Foliar Endophytic Fungi from the Endangered Eastern Mountain Avens (Geum peckii, Rosaceae) in Canada
Abstract
1. Introduction
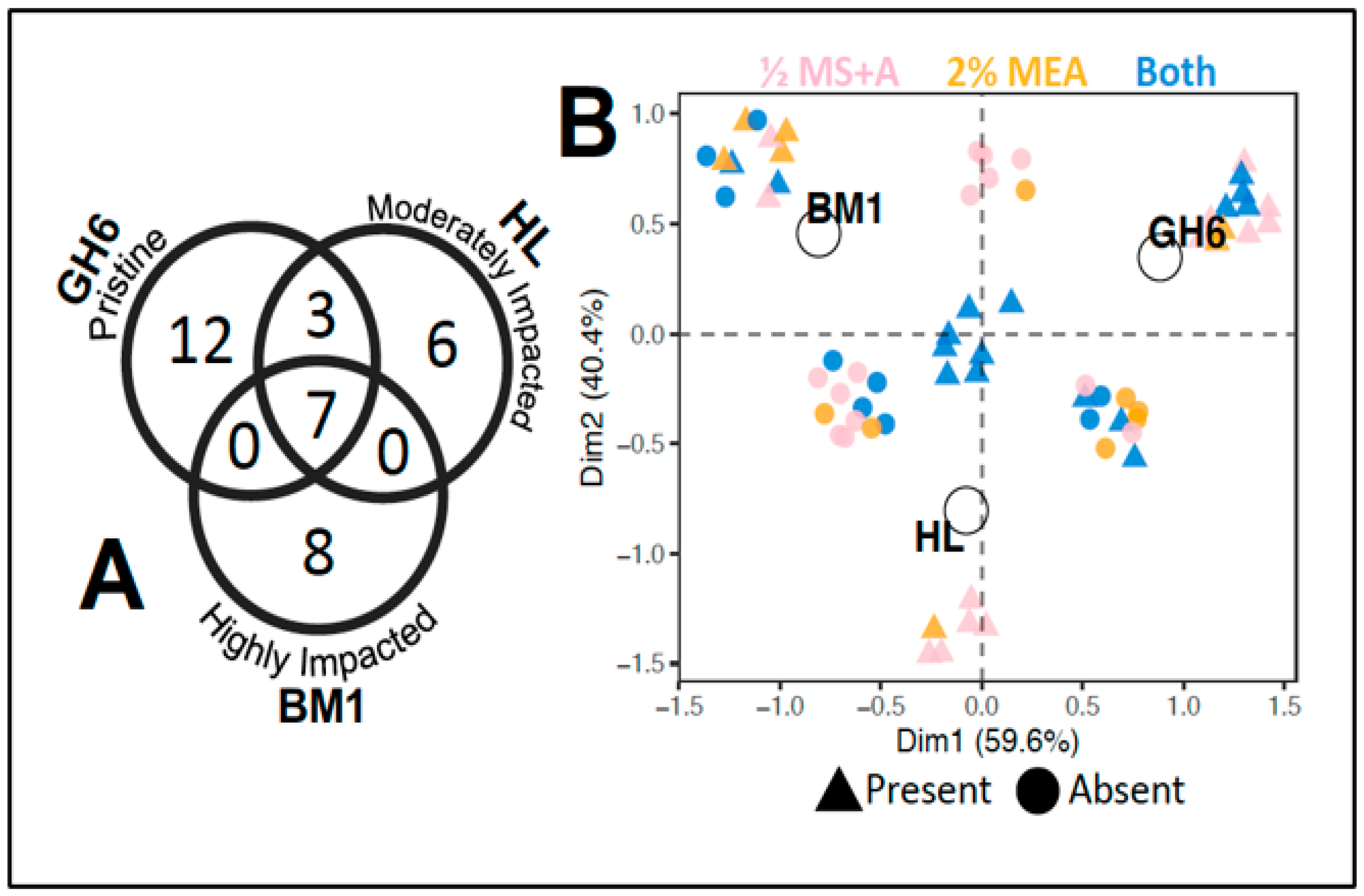
2. Results
3. Discussion
3.1. Trends in the Endophytic Fungi from Geum peckii
3.2. Known Ecologies of Sordariomycetes Recovered from Geum peckii
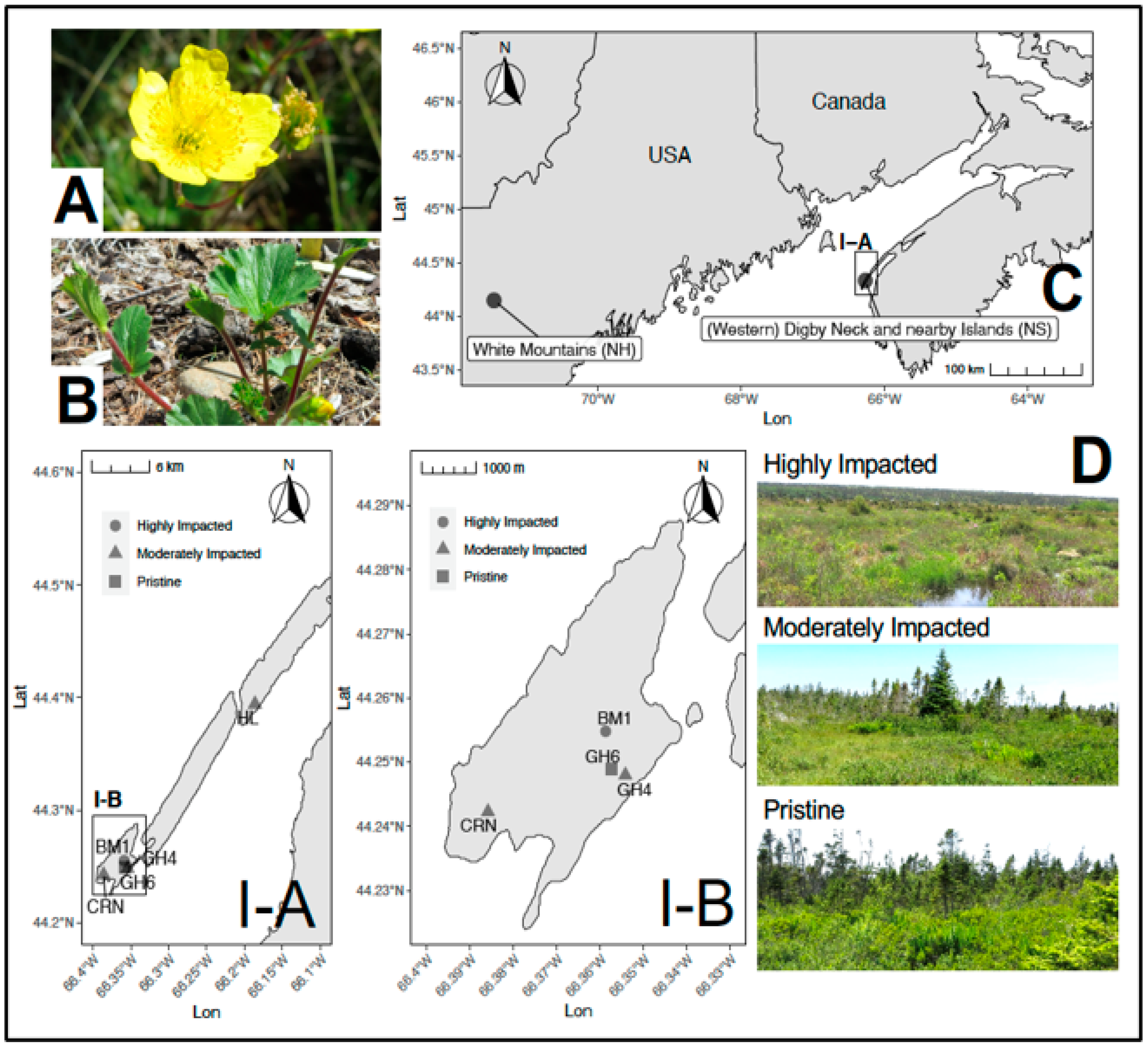
4. Materials and Methods
4.1. Field Collection
4.2. Endophyte Culturing and Molecular Identifications
4.3. Data Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, M.; Arato, M.; Borghi, L.; Nouri, E.; Reinhardt, D. Beneficial Services of Arbuscular Mycorrhizal Fungi—From Ecology to Application. Front. Plant Sci. 2018, 9, 1270. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Kohler, A.; Murat, C.; Veneault-Fourrey, C.; Hibbett, D.S. Unearthing the roots of ectomycorrhizal symbioses. Nat. Rev. Microbiol. 2016, 14, 760–773. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; White, J.F., Jr.; Arnold, A.E.; Redman, A.R.A. Fungal endophytes: Diversity and functional roles. Tan. Rev. N. Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhu, J.; Zhao, X.; Shi, J.; Jiang, C.; Shao, D. Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 2019, 103, 3327–3340. [Google Scholar] [CrossRef] [PubMed]
- Rana, K.L.; Kour, D.; Sheikh, I.; Yadav, N.; Yadav, A.N.; Kumar, V.; Singh, B.P.; Dhaliwal, H.S.; Saxena, A.K. Biodiversity of Endophytic Fungi from Diverse Niches and Their Biotechnological Applications. In Advances in Endophytic Fungal Research: Present Status and Future Challenges; Singh, B.P., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 105–144. ISBN 978-3-030-03589-1. [Google Scholar]
- Gooden, B.; Thompson, E.R.; French, K. Do native plant associations with arbuscular mycorrhizal fungi and dark septate endophytes differ between reconstructed and remnant coastal dunes? Plant. Ecol. 2020, 221, 757–771. [Google Scholar] [CrossRef]
