J.M.; Hellmuth, A.; Lanctot, A.; Feldman, T.P.; Moss, B.L.; Klavins, E.; Calderón Villalobos, L.I.; Nemhauser, J.L. Auxin-induced degradation dynamics set the pace for lateral root development. Development 2015, 142, 905–909. [Google Scholar]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, V.; Parasuraman, B.; Laxmi, A.; Chattopadhyay, D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 2009, 58, 778–790. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Q.Q.; Zhou, L.; Ren, F.; Li, D.D.; Li, X.B. Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol. Biol. Rep. 2013, 40, 4759–4767. [Google Scholar] [CrossRef]
- Hu, W.; Xia, Z.; Yan, Y.; Ding, Z.; Tie, W.; Wang, L.; Zou, M.; Wei, Y.; Lu, C.; Hou, X.; et al. Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Front. Plant Sci. 2015, 6, 914. [Google Scholar] [CrossRef] [Green Version]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharsallah, C.; Fakhfakh, H.; Grubb, D.; Gorsane, F. Effect of salt stress on ion concentration, proline content, antioxidant enzyme activities and gene expression in tomato cultivars. AoB Plants 2016, 8, plw055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.H.; Ma, T.; Luo, W.C.; Xu, J.M.; Liu, J.Q.; Wan, D.S. Identification of 4CL Genes in Desert Poplars and Their Changes in Expression in Response to Salt Stress. Genes 2015, 6, 901–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafi, A.; Chauhan, R.; Gill, T.; Swarnkar, M.K.; Sreenivasulu, Y.; Kumar, S.; Kumar, N.; Shankar, R.; Ahuja, P.S.; Singh, A.K. Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol. Biol. 2015, 87, 615–631. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Nveawiah-Yoho, P.; Zhou, J.; Palmer, M.; Sauve, R.; Zhou, S. Identification of Proteins for Salt Tolerance Using a Comparative Proteomics Analysis of Tomato Accessions with Contrasting Salt Tolerance. J. Am. Soc. Hortic. Sci. 2013, 138, 382–394. [Google Scholar] [CrossRef] [Green Version]
- Maršálová, L.; Vítámvás, P.; Hynek, R.; Prášil, I.T.; Kosová, K. Proteomic Response of Hordeum vulgare cv. Tadmor and Hordeum marinum to Salinity Stress: Similarities and Differences between a Glycophyte and a Halophyte. Front. Plant Sci. 2016, 7, 1154. [Google Scholar] [CrossRef] [PubMed]
