Lessons from Comparison of Hypoxia Signaling in Plants and Mammals
Abstract
1. Introduction
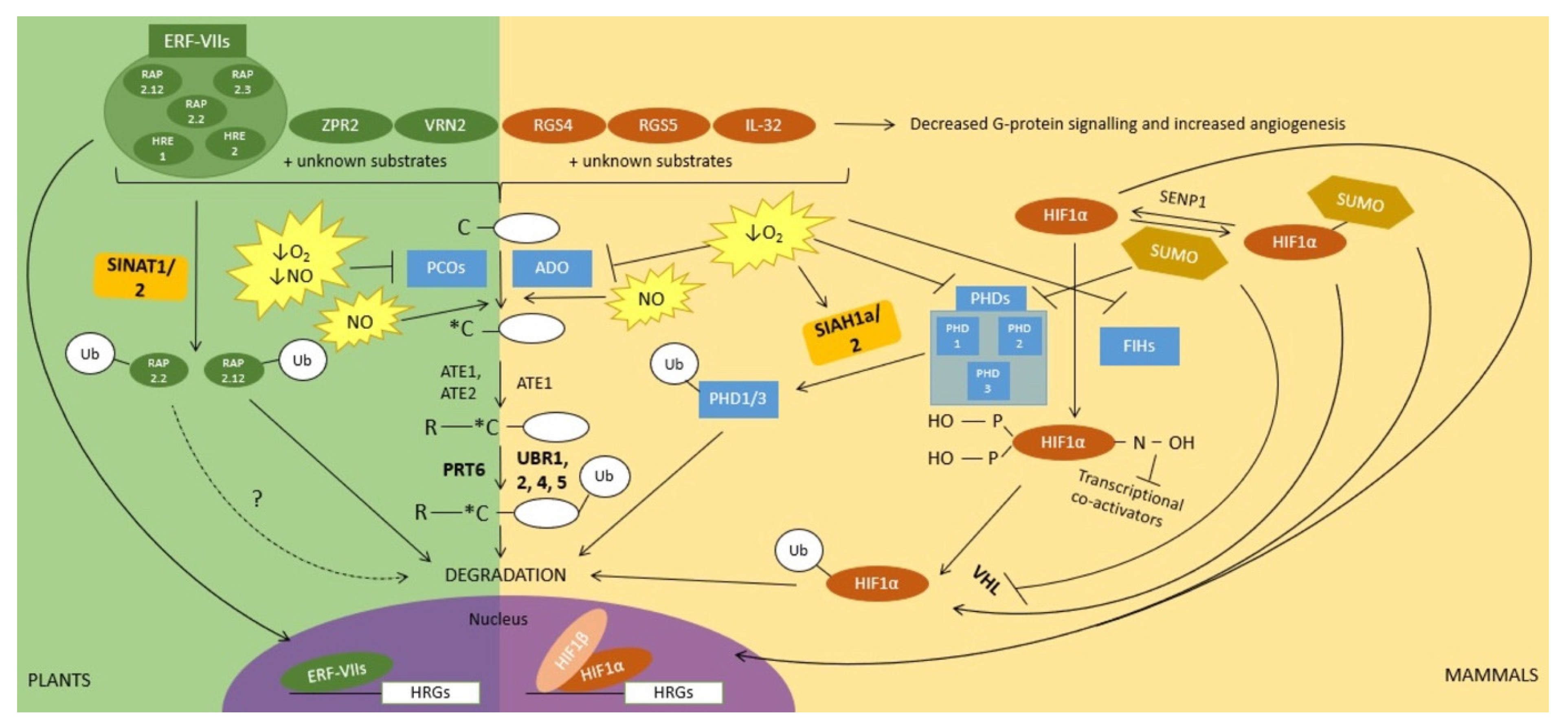
2. Ubiquitin and Sumoylation Function in Oxygen Sensing and in Downstream Signal Transduction
3. Ubiquitin and Sumoylation Function in Sugar/Energy Sensing and Downstream Signal Transduction
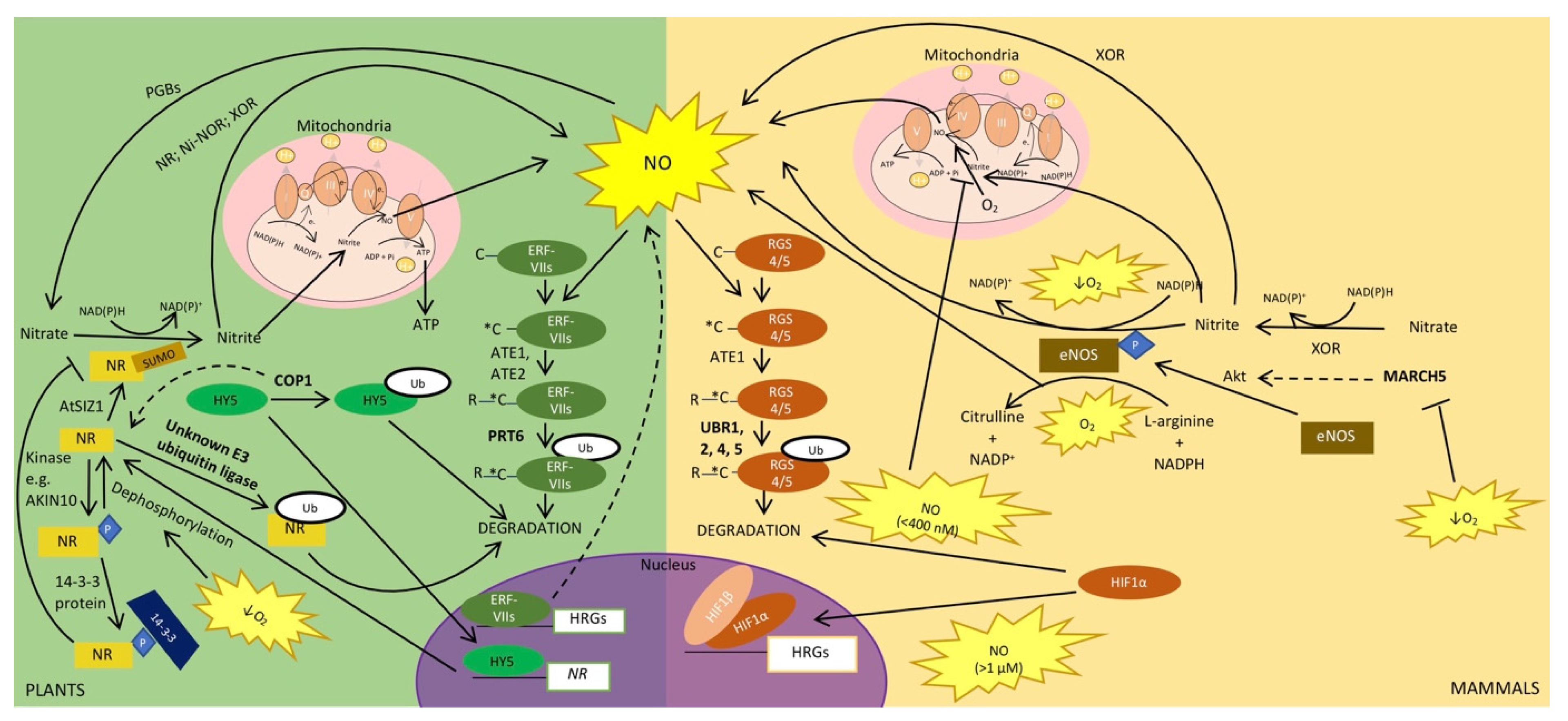
4. Ubiquitin and SUMO in the Regulation of NO Signalling during Hypoxia
4.1. NO Production and Regulation
4.2. Plant NRs and Mammalian eNOS Are Regulated by Sumoylation and Ubiquitination
4.3. Regulation of NO and Oxygen Sensing Pathways
4.4. Downstream Effects of NO also Require the Ubiquitin System and Ubiquitin-Like Proteins
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holdsworth, M.J.; Gibbs, D.J. Comparative Biology of Oxygen Sensing in Plants and Animals. Curr. Biol. 2020, 30, R362–R369. [Google Scholar] [CrossRef] [PubMed]
- Hammarlund, E.U.; Flashman, E.; Mohlin, S.; Licausi, F. Oxygen-sensing mechanisms across eukaryotic kingdoms and their roles in complex multicellularity. Science 2020, 370, eaba3512. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Barrera-Figueroa, B.E.; Juntawong, P.; Peña-Castro, J.M. Submergence and Waterlogging Stress in Plants: A Review Highlighting Research Opportunities and Understudied Aspects. Front. Plant Sci. 2019, 10, 340. [Google Scholar] [CrossRef] [PubMed]
- Weits, D.A.; Van Dongen, J.T.; Licausi, F. Molecular oxygen as a signaling component in plant development. New Phytol. 2021, 229, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Weits, D.; Kunkowska, A.B.; Kamps, N.C.W.; Portz, K.M.S.; Packbier, N.K.; Venza, Z.N.; Gaillochet, C.; Lohmann, J.U.; Pedersen, O.; Van Dongen, J.T.; et al. An apical hypoxic niche sets the pace of shoot meristem activity. Nat. Cell Biol. 2019, 569, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.; Lombardi, L.; Iacopino, S.; Pencik, A.; Novak, O.; Perata, P.; Giuntoli, B.; Licausi, F. Endogenous Hypoxia in Lateral Root Primordia Controls Root Architecture by Antagonizing Auxin Signaling in Arabidopsis. Mol. Plant 2019, 12, 538–551. [Google Scholar] [CrossRef]
- Rolletschek, H.; Stangelmayer, A.; Borisjuk, L. Methodology and Significance of Microsensor-based Oxygen Mapping in Plant Seeds—An Overview. Sensors 2009, 9, 3218–3227. [Google Scholar] [CrossRef]
- Mohyeldin, A.; Garzón-Muvdi, T.; Quiñones-Hinojosa, A. Oxygen in Stem Cell Biology: A Critical Component of the Stem Cell Niche. Cell Stem Cell 2010, 7, 150–161. [Google Scholar] [CrossRef]
- Voesenek, L.A.C.J.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.; van Dongen, J.T. Making sense of low oxygen sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef]
- Saxena, K.; Jolly, M.K. Acute vs. Chronic vs. Cyclic Hypoxia: Their Differential Dynamics, Molecular Mechanisms, and Effects on Tumor Progression. Biomolecules 2019, 9, 339. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Loreti, E.; Shih, M.; Perata, P. Energy and sugar signaling during hypoxia. New Phytol. 2021, 229, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Chandel, N.S.; Simon, M.C. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat. Rev. Mol. Cell Biol. 2020, 21, 268–283. [Google Scholar] [CrossRef]
- Pucciariello, C.; Perata, P. The Oxidative Paradox in Low Oxygen Stress in Plants. Antioxidants 2021, 10, 332. [Google Scholar] [CrossRef]
- Farnese, F.D.S.; Menezes-Silva, P.E.; Gusman, G.S.; Oliveira, J.A. When Bad Guys Become Good Ones: The Key Role of Reactive Oxygen Species and Nitric Oxide in the Plant Responses to Abiotic Stress. Front. Plant Sci. 2016, 7, 471. [Google Scholar] [CrossRef]
- Steffens, B.; Steffen-Heins, A.; Sauter, M. Reactive oxygen species mediate growth and death in submerged plants. Front. Plant Sci. 2013, 4, 179. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef]
- Akter, S.; Huang, J.; Waszczak, C.; Jacques, S.; Gevaert, K.; Van Breusegem, F.; Messens, J. Cysteines under ROS attack in plants: A proteomics view. J. Exp. Bot. 2015, 66, 2935–2944. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.A.; Bailey-Serres, J. Conserved and nuanced hierarchy of gene regulatory response to hypoxia. New Phytol. 2021, 229, 71–78. [Google Scholar] [CrossRef]
- Reynoso, M.A.; Kajala, K.; Bajic, M.; West, D.A.; Pauluzzi, G.; Yao, A.I.; Hatch, K.; Zumstein, K.; Woodhouse, M.; Rodriguez-Medina, J.; et al. Evolutionary flexibility in flooding response circuitry in angiosperms. Science 2019, 365, 1291–1295. [Google Scholar] [CrossRef]
- Filippopoulou, C.; Simos, G.; Chachami, G. The Role of Sumoylation in the Response to Hypoxia: An Overview. Cells 2020, 9, 2359. [Google Scholar] [CrossRef] [PubMed]
- Tomanov, K.; Nukarinen, E.; Vicente, J.; Mendiondo, G.M.; Winter, N.; Nehlin, L.; Weckwerth, W.; Holdsworth, M.J.; Teige, M.; Bachmair, A. Sumoylation and phosphorylation: Hidden and overt links. J. Exp. Bot. 2018, 69, 4583–4590. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Song, J.T.; Seo, H.S. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat. Commun. 2011, 2, 1–10. [Google Scholar] [CrossRef]