- Sikes, B.A.; Hawkes, C.V.; Fukami, T. Plant and root endophyte assembly history: Interactive effects on native and exotic plants. Ecology 2016, 97, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Frasz, S.L.; Walker, A.K.; Nsiama, T.K.; Adams, G.W.; Miller, J.D. Distribution of the foliar fungal endophyte Phialocephala scopiformis and its toxin in the crown of a mature white spruce tree as revealed by chemical and qPCR analyses. Can. J. For. Res. 2014, 44, 1138–1143. [Google Scholar] [CrossRef]
- LaRue, D. In situ and ex situ propagation of the globally rare Geum peckii (Rosaceae). Rhodora 2016, 118, 315–319. [Google Scholar] [CrossRef]
- Ulrey, C.; Quintana-Ascencio, P.F.; Kauffman, G.; Smith, A.B.; Menges, E.S. Life at the top: Long-term demography, microclimatic refugia, and responses to climate change for a high-elevation southern Appalachian endemic plant. Biol. Conserv. 2016, 200, 80–92. [Google Scholar] [CrossRef]
- Paterson, I.G.; Snyder, M. Genetic evidence supporting the taxonomy of Geum peckii (Rosaceae) and G. radiatum as separate species. Rhodora 1999, 101, 325–340. [Google Scholar]
- Atlantic Geoscience Society. The Last Billion Years: A Geological History of the Maritime Provinces of Canada; Nimbus Publishing: Halifax, NS, Canada, 2001; ISBN 978-1-55109-351-2. [Google Scholar]
- LaRue, D. Natural regeneration of the globally rare Geum peckii (Rosaceae) on Brier Island, Nova Scotia. Rhodora 2016, 118, 310–314. [Google Scholar] [CrossRef]
- Pluess, A.R.; Stöcklin, J. The importance of population origin and environment on clonal and sexual reproduction in the alpine plant Geum reptans. Funct. Ecol. 2005, 19, 228–237. [Google Scholar] [CrossRef]
- Fancy, S.; López-Gutiérrez, J.C.; Walker, A.K.; la Rue, D.; Browne, R. Evaluating out-planting success and mycorrhizal status of endangered Geum peckii Pursh (Rosaceae), the Eastern Mountain Avens, in Nova Scotia. PNSIS 2020, 50, 269. [Google Scholar] [CrossRef]
- Borchardt, J.R.; Wyse, D.L.; Sheaffer, C.C.; Kauppi, K.L.; Fulcher, R.G.; Ehlke, N.J.; Biesboer, D.D.; Bey, R.F. Antimicrobial activity of native and naturalized plants of Minnesota and Wisconsin. J. Med. Plant. Res. 2008, 2, 5. [Google Scholar]