- Lorbiecke, R.; Steffens, M.; Tomm, J.M.; Scholten, S.; Wiegen, P.; Kranz, E.; Wienand, U.; Sauter, M. Phytosulphokine gene regulation during maize (Zea mays L.) reproduction. J. Exp. Bot. 2005, 56, 1805–1819. [Google Scholar] [CrossRef]
- Sauter, M. Phytosulfokine peptide signaling. J. Exp. Bot. 2015, 66, 5161–5169. [Google Scholar] [CrossRef]
- Kutschmar, A.; Rzewuski, G.; Stührwohldt, N.; Beemster, G.T.; Inzé, D.; Sauter, M. PSK-α promotes root growth in Arabidopsis. New Phytol. 2009, 181, 820–831. [Google Scholar] [CrossRef]
- Macovei, A.; Vaid, N.; Tula, S.; Tuteja, N. A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signal. Behav. 2012, 7, 1138–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuteja, N.; Banu, M.S.; Huda, K.M.; Gill, S.S.; Jain, P.; Pham, X.H.; Tuteja, R. Pea p68, a DEAD-box helicase, provides salinity stress tolerance in transgenic tobacco by reducing oxidative stress and improving photosynthesis machinery. PLoS ONE 2014, 30, e98287. [Google Scholar] [CrossRef] [Green Version]
- Dietz, K.J.; Sauter, A.; Wichert, K.; Messdaghi, D.; Hartung, W. Extracellular beta-glucosidase activity in barley involved in the hydrolysis of ABA glucose conjugate in leaves. J. Exp. Bot. 2000, 51, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhao, F.; Fang, W.; Xie, D.; Hou, J.; Yang, X.; Zhao, Y.; Tang, Z.; Nie, L.; Lv, S. Identification of early salt stress responsive proteins in seedling roots of upland cotton (Gossypium hirsutum L.) employing iTRAQ-based proteomic technique. Front. Plant Sci. 2015, 6, 732. [Google Scholar] [CrossRef] [Green Version]
- Dang, H.Q.; Tran, N.Q.; Gill, S.S.; Tuteja, R.; Tuteja, N. A single subunit MCM6 from pea promotes salinity stress tolerance without affecting yield. Plant Mol. Biol. 2011, 76, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Xiong, T.F.; Ni, Z.Y.; Chen., X.P.; Chen, M.; Li, L.C.; Gao, D.Y.; Yu, X.D.; Liu, P.; Ma, Y.Z. Isolation and identification of two genes encoding leucine-rich repeat (LRR) proteins differentially responsive to pathogen attack and salt stress in tobacco. Plant Sci. 2009, 176, 38–45. [Google Scholar] [CrossRef]