- Crozet, P.; Margalha, L.; Butowt, R.; Fernandes, N.; Elias, C.A.; Orosa, B.; Tomanov, K.; Teige, M.; Bachmair, A.; Sadanandom, A.; et al. SUMOylation represses SnRK1 signaling in Arabidopsis. Plant J. 2016, 85, 120–133. [Google Scholar] [CrossRef]
- Kunz, K.; Wagner, K.; Mendler, L.; Hölper, S.; Dehne, N.; Müller, S. SUMO Signaling by Hypoxic Inactivation of SUMO-Specific Isopeptidases. Cell Rep. 2016, 16, 3075–3086. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Rus, A.; Sharkhuu, A.; Yokoi, S.; Karthikeyan, A.S.; Raghothama, K.G.; Baek, D.; Koo, Y.D.; Jin, J.B.; Bressan, R.A.; et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 2005, 102, 7760–7765. [Google Scholar] [CrossRef]
- Miura, K.; Jin, J.B.; Lee, J.; Yoo, C.Y.; Stirm, V.; Miura, T.; Ashworth, E.N.; Bressan, R.A.; Yun, D.-J.; Hasegawa, P.M. SIZ1-Mediated Sumoylation of ICE1 Controls CBF3/DREB1A Expression and Freezing Tolerance in Arabidopsis. Plant Cell 2007, 19, 1403–1414. [Google Scholar] [CrossRef]
- Catala, R.; Ouyang, J.; Abreu, I.A.; Hu, Y.; Seo, H.; Zhang, X.; Chua, N.-H. The Arabidopsis E3 SUMO Ligase SIZ1 Regulates Plant Growth and Drought Responses. Plant Cell 2007, 19, 2952–2966. [Google Scholar] [CrossRef]
- Conti, L.; Price, G.; O’Donnell, E.; Schwessinger, B.; Dominy, P.; Sadanandom, A. Small Ubiquitin-Like Modifier Proteases Overly Tolerant to Salt1 and -2 Regulate Salt Stress Responses in Arabidopsis. Plant Cell 2008, 20, 2894–2908. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Zhang, C.; Yates, G.; Bailey, M.; Brown, A.; Sadanandom, A. SUMO Is a Critical Regulator of Salt Stress Responses in Rice. Plant Physiol. 2016, 170, 2378–2391. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.C.; York, S.L.; Rytz, T.C.; Vierstra, R.D. Defining the SUMO System in Maize: SUMOylation Is Up-Regulated during Endosperm Development and Rapidly Induced by Stress. Plant Physiol. 2016, 171, 2191–2210. [Google Scholar] [CrossRef]
- Li, Y.; Wang, G.; Xu, Z.; Li, J.; Sun, M.; Guo, J.; Ji, W. Organization and Regulation of Soybean SUMOylation System under Abiotic Stress Conditions. Front. Plant Sci. 2017, 8, 1458. [Google Scholar] [CrossRef] [PubMed]
- Tomanov, K.; Zeschmann, A.; Hermkes, R.; Eifler, K.; Ziba, I.; Grieco, M.; Novatchkova, M.; Hofmann, K.; Hesse, H.; Bachmair, A. Arabidopsis PIAL1 and 2 Promote SUMO Chain Formation as E4-Type SUMO Ligases and Are Involved in Stress Responses and Sulfur Metabolism. Plant Cell 2014, 26, 4547–4560. [Google Scholar] [CrossRef]
- Orosa, B.; Yates, G.; Verma, V.; Srivastava, A.K.; Srivastava, M.; Campanaro, A.; De Vega, D.; Fernandes, A.; Zhang, C.; Lee, J.; et al. SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Orosa, B.; Singh, P.; Cummins, I.; Walsh, C.; Zhang, C.; Grant, M.; Roberts, M.R.; Anand, G.S.; Fitches, E.; et al. SUMO Suppresses the Activity of the Jasmonic Acid Receptor Coronatine Insensitive1. Plant Cell 2018, 30, 2099–2115. [Google Scholar] [CrossRef] [PubMed]
- Morrell, R.; Sadanandom, A. Dealing with Stress: A Review of Plant SUMO Proteases. Front. Plant Sci. 2019, 10, 1122. [Google Scholar] [CrossRef]
- Srivastava, M.; Sadanandom, A. An Insight into the Factors Influencing Specificity of the SUMO System in Plants. Plants 2020, 9, 1788. [Google Scholar] [CrossRef] [PubMed]
- Miricescu, A.; Goslin, K.; Graciet, E. Ubiquitylation in plants: Signaling hub for the integration of environmental signals. J. Exp. Bot. 2018, 69, 4511–4527. [Google Scholar] [CrossRef]
- Roy, D.; Sadanandom, A. SUMO mediated regulation of transcription factors as a mechanism for transducing environmental cues into cellular signaling in plants. Cell. Mol. Life Sci. 2021, 78, 2641–2664. [Google Scholar] [CrossRef] [PubMed]
- Elrouby, N. Extent and significance of non-covalent SUMO interactions in plant development. Plant Signal. Behav. 2014, 9, e27948. [Google Scholar] [CrossRef]
- Elrouby, N.; Bonequi, M.V.; Porri, A.; Coupland, G. Identification of Arabidopsis SUMO-interacting proteins that regulate chromatin activity and developmental transitions. Proc. Natl. Acad. Sci. USA 2013, 110, 19956–19961. [Google Scholar] [CrossRef] [PubMed]
- Isono, E.; Nagel, M.-K. Deubiquitylating enzymes and their emerging role in plant biology. Front. Plant Sci. 2014, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbé, S. Deubiquitylases From Genes to Organism. Physiol. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X. SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes. Mol. Cell 2018, 71, 409–418. [Google Scholar] [CrossRef] [PubMed]
- White, M.D.; Klecker, M.; Hopkinson, R.J.; Weits, D.; Mueller, C.; Naumann, C.; O’Neill, R.; Wickens, J.; Yang, J.; Brooks-Bartlett, J.C.; et al. Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat. Commun. 2017, 8, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Weits, D.; Giuntoli, B.; Kosmacz, M.; Parlanti, S.; Hubberten, H.-M.; Riegler, H.; Hoefgen, R.; Perata, P.; Van Dongen, J.T.; Licausi, F. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef]