- Borchardt, J.R.; Wyse, D.L.; Sheaffer, C.C.; Kauppi, K.L.; Fulcher, R.G.; Ehlke, N.J.; Biesboer, D.D.; Bey, R.F. Antioxidant and antimicrobial activity of seed from plants of the Mississippi River Basin. J. Med. Plant. Res. 2008, 2, 4. [Google Scholar]
- Dimitrova, L.; Zaharieva, M.M.; Popova, M.; Kostadinova, N.; Tsvetkova, I.; Bankova, V.; Najdenski, H. Antimicrobial and antioxidant potential of different solvent extracts of the medicinal plant Geum urbanum L. Chem. Central J. 2017, 11, 113. [Google Scholar] [CrossRef]
- Panizzi, L.; Catalano, S.; Miarelli, C.; Cioni, P.L.; Campeol, E. In vitro antimicrobial activity of extracts and isolated constituents of Geum rivale. Phytothe. Res. 2000, 14, 561–563. [Google Scholar] [CrossRef]
- Henning, J.A.; Kinkel, L.; May, G.; Lumibao, C.Y.; Seabloom, E.W.; Borer, E.T. Plant diversity and litter accumulation mediate the loss of foliar endophyte fungal richness following nutrient addition. Ecology 2020, 102, 03210. [Google Scholar] [CrossRef]
- Lumibao, C.Y.; Borer, E.T.; Condon, B.; Kinkel, L.; May, G.; Seabloom, E.W. Site-specific responses of foliar fungal microbiomes to nutrient addition and herbivory at different spatial scales. Ecol. Evol. 2019, 9, 12231–12244. [Google Scholar] [CrossRef]
- Seabloom, E.W.; Condon, B.; Kinkel, L.; Komatsu, K.J.; Lumibao, C.Y.; May, G.; McCulley, R.L.; Borer, E.T. Effects of nutrient supply, herbivory, and host community on fungal endophyte diversity. Ecology 2019, 100, e02758. [Google Scholar] [CrossRef]
- Bradbury, J. Contributions of biochar and arbuscular mycorrhizal fungi to the growth of Geum peckii (Eastern Mountian avens). Honours thesis, Acadia University, Wolfville, NS, Canada, 2018. [Google Scholar]
- Crocker, E.V.; Lanzafane, J.J.; Karp, M.A.; Nelson, E.B. Overwintering seeds as reservoirs for seedling pathogens of wetland plant species. Ecosphere 2016, 7, e01281. [Google Scholar] [CrossRef]
- Mirhosseini, H.; Rahimian, H.; Babaeizad, V.; Hashemi, L. Outbreak of leafspot on blackberry (Rubus fruticosus) caused by Gnomoniopsis sp. in Iran. N. Dis. Rep. 2015, 31, 9. [Google Scholar] [CrossRef]