- Li, X.J.; Li, M.; Zhou, Y.; Hu, S.; Hu, R.; Chen, Y.; Li, X.B. Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS ONE 2015, 10, e0118056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miura, K.; Hasegawa, P.M. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 2010, 20, 223–232. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Zhang, C.; Yates, G.; Bailey, M.; Brown, A.; Sadanandom, A. SUMO is a Critical Regulator of Salt Stress Responses in Rice. Plant Physiol. 2016, 170, 2378–2391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemaire, K.; Moura, R.F.; Granvik, M.; Igoillo-Esteve, M.; Hohmeier, H.E.; Hendrickx, N.; Newgard, C.B.; Waelkens, E.; Cnop, M.; Schuit, F. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS ONE 2011, 6, e18517. [Google Scholar] [CrossRef] [Green Version]
- Nagar, P.K.; Sood, S. Changes in endogenous auxins during winter dormancy in tea (Camellia sinensis L.) O. Kuntze. Acta Physiol. Plant 2006, 28, 165–169. [Google Scholar] [CrossRef]
- Faurobert, M.; Pelpoir, E.; Chaïb, J. Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. Methods Mol. Biol. 2007, 355, 9–14. [Google Scholar] [PubMed]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kaur, D.; Dogra, V.; Thapa, P.; Bhattacharya, A.; Sood, A.; Sreenivasulu, Y. In vitro flowering associated protein changes in Dendrocalamus hamiltonii. Proteomics 2015, 15, 1291–1306. [Google Scholar] [CrossRef] [PubMed]
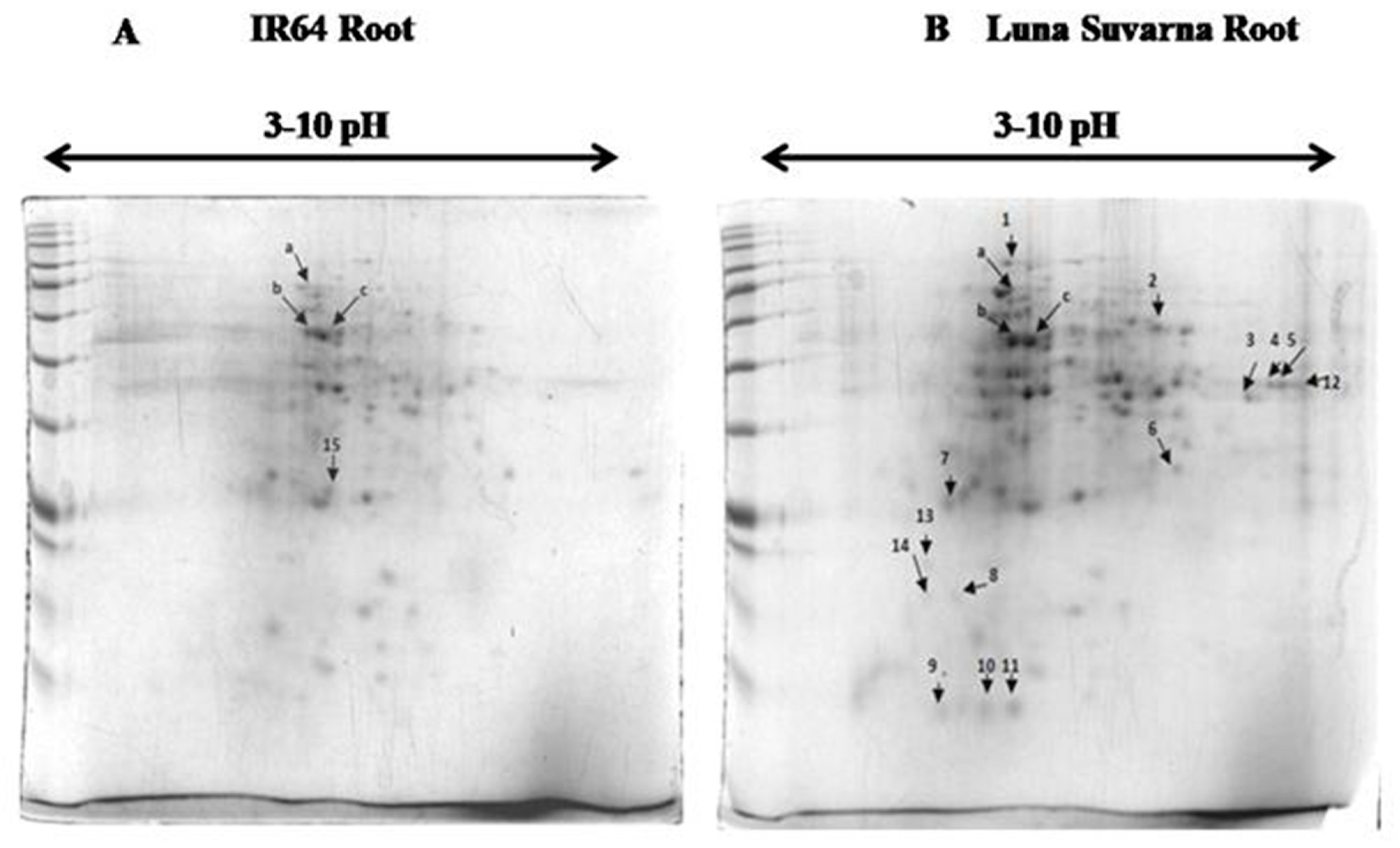
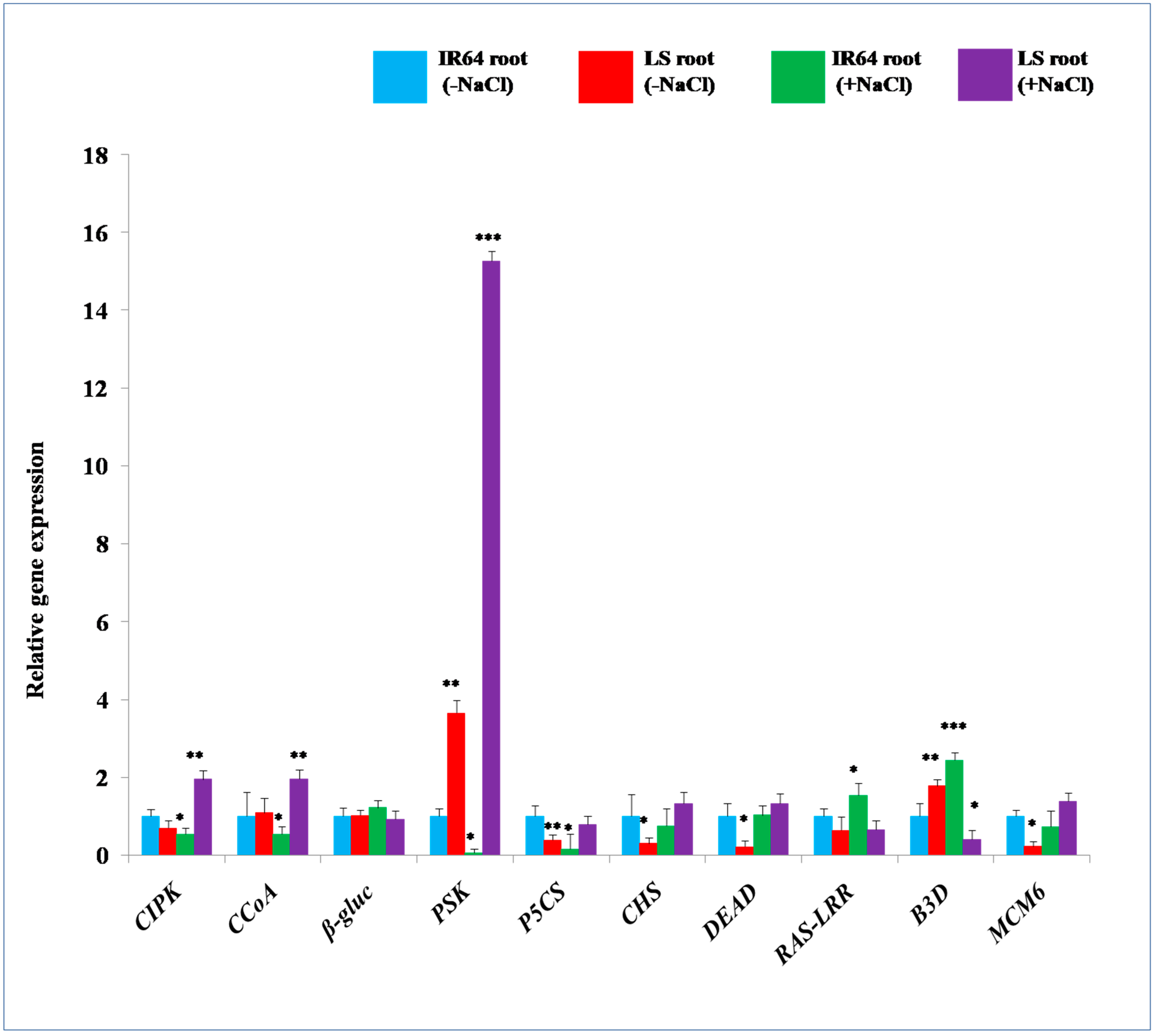
| Spot | Protein Name | Accession Number | MSU Number | Reference Organism | Mrpi (Theoretical) | Score | Coverage | Function |
|---|---|---|---|---|---|---|---|---|
| 1 | Histone H2B.10 | H2B10_ORYSI | LOC_Os01g05610.1 | Oryza sativa subsp. Indica | 16522 (10.02) | 41 | 36% | Core nucleosome component |
| 2 | DEAD-box ATP-dependent RNA helicase 53 | RH53_ORYSJ | LOC_Os07g05050.1 | Oryza sativa subsp. Japonica | 65429 (9.44) | 47 | 15% | Abiotic stress responses |