- White, M.D.; Carbonare, L.D.; Puerta, M.L.; Iacopino, S.; Edwards, M.; Dunne, K.; Pires, E.; Levy, C.; McDonough, M.A.; Licausi, F.; et al. Structures of Arabidopsis thaliana oxygen-sensing plant cysteine oxidases 4 and 5 enable targeted manipulation of their activity. Proc. Natl. Acad. Sci. USA 2020, 117, 23140–23147. [Google Scholar] [CrossRef]
- White, M.D.; Kamps, J.J.A.G.; East, S.; Kearney, L.J.T.; Flashman, E. The plant cysteine oxidases from Arabidopsis thaliana are kinetically tailored to act as oxygen sensors. J. Biol. Chem. 2018, 293, 11786–11795. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Tedds, H.M.; Labandera, A.-M.; Bailey, M.; White, M.D.; Hartman, S.; Sprigg, C.; Mogg, S.L.; Osborne, R.; Dambire, C.; et al. Oxygen-dependent proteolysis regulates the stability of angiosperm polycomb repressive complex 2 subunit VERNALIZATION 2. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Lee, S.C.; Isa, N.M.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodoulou, F.L.; Bailey-Serres, J.; et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nat. Cell Biol. 2011, 479, 415–418. [Google Scholar] [CrossRef]
- Licausi, F.; Kosmacz, M.; Weits, D.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.C.J.; Perata, P.; Van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nat. Cell Biol. 2011, 479, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Conde, J.V.; Berckhan, S.; Prasad, G.; Mendiondo, G.M.; Holdsworth, M.J. Group VII Ethylene Response Factors Coordinate Oxygen and Nitric Oxide Signal Transduction and Stress Responses in Plants. Plant Physiol. 2015, 169, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Ito, M.; Callis, J.; Nishida, I.; Watanabe, A. A delayed leaf senescence mutant is defective in arginyl-tRNA:protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 2002, 32, 129–137. [Google Scholar] [CrossRef]
- Garzón, M.; Eifler, K.; Faust, A.; Scheel, H.; Hofmann, K.; Koncz, C.; Yephremov, A.; Bachmair, A. PRT6/At5g02310 encodes anArabidopsisubiquitin ligase of the N-end rule pathway with arginine specificity and is not theCER3locus. FEBS Lett. 2007, 581, 3189–3196. [Google Scholar] [CrossRef] [PubMed]
- Graciet, E.; Mesiti, F.; Wellmer, F. Structure and evolutionary conservation of the plant N-end rule pathway. Plant J. 2010, 61, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Graciet, E.; Wellmer, F. The plant N-end rule pathway: Structure and functions. Trends Plant Sci. 2010, 15, 447–453. [Google Scholar] [CrossRef]
- Graciet, E.; Walter, F.; Maoileidigh, D.O.; Pollmann, S.; Meyerowitz, E.M.; Varshavsky, A.; Wellmer, F. The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc. Natl. Acad. Sci. USA 2009, 106, 13618–13623. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.R.; Fulda, M.; Paul, M.V.; Anders, M.; Plum, F.; Weits, D.A.; Kosmacz, M.; Larson, T.R.; Graham, I.A.; Beemster, G.T.S.; et al. Low-oxygen response is triggered by an ATP-dependent shift in oleoyl-CoA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E12101–E12110. [Google Scholar] [CrossRef]
- Dissmeyer, N. Conditional Protein Function via N-Degron Pathway–Mediated Proteostasis in Stress Physiology. Annu. Rev. Plant Biol. 2019, 70, 83–117. [Google Scholar] [CrossRef]
- Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef]
- Masson, N.; Keeley, T.P.; Giuntoli, B.; White, M.D.; Puerta, M.L.; Perata, P.; Hopkinson, R.J.; Flashman, E.; Licausi, F.; Ratcliffe, P.J. Conserved N-terminal cysteine dioxygenases transduce responses to hypoxia in animals and plants. Science 2019, 365, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.-G.; Sheng, J.; Qi, X.; Xu, Z.; Takahashi, T.T.; Varshavsky, A. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nat. Cell Biol. 2005, 437, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Tasaki, T.; Moroi, K.; An, J.Y.; Kimura, S.; Davydov, I.V.; Kwon, Y.T. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 15030–15035. [Google Scholar] [CrossRef]
- Huang, L.E.; Gu, J.; Schau, M.; Bunn, H.F. Regulation of hypoxia-inducible factor 1 is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 7987–7992. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.-W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nat. Cell Biol. 1999, 399, 271–275. [Google Scholar] [CrossRef]
- Tanimoto, K.; Makino, Y.; Pereira, T.; Poellinger, L. Mechanism of regulation of the hypoxia-inducible factor-1alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 2000, 19, 4298–4309. [Google Scholar] [CrossRef]
- Jaakkola, P.; Mole, D.R.; Tian, Y.-M.; Wilson, M.I.; Gielbert, J.; Gaskell, S.J.; Von Kriegsheim, A.; Hebestreit, H.F.; Mukherji, M.; Schofield, C.J.; et al. Targeting of HIF-alpha to the von Hippel-Lindau Ubiquitylation Complex by O2-Regulated Prolyl Hydroxylation. Science 2001, 292, 468–472. [Google Scholar] [CrossRef]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, W.G. HIFalpha Targeted for VHL-Mediated Destruction by Proline Hydroxylation: Implications for O2 Sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Lando, D.; Peet, D.J.; Gorman, J.J.; Whelan, D.A.; Whitelaw, M.L.; Bruick, R.K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002, 16, 1466–1471. [Google Scholar] [CrossRef]