- Walker, D.M. Taxonomy, Systematics, Ecology, and Evolutionary Biology of the Gnomoniaceae (Diaporthales), with Emphasis on Gnomoniopsis and Ophiognomonia. Ph.D. Thesis, Rutgers The State University of New Jersey, New Brunswick, NJ, USA, 2012. [Google Scholar]
- Walker, D.M.; Castlebury, L.A.; Rossman, A.Y.; Struwe, L. Host conservatism or host specialization? Patterns of fungal diversification are influenced by host plant specificity in Ophiognomonia (Gnomoniaceae: Diaporthales). Biol. J. Linn. Soc. 2013, 111, 1–16. [Google Scholar] [CrossRef]
- Żukiewicz-Sobczak, W.; Cholewa, G.; Krasowska, E.; Zwoliński, J.; Sobczak, P.; Zawiślak, K.; Chmielewska-Badora, J.; Piątek, J.; Wojtyła, A. Pathogenic fungi in the work environment of organic and conventional farmers. Adv. Dermatol. Allergol. 2012, 4, 252–262. [Google Scholar] [CrossRef]
- Filippova, N.V.; Thormann, M.N. The fungal consortium of Andromeda polifolia in bog habitats. Mires. Peat. 2015, 16, 29. [Google Scholar]
- Karolina, G.; Ewa, W.; Rafał, S.; Marlena, L. Endophytic fungi and latent pathogens in the sedge Carex secalina (Cyperaceae), a critically endangered species in Europe. Plant Prot. Sci. 2019, 55, 102–108. [Google Scholar] [CrossRef]
- Vašutová, M.; Jiroušek, M.; Hájek, M. High fungal substrate specificity limits the utility of environmental DNA to detect fungal diversity in bogs. Ecol. Indic. 2021, 121, 107009. [Google Scholar] [CrossRef]
- Arnold, A.E. Understanding the diversity of foliar endophytic fungi: Progress, challenges, and frontiers. Fungal Biol. Rev. 2007, 21, 51–66. [Google Scholar] [CrossRef]
- Albrectsen, B.R.; Björkén, L.; Varad, A.; Hagner, Å.; Wedin, M.; Karlsson, J.; Jansson, S. Endophytic fungi in European aspen (Populus tremula) leaves—Diversity, detection, and a suggested correlation with herbivory resistance. Fungal Divers. 2010, 41, 17–28. [Google Scholar] [CrossRef]