| 3 | Glyceraldehyde-3-phosphate dehydrogenase 3 cytosolic | G3PC3_ORYSJ | LOC_Os02g38920 | Oryza sativa subsp. Japonica | 36716 (7.68) | 37 | 38% | Glycolysis enzyme |
| 4 | Calcineurin B-like interacting protein kinase 21 | CIPKL_ORYSJ | LOC_Os07g44290 | Oryza sativa subsp. Japonica | 59592 (9.26) | 39 | 15% | Abiotic stress tolerance |
| 5 | 4-Coumarate-CoA ligase-like 9 | 4CLL9_ORYSJ | LOC_Os04g24530 | Oryza sativa subsp. Japonica | 59782 (5.69) | 46 | 22% | Abiotic stress tolerance |
| 6 | β-glucosidases 12 | BGL12_ORYSJ | LOC_Os04g39880 | Oryza sativa subsp. Japonica | 57713 (8.75) | 47 | 11% | Abiotic stress tolerance |
| 7 | Ubiquitin-like protein autophagy-related | ATG12_ORYSI | LOC_Os06g10340 | Oryza sativa subsp. Indica | 10454/9.05 | 34 | 32% | Autophagy and protein transport |
| 8 | Phytosulfokines 3 | PSK3_ORYSJ | LOC_Os03g47230 | Oryza sativa subsp. Japonica | 8341/5.83 | 34 | 29% | Abiotic stress tolerance |
| 9 | Cyanate hydratase | CYNS_ORYSI | LOC_Os10g33270 | Oryza sativa subsp. Indica | 18653/5.61 | 33 | 44% | Abiotic stress tolerance |
| 10 | Probable protein arginine N-methyltransferase | ANM3_ORYSI | LOC_Os07g44640 | Oryza sativa subsp. Indica | 69127/4.49 | 29 | 13% | Mediates methylation process |
| 11 | Heat stress transcription factor B 1 | HSFB1_ORYSJ | LOC_Os09g28354 | Oryza sativa subsp. Japonica | 33121/9.35 | 33 | 43% | Abiotic stress responses |
| 12 | Delta 1-pyrroline-5-carboxylate synthase | P5CS_ORYSJ | LOC_Os05g38150 | Oryza sativa subsp. Japonica | 78153/6.37 | 53 | 14% | Abiotic stress tolerance |
| 13 | Calcineurin B-like-interacting protein kinase 29 | CIPKT_ORYSJ | LOC_Os07g48090 | Oryza sativa subsp. Japonica | 48581/8.69 | 38 | 44% | Abiotic stress tolerance |
| 14 | Minichromosome maintenance 6 | MCM6_ORYSI | LOC_Os05g14590 | Oryza sativa subsp. Indica | 93168/5.55 | 43 | 17% | Abiotic stress tolerance |
| 15 | UDP-glucose 6-dehydrogenase 1 | UGDH1_ORYSJ | LOC_Os03g31210 | Oryza sativa subsp. Japonica | 52834 (5.75) | 39 | 13% | Enzymatic function |
| Spot | Protein Name | Accession Number | MSU Number | Reference Organism | Mrpi (Theoretical) | Mrpi (Experimental) | Score | Coverage | Function |