- Lando, D.; Peet, D.J.; Whelan, D.A.; Gorman, J.J.; Whitelaw, M.L. Asparagine Hydroxylation of the HIF Transactivation Domain: A Hypoxic Switch. Science 2002, 295, 858–861. [Google Scholar] [CrossRef] [PubMed]
- McNeill, L.A.; Hewitson, K.S.; Claridge, T.D.; Seibel, J.F.; Horsfall, L.E.; Schofield, C.J. Hypoxia-inducible factor asparaginyl hydroxylase (FIH-1) catalyses hydroxylation at the β-carbon of asparagine-803. Biochem. J. 2002, 367, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Hewitson, K.S.; McNeill, L.A.; Riordan, M.V.; Tian, Y.-M.; Bullock, A.N.; Welford, R.W.; Elkins, J.M.; Oldham, N.J.; Bhattacharya, S.; Gleadle, J.M.; et al. Hypoxia-inducible Factor (HIF) Asparagine Hydroxylase Is Identical to Factor Inhibiting HIF (FIH) and Is Related to the Cupin Structural Family. J. Biol. Chem. 2002, 277, 26351–26355. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Kang, X.; Zhang, S.; Yeh, E.T. SUMO-Specific Protease 1 Is Essential for Stabilization of HIF1α during Hypoxia. Cell 2007, 131, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.-H.; Jeong, J.-W.; Park, J.A.; Kim, S.-H.; Bae, M.-K.; Choi, S.-J.; Kim, K.-W. Sumoylation increases HIF-1α stability and its transcriptional activity. Biochem. Biophys. Res. Commun. 2004, 324, 394–400. [Google Scholar] [CrossRef]
- Carbia-Nagashima, A.; Gerez, J.; Perez-Castro, C.; Paez-Pereda, M.; Silberstein, S.; Stalla, G.K.; Holsboer, F.; Arzt, E. RSUME, a Small RWD-Containing Protein, Enhances SUMO Conjugation and Stabilizes HIF-1α during Hypoxia. Cell 2007, 131, 309–323. [Google Scholar] [CrossRef]
- Berta, M.A.; Mazure, N.; Hattab, M.; Pouysségur, J.; Brahimi-Horn, M.C. SUMOylation of hypoxia-inducible factor-1α reduces its transcriptional activity. Biochem. Biophys. Res. Commun. 2007, 360, 646–652. [Google Scholar] [CrossRef]
- Kang, X.; Li, J.; Zou, Y.; Yi, J.; Zhang, H.; Cao, M.; Yeh, E.T.H.; Cheng, J. PIASy stimulates HIF1α SUMOylation and negatively regulates HIF1α activity in response to hypoxia. Oncogene 2010, 29, 5568–5578. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Long, X.-D.; Wang, W.; Jiao, H.-K.; Mei, Z.; Yin, Q.-Q.; Ma, L.-N.; Zhou, A.-W.; Wang, L.-S.; et al. Cbx4 Governs HIF-1α to Potentiate Angiogenesis of Hepatocellular Carcinoma by Its SUMO E3 Ligase Activity. Cancer Cell 2014, 25, 118–131. [Google Scholar] [CrossRef]
- Nakagawa, K.; Kohara, T.; Uehata, Y.; Miyakawa, Y.; Sato-Ueshima, M.; Okubo, N.; Asaka, M.; Takeda, H.; Kobayashi, M. PIAS3 enhances the transcriptional activity of HIF-1α by increasing its protein stability. Biochem. Biophys. Res. Commun. 2016, 469, 470–476. [Google Scholar] [CrossRef]
- Gerez, J.; Tedesco, L.; Bonfiglio, J.J.; Fuertes, M.A.; Barontini, M.; Silberstein, S.; Wu, Y.; Renner, U.; Paezpereda, M.; Holsboer, F.; et al. RSUME inhibits VHL and regulates its tumor suppressor function. Oncogene 2015, 34, 4855–4866. [Google Scholar] [CrossRef]
- Cai, Q.; Verma, S.C.; Kumar, P.; Ma, M.; Robertson, E.S. Hypoxia Inactivates the VHL Tumor Suppressor through PIASy-Mediated SUMO Modification. PLoS ONE 2010, 5, e9720. [Google Scholar] [CrossRef]
- Núñez-O’Mara, A.; Gerpe-Pita, A.; Pozo, S.; Carlevaris, O.; Urzelai, B.; Lopitz-Otsoa, F.; Rodríguez, M.S.; Berra, E. PHD3–SUMO conjugation represses HIF1 transcriptional activity independently of PHD3 catalytic activity. J. Cell Sci. 2015, 128, 40–49. [Google Scholar] [CrossRef]
- Papdi, C.; Pérez-Salamó, I.; Joseph, M.P.; Giuntoli, B.; Bögre, L.; Koncz, C.; Szabados, L. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genesRAP2.12, RAP2.2andRAP2.3. Plant J. 2015, 82, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.; Mendiondo, G.M.; Movahedi, M.; Peirats-Llobet, M.; Juan, Y.T.; Shen, Y.Y.; Dambire, C.; Smart, K.; Rodriguez, P.L.; Charng, Y.Y.; et al. The Cys-Arg/N-End Rule Pathway Is a General Sensor of Abiotic Stress in Flowering Plants. Curr. Biol. 2017, 27, 3183–3190. e3184. [Google Scholar] [CrossRef] [PubMed]
- Gravot, A.; Richard, G.; Lime, T.; Lemarié, S.; Jubault, M.; Lariagon, C.; Lemoine, J.; Vicente, J.; Robert-Seilaniantz, A.; Holdsworth, M.J.; et al. Hypoxia response in Arabidopsis roots infected by Plasmodiophora brassicae supports the development of clubroot. BMC Plant Biol. 2016, 16, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wei, T.; Yin, K.; Chen, Z.; Gu, H.; Qu, L.; Qin, G. Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol. 2012, 195, 450–460. [Google Scholar] [CrossRef]
- Giuntoli, B.; Perata, P. Group VII Ethylene Response Factors in Arabidopsis: Regulation and Physiological Roles. Plant Physiol. 2018, 176, 1143–1155. [Google Scholar] [CrossRef]
- Welsch, R.; Maass, D.; Voegel, T.; DellaPenna, D.; Beyer, P. Transcription Factor RAP2.2 and Its Interacting Partner SINAT2: Stable Elements in the Carotenogenesis of Arabidopsis Leaves. Plant Physiol. 2007, 145, 1073–1085. [Google Scholar] [CrossRef]
- Zhang, C.; Hao, Z.; Ning, Y.; Wang, G.-L. SINA E3 Ubiquitin Ligases: Versatile Moderators of Plant Growth and Stress Response. Mol. Plant 2019, 12, 610–612. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.-N.; Zeng, B.; Liu, H.-S.; Qi, H.; Xie, L.-J.; Yu, L.-J.; Chen, Q.-F.; Li, J.-F.; Chen, Y.-Q.; Jiang, L.; et al. SINAT E3 Ubiquitin Ligases Mediate FREE1 and VPS23A Degradation to Modulate Abscisic Acid Signaling. Plant Cell 2020, 32, 3290–3310. [Google Scholar] [CrossRef]