- Luo, B.; Sun, H.; Zhang, Y.; Gu, Y.; Yan, W.; Zhang, R.; Ni, Y. Habitat-specificity and diversity of culturable cold-adapted yeasts of a cold-based glacier in the Tianshan Mountains, northwestern China. Appl. Microbiol. Biotechnol. 2018, 103, 2311–2327. [Google Scholar] [CrossRef]
- Lau, M.K.; Arnold, A.E.; Johnson, N.C. Factors influencing communities of foliar fungal endophytes in riparian woody plants. Fungal Ecol. 2013, 6, 365–378. [Google Scholar] [CrossRef]
- Raja, H.A.; Schmit, J.P.; Shearer, C.A. Latitudinal, habitat and substrate distribution patterns of freshwater ascomycetes in the Florida Peninsula. Biodivers. Conserv. 2008, 18, 419–455. [Google Scholar] [CrossRef]
- Bomble, Y.J.; Lin, C.-Y.; Amore, A.; Wei, H.; Holwerda, E.K.; Ciesielski, P.N.; Donohoe, B.S.; Decker, S.R.; Lynd, L.R.; Himmel, M.E. Lignocellulose deconstruction in the biosphere. Curr. Opin. Chem. Biol. 2017, 41, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.M.; Castlebury, L.A.; Rossman, A.Y.; Sogonov, M.V.; White, J.F. Systematics of genus Gnomoniopsis (Gnomoniaceae, Diaporthales) based on a three gene phylogeny, host associations and morphology. Mycology 2010, 102, 1479–1496. [Google Scholar] [CrossRef]
- Shestibratov, K.A.; Baranov, O.Y.; Subbotina, N.M.; Lebedev, V.G.; Panteleev, S.V.; Krutovsky, K.V.; Padutov, V.E. Early Detection and Identification of the Main Fungal Pathogens for Resistance Evaluation of New Genotypes of Forest Trees. Forests 2018, 9, 732. [Google Scholar] [CrossRef]
- Walker, A.; Hirooka, Y.; Walker, D. Ophiognomonia acadiensis. Fungal planet 274. Persoonia 2014, 32, 290–291. [Google Scholar]
- Hüseyin, E.; Selçuk, F. Coelomycetous fungi in several forest ecosystems of Black Sea provinces of Turkey. Agric. For. 2014, 60, 19–32. [Google Scholar]
- Holm, K.; Holm, L. Ascomycetes on Myrica gale in Sweden. Nord. J. Bot. 1991, 11, 675–687. [Google Scholar] [CrossRef]
- Tobias, A.; Balashova, N.; Kiselev, G. The Materials to Study of Lebyazhy Nature Reserve Microfungi (Lomonosov District of Leningrad Region). Available online: https://biocomm.spbu.ru (accessed on 12 November 2020).