|---|---|---|---|---|---|---|---|---|---|
| a | Plant intracellular Ras group related LRR protein 2 | PIRL2_ORYSJ | LOC_Os02g38040 | Oryza sativa subsp. Japonica | 55345/5.51 | 114.7/5.6 | 30 | 27% | Abiotic stress tolerant |
| b | Ubiquitin-fold modifier 1 | UFM1_ORYSJ | LOC_Os01g73140 | Oryza sativa subsp. Japonica | 10356/9.60 | 81.67/5.98 | 30 | 44% | Abiotic stress tolerant |
| c | B3 domain-containing protein | Y1237_ORYSJ | LOC_Os01g52540 | Oryza sativa subsp. Japonica | 83749/5.81 | 118.37/5.71 | 38 | 34% | Abiotic stress tolerant |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saini, S.; Kaur, N.; Marothia, D.; Singh, B.; Singh, V.; Gantet, P.; Pati, P.K. Morphological Analysis, Protein Profiling and Expression Analysis of Auxin Homeostasis Genes of Roots of Two Contrasting Cultivars of Rice Provide Inputs on Mechanisms Involved in Rice Adaptation towards Salinity Stress. Plants 2021, 10, 1544. https://doi.org/10.3390/plants10081544
Saini S, Kaur N, Marothia D, Singh B, Singh V, Gantet P, Pati PK. Morphological Analysis, Protein Profiling and Expression Analysis of Auxin Homeostasis Genes of Roots of Two Contrasting Cultivars of Rice Provide Inputs on Mechanisms Involved in Rice Adaptation towards Salinity Stress. Plants. 2021; 10(8):1544. https://doi.org/10.3390/plants10081544
Chicago/Turabian StyleSaini, Shivani, Navdeep Kaur, Deeksha Marothia, Baldev Singh, Varinder Singh, Pascal Gantet, and Pratap Kumar Pati. 2021. "Morphological Analysis, Protein Profiling and Expression Analysis of Auxin Homeostasis Genes of Roots of Two Contrasting Cultivars of Rice Provide Inputs on Mechanisms Involved in Rice Adaptation towards Salinity Stress" Plants 10, no. 8: 1544. https://doi.org/10.3390/plants10081544
APA StyleSaini, S., Kaur, N., Marothia, D., Singh, B., Singh, V., Gantet, P., & Pati, P. K. (2021). Morphological Analysis, Protein Profiling and Expression Analysis of Auxin Homeostasis Genes of Roots of Two Contrasting Cultivars of Rice Provide Inputs on Mechanisms Involved in Rice Adaptation towards Salinity Stress. Plants, 10(8), 1544. https://doi.org/10.3390/plants10081544