- Qi, H.; Li, J.; Xia, F.-N.; Chen, J.-Y.; Lei, X.; Han, M.-Q.; Xie, L.-J.; Zhou, Q.-M.; Xiao, S. Arabidopsis SINAT Proteins Control Autophagy by Mediating Ubiquitylation and Degradation of ATG13. Plant Cell 2020, 32, 263–284. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Xia, F.-N.; Xie, L.-J.; Yu, L.-J.; Chen, Q.-F.; Zhuang, X.-H.; Wang, Q.; Li, F.; Jiang, L.; Xie, Q.; et al. TRAF Family Proteins Regulate Autophagy Dynamics by Modulating AUTOPHAGY PROTEIN6 Stability in Arabidopsis. Plant Cell 2017, 29, 890–911. [Google Scholar] [CrossRef] [PubMed]
- Calzado, M.A.; De La Vega, L.; Möller, A.; Bowtell, D.D.L.; Schmitz, M.L. An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat. Cell Biol. 2008, 11, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Park, S.; Zhang, X.; Dai, W.; Xu, D. Mutual regulation between Polo-like kinase 3 and SIAH2 E3 ubiquitin ligase defines a regulatory network that fine-tunes the cellular response to hypoxia and nickel. J. Biol. Chem. 2017, 292, 11431–11444. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Chen, Y.; Chen, L.; Cheng, H.; Mu, C.; Li, J.; Gao, R.; Zhou, C.; Cao, L.; Liu, J.; et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat. Cell Biol. 2014, 17, 95–103. [Google Scholar] [CrossRef]
- Nakayama, K.; Frew, I.J.; Hagensen, M.; Skals, M.; Habelhah, H.; Bhoumik, A.; Kadoya, T.; Erdjument-Bromage, H.; Tempst, P.; Frappell, P.B.; et al. Siah2 Regulates Stability of Prolyl-Hydroxylases, Controls HIF1α Abundance, and Modulates Physiological Responses to Hypoxia. Cell 2004, 117, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Polge, C.; Thomas, M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007, 12, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Srivastava, V. SNF1-related protein kinase 1: The many-faced signaling hub regulating developmental plasticity in plants. J. Exp. Bot. 2021. [Google Scholar] [CrossRef]
- Broeckx, T.; Hulsmans, S.; Rolland, F. The plant energy sensor: Evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 2016, 67, 6215–6252. [Google Scholar] [CrossRef]
- Dengler, F. Activation of AMPK under Hypoxia: Many Roads Leading to Rome. Int. J. Mol. Sci. 2020, 21, 2428. [Google Scholar] [CrossRef]
- Hawley, S.A.; Davison, M.; Woods, A.; Davies, S.P.; Beri, R.K.; Carling, D.; Hardie, D.G. Characterization of the AMP-activated Protein Kinase Kinase from Rat Liver and Identification of Threonine 172 as the Major Site at Which It Phosphorylates AMP-activated Protein Kinase. J. Biol. Chem. 1996, 271, 27879–27887. [Google Scholar] [CrossRef]
- Shaw, R.J.; Lamia, K.A.; Vasquez, D.; Koo, S.-H.; Bardeesy, N.; Depinho, R.A.; Montminy, M.; Cantley, L.C. The Kinase LKB1 Mediates Glucose Homeostasis in Liver and Therapeutic Effects of Metformin. Science 2005, 310, 1642–1646. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nat. Cell Biol. 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Fragoso, S.; Espíndola, L.; Páez-Valencia, J.; Gamboa, A.; Camacho, Y.; Martínez-Barajas, E.; Coello, P. SnRK1 Isoforms AKIN10 and AKIN11 Are Differentially Regulated in Arabidopsis Plants under Phosphate Starvation. Plant Physiol. 2009, 149, 1906–1916. [Google Scholar] [CrossRef] [PubMed]
- Emanuelle, S.; Doblin, M.S.; Gooley, P.R.; Gentry, M.S. The UBA domain of SnRK1 promotes activation and maintains catalytic activity. Biochem. Biophys. Res. Commun. 2018, 497, 127–132. [Google Scholar] [CrossRef]
- Cho, Y.-H.; Hong, J.-W.; Kim, E.-C.; Yoo, S.-D. Regulatory Functions of SnRK1 in Stress-Responsive Gene Expression and in Plant Growth and Development. Plant Physiol. 2012, 158, 1955–1964. [Google Scholar] [CrossRef]
- Cho, H.-Y.; Wen, T.-N.; Wang, Y.-T.; Shih, M.-C. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J. Exp. Bot. 2016, 67, 2745–2760. [Google Scholar] [CrossRef]
- Soltani, A.; MafiMoghaddam, S.; Oladzad-Abbasabadi, A.; Walter, K.; Kearns, P.J.; Vasquez-Guzman, J.; Mamidi, S.; Lee, R.; Shade, A.L.; Jacobs, J.L.; et al. Genetic Analysis of Flooding Tolerance in an Andean Diversity Panel of Dry Bean (Phaseolus vulgaris L.). Front. Plant Sci. 2018, 9, 767. [Google Scholar] [CrossRef]
- Ramon, M.; Dang, T.V.T.; Broeckx, T.; Hulsmans, S.; Crepin, N.; Sheen, J.; Rolland, F. Default Activation and Nuclear Translocation of the Plant Cellular Energy Sensor SnRK1 Regulate Metabolic Stress Responses and Development. Plant Cell 2019, 31, 1614–1632. [Google Scholar] [CrossRef]
- Cho, H.-Y.; Lu, M.-Y.J.; Shih, M.-C. The Sn RK 1- eIF iso4G1 signaling relay regulates the translation of specific mRNA s in Arabidopsis under submergence. New Phytol. 2018, 222, 366–381. [Google Scholar] [CrossRef]
- Margalha, L.; Confraria, A.; Baena-González, E. SnRK1 and TOR: Modulating growth–defense trade-offs in plant stress responses. J. Exp. Bot. 2019, 70, 2261–2274. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Reyes, M.I.; Hanley-Bowdoin, L. Arabidopsis Protein Kinases GRIK1 and GRIK2 Specifically Activate SnRK1 by Phosphorylating Its Activation Loop. Plant Physiol. 2009, 150, 996–1005. [Google Scholar] [CrossRef] [PubMed]