- Mejía, L.C.; Castlebury, L.A.; Rossman, A.Y.; Sogonov, M.V.; White, J.F. A systematic account of the genus Plagiostoma (Gnomoniaceae, Diaporthales) based on morphology, host-associations, and a four-gene phylogeny. Stud. Mycol. 2011, 68, 211–235. [Google Scholar] [CrossRef]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Chukeatirote, E.; Hyde, K.D. Insights into the genus Diaporthe: Phylogenetic species delimitation in the D. eres species complex. Fungal Divers. 2014, 67, 203–229. [Google Scholar] [CrossRef]
- Becker, R.; Ulrich, K.; Behrendt, U.; Kube, M.; Ulrich, A. Analyzing Ash Leaf-Colonizing Fungal Communities for Their Biological Control of Hymenoscyphus fraxineus. Front. Microbiol. 2020, 11, 590944. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, T.; Kraj, W.; Bednarz, B. Fungi on stems and twigs in initial and advanced stages of dieback of European ash (Fraxinus excelsior) in Poland. Eur. J. For. Res. 2016, 135, 565–579. [Google Scholar] [CrossRef]
- Melo, R.F.R.; Gondim, N.H.D.B.; Santiago, A.L.C.M.D.A.; Maia, L.C.; Miller, A.N. Coprophilous fungi from Brazil: Updated identification keys to all recorded species. Phytotaxa 2020, 436, 104–124. [Google Scholar] [CrossRef]
- Johnston, P.R.; Quijada, L.; Smith, C.A.; Baral, H.-O.; Hosoya, T.; Baschien, C.; Pärtel, K.; Zhuang, W.-Y.; Haelewaters, D.; Park, D.; et al. A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 2019, 10, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Polashock, J.J.; Caruso, F.L.; Oudemans, P.V.; McManus, P.S.; Crouch, J.A. The North American cranberry fruit rot fungal community: A systematic overview using morphological and phylogenetic affinities. Plant Pathol. 2009, 58, 1116–1127. [Google Scholar] [CrossRef]
- Wu, W.; Sutton, B.C.; Gange, A.C. Coleophoma fusiformis sp. nov. from leaves of Rhododendron, with notes on the genus Coleophoma. Mycol. Res. 1996, 100, 943–947. [Google Scholar] [CrossRef]
- Filippova, N.V. Discomycetes from plant, leave and sphagnum litter in ombrotrophic bog (West Siberia). Environ. Dyn. Glob. Clim. Chang. 2012, 3, 1–20. [Google Scholar] [CrossRef]
- Tanney, J.; Seifert, K. Mollisiaceae: An overlooked lineage of diverse endophytes. Stud. Mycol. 2020, 95, 293–380. [Google Scholar] [CrossRef]
- Walsh, E.; Duan, W.; Mehdi, M.; Naphri, K.; Khiste, S.; Scalera, A.; Zhang, N. Cadophora meredithiae and C. interclivum, new species from roots of sedge and spruce in a western Canada subalpine forest. Mycology 2018, 110, 201–214. [Google Scholar] [CrossRef]
- Asemaninejad, A.; Thorn, R.G.; Lindo, Z. Vertical distribution of fungi in hollows and hummocks of boreal peatlands. Fungal Ecol. 2017, 27, 59–68. [Google Scholar] [CrossRef]