- Glab, N.; Oury, C.; Guérinier, T.; Domenichini, S.; Crozet, P.; Thomas, M.; Vidal, J.; Hodges, M. The impact of Arabidopsis thaliana SNF 1-related-kinase 1 (Sn RK 1)-activating kinase 1 (Sn AK 1) and Sn AK 2 on Sn RK 1 phosphorylation status: Characterization of a Sn AK double mutant. Plant J. 2017, 89, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Oakhill, J.S.; Steel, R.; Chen, Z.-P.; Scott, J.W.; Ling, N.; Tam, S.; Kemp, B.E. AMPK Is a Direct Adenylate Charge-Regulated Protein Kinase. Science 2011, 332, 1433–1435. [Google Scholar] [CrossRef]
- Wurzinger, B.; Mair, A.; Fischer-Schrader, K.; Nukarinen, E.; Roustan, V.; Weckwerth, W.; Teige, M. Redox state-dependent modulation of plant SnRK1 kinase activity differs from AMPK regulation in animals. FEBS Lett. 2017, 591, 3625–3636. [Google Scholar] [CrossRef]
- Zhang, Y.; Primavesi, L.F.; Jhurreea, D.; Andralojc, P.J.; Mitchell, R.A.; Powers, S.J.; Schluepmann, H.; Delatte, T.; Wingler, A.; Paul, M.J. Inhibition of SNF1-Related Protein Kinase1 Activity and Regulation of Metabolic Pathways by Trehalose-6-Phosphate. Plant Physiol. 2009, 149, 1860–1871. [Google Scholar] [CrossRef]
- Zhai, Z.; Keereetaweep, J.; Liu, H.; Feil, R.; Lunn, J.E.; Shanklin, J. Trehalose 6-Phosphate Positively Regulates Fatty Acid Synthesis by Stabilizing WRINKLED1. Plant Cell 2018, 30, 2616–2627. [Google Scholar] [CrossRef]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Glinski, M.; Weckwerth, W. Differential Multisite Phosphorylation of the Trehalose-6-phosphate Synthase Gene Family in Arabidopsis thaliana. Mol. Cell. Proteom. 2005, 4, 1614–1625. [Google Scholar] [CrossRef]
- Harthill, J.E.; Meek, S.E.M.; Morrice, N.; Peggie, M.W.; Borch, J.; Wong, B.H.C.; Mackintosh, C. Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J. 2006, 47, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Nukarinen, E.; Nägele, T.; Pedrotti, L.; Wurzinger, B.; Mair, A.; Landgraf, R.; Börnke, F.; Hanson, J.; Teige, M.; Baena-Gonzalez, E.; et al. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 2016, 6, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ecrozet, P.; Emargalha, L.; Econfraria, A.; Rodrigues, A.; Martinho, C.; Eadamo, M.; Elias, C.A.; Baena-Gonzã¡lez, E. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front. Plant Sci. 2014, 5, 190. [Google Scholar] [CrossRef]
- Farrás, R.; Ferrando, A.; Jásik, J.; Kleinow, T.; Ökrész, L.; Tiburcio, A.; Salchert, K.; Del Pozo, C.; Schell, J.; Koncz, C. SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 2001, 20, 2742–2756. [Google Scholar] [CrossRef]
- Bhalerao, R.; Salchert, K.; Bakó, L.; Ökrész, L.; Szabados, L.; Muranaka, T.; Machida, Y.; Schell, J.; Koncz, C. Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. USA 1999, 96, 5322–5327. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Terzaghi, W.; Gusmaroli, G.; Charron, J.-B.F.; Yoon, H.-J.; Chen, H.; He, Y.J.; Xiong, Y.; Deng, X.W. Characterization of Arabidopsis and Rice DWD Proteins and Their Roles as Substrate Receptors for CUL4-RING E3 Ubiquitin Ligases. Plant Cell 2008, 20, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Ananieva, E.A.; Gillaspy, G.E.; Ely, A.; Burnette, R.N.; Erickson, F.L. Interaction of the WD40 Domain of a Myoinositol Polyphosphate 5-Phosphatase with SnRK1 Links Inositol, Sugar, and Stress Signaling. Plant Physiol. 2008, 148, 1868–1882. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E.; Li, X.; Deng, Y.; Chang, H.W.; Kim, D.Y. AMPK is down-regulated by the CRL4A-CRBN axis through the polyubiquitination of AMPKα isoforms. FASEB J. 2019, 33, 6539–6550. [Google Scholar] [CrossRef] [PubMed]
- Moreno, D.; Towler, M.C.; Hardie, D.G.; Knecht, E.; Sanz, P. The Laforin–Malin Complex, Involved in Lafora Disease, Promotes the Incorporation of K63-linked Ubiquitin Chains into AMP-activated Protein Kinase β Subunits. Mol. Biol. Cell 2010, 21, 2578–2588. [Google Scholar] [CrossRef] [PubMed]
- Al-Hakim, A.K.; Zagorska, A.; Chapman, L.; Deak, M.; Peggie, M.; Alessi, D.R. Control of AMPK-related kinases by USP9X and atypical Lys29/Lys33-linked polyubiquitin chains. Biochem. J. 2008, 411, 249–260. [Google Scholar] [CrossRef]
- Liu, H.; Ding, J.; Köhnlein, K.; Urban, N.; Ori, A.; Villavicencio-Lorini, P.; Walentek, P.; Klotz, L.-O.; Hollemann, T.; Pfirrmann, T. The GID ubiquitin ligase complex is a regulator of AMPK activity and organismal lifespan. Autophagy 2020, 16, 1618–1634. [Google Scholar] [CrossRef]
- Ovens, A.; Scott, J.; Langendorf, C.; Kemp, B.; Oakhill, J.; Smiles, W. Post-Translational Modifications of the Energy Guardian AMP-Activated Protein Kinase. Int. J. Mol. Sci. 2021, 22, 1229. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Yang, X.; Qin, B.; Liu, T.; Zhang, H.; Guo, W.; Lee, S.B.; Kim, J.J.; Yuan, J.; Pei, H.; et al. Deubiquitination and Activation of AMPK by USP10. Mol. Cell 2016, 61, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-J.; Jeon, S.-J.; Van Nguyen, T.; Deshaies, R.J.; Park, C.-S.; Lee, K.M. Ubiquitin-dependent proteasomal degradation of AMPK gamma subunit by Cereblon inhibits AMPK activity. Biochim. Biophys. Acta Bioenerg. 2020, 1867, 118729. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.O.; Lee, S.K.; Kim, N.; Kim, J.H.; You, G.Y.; Moon, J.W.; Jie, S.; Kim, S.J.; Lee, Y.W.; Kang, H.J.; et al. E3 Ubiquitin Ligase, WWP1, Interacts with AMPKα2 and Down-regulates Its Expression in Skeletal Muscle C2C12 Cells. J. Biol. Chem. 2013, 288, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- León, J.; Castillo, M.C.; Gayubas, B. The hypoxia–reoxygenation stress in plants. J. Exp. Bot. 2020. [Google Scholar] [CrossRef] [PubMed]
- Elrouby, N.; Coupland, G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. USA 2010, 107, 17415–17420. [Google Scholar] [CrossRef]
- Rubio, T.; Vernia, S.; Sanz, P. Sumoylation of AMPKβ2 subunit enhances AMP-activated protein kinase activity. Mol. Biol. Cell 2013, 24, 1801–1811. [Google Scholar] [CrossRef]
- Yan, Y.; Ollila, S.; Wong, I.P.L.; Vallenius, T.; Palvimo, J.J.; Vaahtomeri, K.; Mäkelä, T.P. SUMOylation of AMPKα1 by PIAS4 specifically regulates mTORC1 signalling. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef]
- Bogdan, C. Nitric oxide and the regulation of gene expression. Trends Cell Biol. 2001, 11, 66–75. [Google Scholar] [CrossRef]
- Mugnai, S.; Azzarello, E.; Baluška, F.; Mancuso, S. Local Root Apex Hypoxia Induces NO-Mediated Hypoxic Acclimation of the Entire Root. Plant Cell Physiol. 2012, 53, 912–920. [Google Scholar] [CrossRef] [PubMed]
- Zhan, N.; Wang, C.; Chen, L.; Yang, H.; Feng, J.; Gong, X.; Ren, B.; Wu, R.; Mu, J.; Li, Y.; et al. S-Nitrosylation Targets GSNO Reductase for Selective Autophagy during Hypoxia Responses in Plants. Mol. Cell 2018, 71, 142–154.e6. [Google Scholar] [CrossRef]
- Wany, A.; Gupta, A.K.; Brotman, Y.; Pandey, S.; Vishwakarma, A.P.; Kumari, A.; Singh, P.; Pathak, P.K.; Igamberdiev, A.U.; Gupta, K.J. Nitric oxide is important for sensing and survival under hypoxia in Arabidopsis. bioRxiv 2018, 462218. [Google Scholar] [CrossRef]
- Gupta, K.J.; Mur, L.A.J.; Wany, A.; Kumari, A.; Fernie, A.R.; Ratcliffe, R.G. The role of nitrite and nitric oxide under low oxygen conditions in plants. New Phytol. 2019, 225, 1143–1151. [Google Scholar] [CrossRef]
- Förstermann, U.; Sessa, W.C. Nitric oxide synthases: Regulation and function. Eur. Heart J. 2012, 33, 829–837. [Google Scholar] [CrossRef]
- Krumenacker, J.S.; Hanafy, K.A.; Murad, F. Regulation of nitric oxide and soluble guanylyl cyclase. Brain Res. Bull. 2004, 62, 505–515. [Google Scholar] [CrossRef]
- Lei, W.; Li, J.; Li, C.; Chen, L.; Huang, F.; Xiao, D.; Zhang, J.; Zhao, J.; Li, G.; Qu, T.; et al. MARCH5 restores endothelial cell function against ischaemic/hypoxia injury via Akt/eNOS pathway. J. Cell. Mol. Med. 2021, 25, 3182–3193. [Google Scholar] [CrossRef] [PubMed]
- Wany, A.; Kumari, A.; Gupta, K.J. Nitric oxide is essential for the development of aerenchyma in wheat roots under hypoxic stress. Plant Cell Environ. 2017, 40, 3002–3017. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Isa, N.M.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; La Rosa, N.M.-D.; Conde, J.V.; Correia, C.S.; Pearce, S.P.; et al. Nitric Oxide Sensing in Plants Is Mediated by Proteolytic Control of Group VII ERF Transcription Factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef]
- Gautier, C.; Van Faassen, E.; Mikula, I.; Martasek, P.; Slama-Schwok, A. Endothelial nitric oxide synthase reduces nitrite anions to NO under anoxia. Biochem. Biophys. Res. Commun. 2006, 341, 816–821. [Google Scholar] [CrossRef]
- Mikula, I.; Durocher, S.; Martásek, P.; Mutus, B.; Slama-Schwok, A. Isoform-specific differences in the nitrite reductase activity of nitric oxide synthases under hypoxia. Biochem. J. 2009, 418, 673–682. [Google Scholar] [CrossRef]
- Vanin, A.; Bevers, L.M.; Slama-Schwok, A.; Van Faassen, E.E. Nitric oxide synthase reduces nitrite to NO under anoxia. Cell. Mol. Life Sci. 2006, 64, 96–103. [Google Scholar] [CrossRef]
- Jansson, E.Å.; Huang, L.; Malkey, R.; Govoni, M.; Nihlén, C.; Olsson, A.; Stensdotter, M.; Petersson, J.; Holm, L.; Weitzberg, E.; et al. A mammalian functional nitrate reductase that regulates nitrite and nitric oxide homeostasis. Nat. Chem. Biol. 2008, 4, 411–417. [Google Scholar] [CrossRef]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E.P.-A. Nitrate Reductase Regulates Plant Nitric Oxide Homeostasis. Trends Plant Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef]
- Lundberg, J.O.; Weitzberg, E.; Gladwin, M.T. The nitrate–nitrite–nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 2008, 7, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Stoimenova, M.; Kaiser, W.M. In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. J. Exp. Bot. 2005, 56, 2601–2609. [Google Scholar] [CrossRef] [PubMed]
- Allègre, A.; Silvestre, J.; Morard, P.; Kallerhoff, J.; Pinelli, E. Nitrate reductase regulation in tomato roots by exogenous nitrate: A possible role in tolerance to long-term root anoxia. J. Exp. Bot. 2004, 55, 2625–2634. [Google Scholar] [CrossRef]