- Visagie, C.; Houbraken, J.; Frisvad, J.; Hong, S.-B.; Klaassen, C.; Perrone, G.; Seifert, K.; Varga, J.; Yaguchi, T.; Samson, R. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Wanke, U.; Draeger, S.; Aust, H.-J. Endophytes from herbaceous plants and shrubs: Effectiveness of surface sterilization methods. Mycol. Res. 1993, 97, 1447–1450. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Plant. Physiol. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.J.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR—Protocols and Applications—A Laboratory Manual; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. ISBN 978-0-12-372180-8. [Google Scholar]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Pebesma, E. Simple Features for R: Standardized Support for Spatial Vector Data. R J. 2018, 10, 439–446. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Dunnington, D. Spatial Data Framework for Ggplot2. Available online: https://paleolimbot.github.io/ggspatial/ (accessed on 17 March 2021).
- Slowikowski, K.; Schep, A.; Hughes, S.; Dang, T.K.; Lukauskas, S.; Irisson, J.-O.; Kamvar, Z.N.; Ryan, T.; Christophe, D.; Hiroaki, Y.; et al. Ggrepel: Automatically Position Non-Overlapping Text Labels with “Ggplot2”; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: AnRPackage for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Bio. Evo. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
| Taxa | Collection Site | No. Sites | Month | Media | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GH6 * | HL * | CRN | GH4 | BM1 * | June | July * | MEA | MS+A | ||
| Dothideomycetes | ||||||||||
| Alternaria sp. | ● | 1 | ● | ● | ● | |||||
| Cladosporium sp. | ● | 1 | ● | ● | ||||||
| Didymocyrtis cladoniicola | ● | 1 | ● | ● | ||||||
| Neostagonospora elegiae | ● | 1 | ● | ● | ||||||
| Phaeosphaeria poagena | ● | 1 | ● | ● | ||||||
| Ramularia sp. | ● | 1 | ● | ● | ||||||
| Stagonospora perfecta | ● | 1 | ● | ● | ||||||
| Eurotiomycetes | ||||||||||
| Penicillium sp. | ● | 1 | ● | ● | ||||||
| Articulospora sp. | ● | 1 | ● | ● | ||||||
| Coccomyces sp. | ● | 1 | ● | ● | ||||||
| Leotiomycetes | ||||||||||
| Godronia sp. | ● | ● | 2 | ● | ● | ● | ● | |||
| Helotiaceae sp. 1 | ● | 1 | ● | ● | ||||||
| Helotiaceae sp. 2 | ● | 1 | ● | ● | ● | |||||
| Lachnum virgineum | ● | 1 | ● | ● | ||||||
| Mollisia melaleuca | ● | 1 | ● | ● | ● | |||||
| Mollisia sp. | ● | ● | ● | ● | 4 | ● | ● | ● | ● | |
| Phacidium sp. | ● | 1 | ● | ● | ||||||
| Phlyctema sp. | ● | 1 | ● | ● | ● | |||||
| Phlyctema phoenicis | ● | 1 | ● | ● | ||||||
| Rhexocercosporidium sp. | ● | ● | 2 | ● | ● | ● | ||||
| Varicosporium elodeae | ● | 1 | ● | ● | ||||||