- Mohn, M.A.; Thaqi, B.; Fischer-Schrader, K. Isoform-Specific NO Synthesis by Arabidopsis thaliana Nitrate Reductase. Plants 2019, 8, 67. [Google Scholar] [CrossRef]
- Stöhr, C.; Strube, F.; Marx, G.; Ullrich, W.R.; Rockel, P. A plasma membrane-bound enzyme of tobacco roots catalyses the formation of nitric oxide from nitrite. Planta 2001, 212, 835–841. [Google Scholar] [CrossRef]
- Stoimenova, M.; Igamberdiev, A.U.; Gupta, K.J.; Hill, R.D. Nitrite-driven anaerobic ATP synthesis in barley and rice root mitochondria. Planta 2007, 226, 465–474. [Google Scholar] [CrossRef]
- Kozlov, A.V.; Staniek, K.; Nohl, H. Nitrite reductase activity is a novel function of mammalian mitochondria. FEBS Lett. 1999, 454, 127–130. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, B.S.; Park, S.W.; Lee, H.Y.; Song, J.T.; Seo, H.S. Nitrate Reductases Are Relocalized to the Nucleus by AtSIZ1 and Their Levels Are Negatively Regulated by COP1 and Ammonium. Int. J. Mol. Sci. 2018, 19, 1202. [Google Scholar] [CrossRef]
- Jonassen, E.M.; Lea, U.S.; Lillo, C. HY5 and HYH are positive regulators of nitrate reductase in seedlings and rosette stage plants. Planta 2007, 227, 559–564. [Google Scholar] [CrossRef]
- Van Veen, H.; Mustroph, A.; Barding, G.A.; Eijk, M.V.-V.; Welschen-Evertman, R.A.M.; Pedersen, O.; Visser, E.J.; Larive, C.K.; Pierik, R.; Bailey-Serres, J.; et al. Two Rumex Species from Contrasting Hydrological Niches Regulate Flooding Tolerance through Distinct Mechanisms. Plant Cell 2013, 25, 4691–4707. [Google Scholar] [CrossRef]
- Hartman, S.; Liu, Z.; Van Veen, H.; Vicente, J.; Reinen, E.; Martopawiro, S.; Zhang, H.; Van Dongen, N.; Bosman, F.; Bassel, G.W.; et al. Ethylene-mediated nitric oxide depletion pre-adapts plants to hypoxia stress. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Perazzolli, M.; Dominici, P.; Romero-Puertas, M.C.; Zago, E.; Zeier, J.; Sonoda, M.; Lamb, C.; Delledonne, M. Arabidopsis Nonsymbiotic Hemoglobin AHb1 Modulates Nitric Oxide Bioactivity. Plant Cell 2004, 16, 2785–2794. [Google Scholar] [CrossRef]
- Hebelstrup, K.H.; Van Zanten, M.; Mandon, J.; Voesenek, L.A.; Harren, F.J.M.; Cristescu, S.M.; Møller, I.M.; Mur, L.A.J. Haemoglobin modulates NO emission and hyponasty under hypoxia-related stress in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 5581–5591. [Google Scholar] [CrossRef]
- Brüne, B.; von Knethen, A.; Sandau, K.B. Transcription factors p53 and HIF-1α as targets of nitric oxide. Cell. Signal. 2001, 13, 525–533. [Google Scholar] [CrossRef]
- Agani, F.H.; Puchowicz, M.; Chavez, J.C.; Pichiule, P.; Lamanna, J. Role of nitric oxide in the regulation of HIF-1α expression during hypoxia. Am. J. Physiol. Physiol. 2002, 283, C178–C186. [Google Scholar] [CrossRef]
- Hagen, T.; Taylor, C.T.; Lam, F.; Moncada, S. Redistribution of Intracellular Oxygen in Hypoxia by Nitric Oxide: Effect on HIF1. Sci. 2003, 302, 1975–1978. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Garcia-Lecea, M.; Cadenas, S.; Hernández, C.; Moncada, S. Regulation of hypoxia-inducible factor-1α by nitric oxide through mitochondria-dependent and -independent pathways. Biochem. J. 2003, 376, 537–544. [Google Scholar] [CrossRef]
- Chen, S.C.; Huang, B.; Liu, Y.C.; Shyu, K.G.; Lin, P.Y.; Wang, D.L. Acute hypoxia enhances proteins’S-nitrosylation in endothelial cells. Biochem. Biophys. Res. Commun. 2008, 377, 1274–1278. [Google Scholar] [CrossRef]
- Albertos, P.; Romero-Puertas, M.C.; Tatematsu, K.; Mateos, I.; Sánchez-Vicente, I.; Nambara, E.; Lorenzo, O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Yun, B.-W.; Feechan, A.; Yin, M.; Saidi, N.B.B.; Le Bihan, T.; Yu, M.; Moore, J.W.; Kang, J.-G.; Kwon, E.; Spoel, S.H.; et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nat. Cell Biol. 2011, 478, 264–268. [Google Scholar] [CrossRef]
- Skelly, M.; Malik, S.I.; Le Bihan, T.; Bo, Y.; Jiang, J.; Spoel, S.H.; Loake, G.J. A role for S-nitrosylation of the SUMO-conjugating enzyme SCE1 in plant immunity. Proc. Natl. Acad. Sci. USA 2019, 116, 17090–17095. [Google Scholar] [CrossRef]
- Chen, L.; Liao, B.; Qi, H.; Xie, L.-J.; Huang, L.; Tan, W.-J.; Zhai, N.; Yuan, L.-B.; Zhou, Y.; Yu, L.-J.; et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 2015, 11, 2233–2246. [Google Scholar] [CrossRef]
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the Integrated Stress Response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doorly, C.M.; Graciet, E. Lessons from Comparison of Hypoxia Signaling in Plants and Mammals. Plants 2021, 10, 993. https://doi.org/10.3390/plants10050993
Doorly CM, Graciet E. Lessons from Comparison of Hypoxia Signaling in Plants and Mammals. Plants. 2021; 10(5):993. https://doi.org/10.3390/plants10050993
Chicago/Turabian StyleDoorly, Catherine M., and Emmanuelle Graciet. 2021. "Lessons from Comparison of Hypoxia Signaling in Plants and Mammals" Plants 10, no. 5: 993. https://doi.org/10.3390/plants10050993
APA StyleDoorly, C. M., & Graciet, E. (2021). Lessons from Comparison of Hypoxia Signaling in Plants and Mammals. Plants, 10(5), 993. https://doi.org/10.3390/plants10050993