| Sordariomycetes | ||||||||||
| Apiognomonia hystrix | ● | 1 | ● | ● | ||||||
| Asteroma alneum | ● | ● | ● | ● | 4 | ● | ● | ● | ||
| Colletotrichum sp. 1 | ● | 1 | ● | ● | ||||||
| Colletotrichum sp. 2 | ● | 1 | ● | ● | ||||||
| Cryptodiaporthe aubertii | ● | ● | ● | ● | ● | 5 | ● | ● | ● | ● |
| Diaporthe sp. 1 | ● | 1 | ● | ● | ||||||
| Diaporthe sp. 2 | ● | 1 | ● | ● | ● | |||||
| Diaporthe sp. 3 | ● | 1 | ● | ● | ||||||
| Diaporthe sp. 4 | ● | ● | ● | ● | 4 | ● | ● | ● | ● | |
| Discula sp. | ● | ● | 2 | ● | ● | ● | ||||
| Fusarium sp. | ● | 1 | ● | ● | ● | |||||
| Gaeumannomycella caricicola | ● | ● | 2 | ● | ● | ● | ||||
| Gnomoniaceae sp. 1 | ● | 1 | ● | ● | ||||||
| Gnomoniaceae sp. 2 | ● | 1 | ● | ● | ||||||
| Gnomoniopsis idaeicola | ● | 1 | ● | ● | ● | |||||
| Gnomoniopsis macounii | ● | 1 | ● | ● | ||||||
| Gnomoniopsis occulta | ● | 1 | ● | ● | ||||||
| Microacsospora sp. | ● | ● | ● | ● | 4 | ● | ● | ● | ● | |
| Ophiognomonia acadiensis | ● | ● | ● | ● | 4 | ● | ● | ● | ||
| Ophiognomonia alni-viridis | ● | 1 | ● | ● | ||||||
| Ophiognomonia aff. gardiennetii | ● | 1 | ● | ● | ● | |||||
| Ophiognomonia intermedia | ● | ● | ● | 3 | ● | ● | ● | |||
| Ophiognomonia ischnostyla | ● | 1 | ● | ● | ||||||
| Ophiognomonia sp. 1 | ● | ● | ● | ● | 4 | ● | ● | ● | ||
| Ophiognomonia sp. 2 | ● | 1 | ● | ● | ||||||
| Ophiognomonia sp. 3 | ● | 1 | ● | ● | ||||||
| Physalospora vaccinii | ● | 1 | ● | ● | ||||||
| Plagiostoma lugubre | ● | ● | ● | ● | ● | 5 | ● | ● | ● | ● |
| Plagiostoma sp. | ● | 1 | ● | ● | ||||||
| Trichoderma sp. | ● | 1 | ● | ● | ||||||
| Totals = | 27 | 16 | 10 | 14 | 16 | 14 | 44 | 31 | 40 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adams, S.J.; Robicheau, B.M.; LaRue, D.; Browne, R.D.; Walker, A.K. Foliar Endophytic Fungi from the Endangered Eastern Mountain Avens (Geum peckii, Rosaceae) in Canada. Plants 2021, 10, 1026. https://doi.org/10.3390/plants10051026
Adams SJ, Robicheau BM, LaRue D, Browne RD, Walker AK. Foliar Endophytic Fungi from the Endangered Eastern Mountain Avens (Geum peckii, Rosaceae) in Canada. Plants. 2021; 10(5):1026. https://doi.org/10.3390/plants10051026
Chicago/Turabian StyleAdams, Sarah J., Brent M. Robicheau, Diane LaRue, Robin D. Browne, and Allison K. Walker. 2021. "Foliar Endophytic Fungi from the Endangered Eastern Mountain Avens (Geum peckii, Rosaceae) in Canada" Plants 10, no. 5: 1026. https://doi.org/10.3390/plants10051026
APA StyleAdams, S. J., Robicheau, B. M., LaRue, D., Browne, R. D., & Walker, A. K. (2021). Foliar Endophytic Fungi from the Endangered Eastern Mountain Avens (Geum peckii, Rosaceae) in Canada. Plants, 10(5), 1026. https://doi.org/10.3390/plants10051026






