Biotechnological Approaches for Genetic Improvement of Lemon (Citrus limon (L.) Burm. f.) against Mal Secco Disease
Abstract
1. Introduction
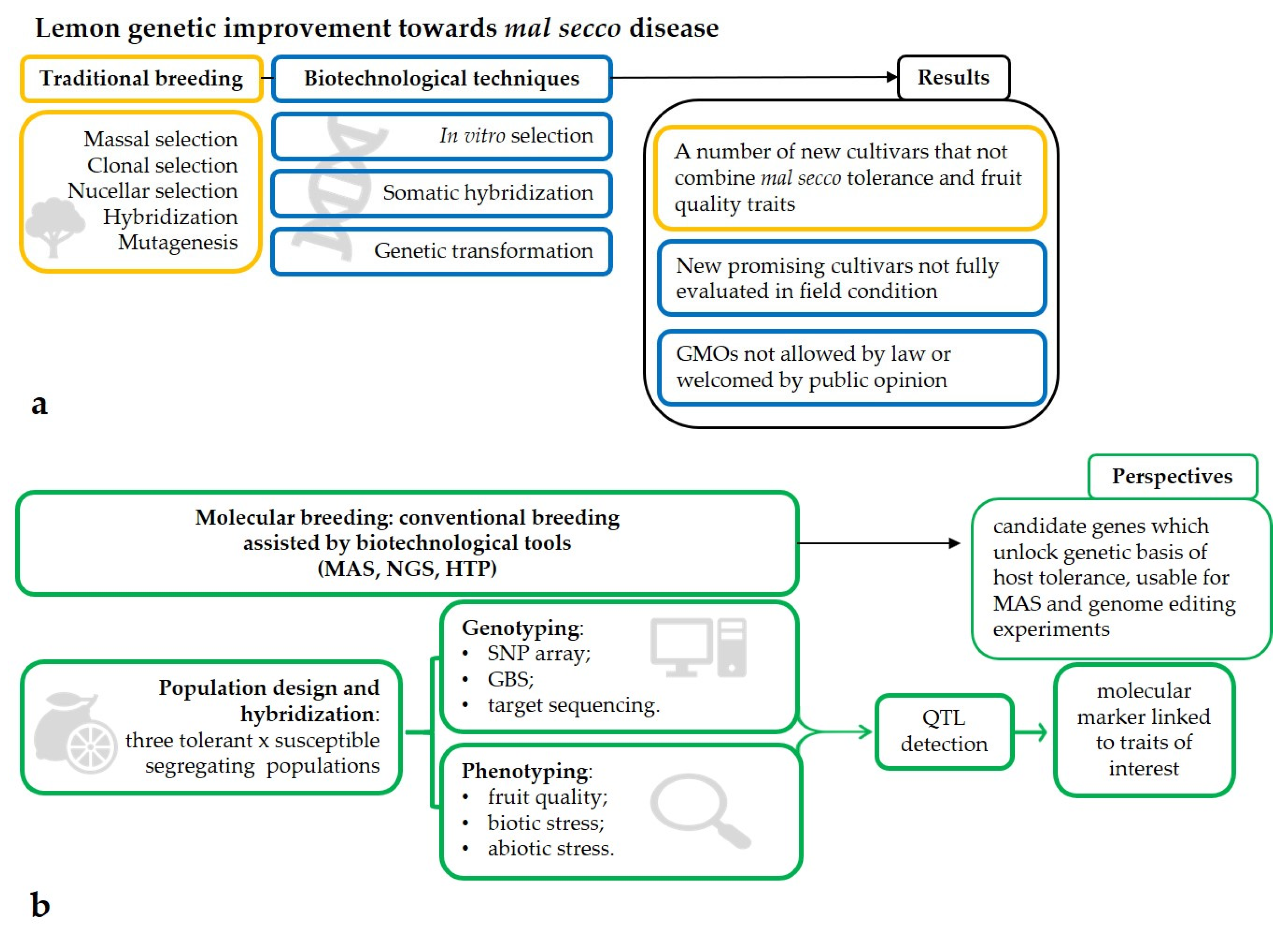
2. Biotechnological Approaches
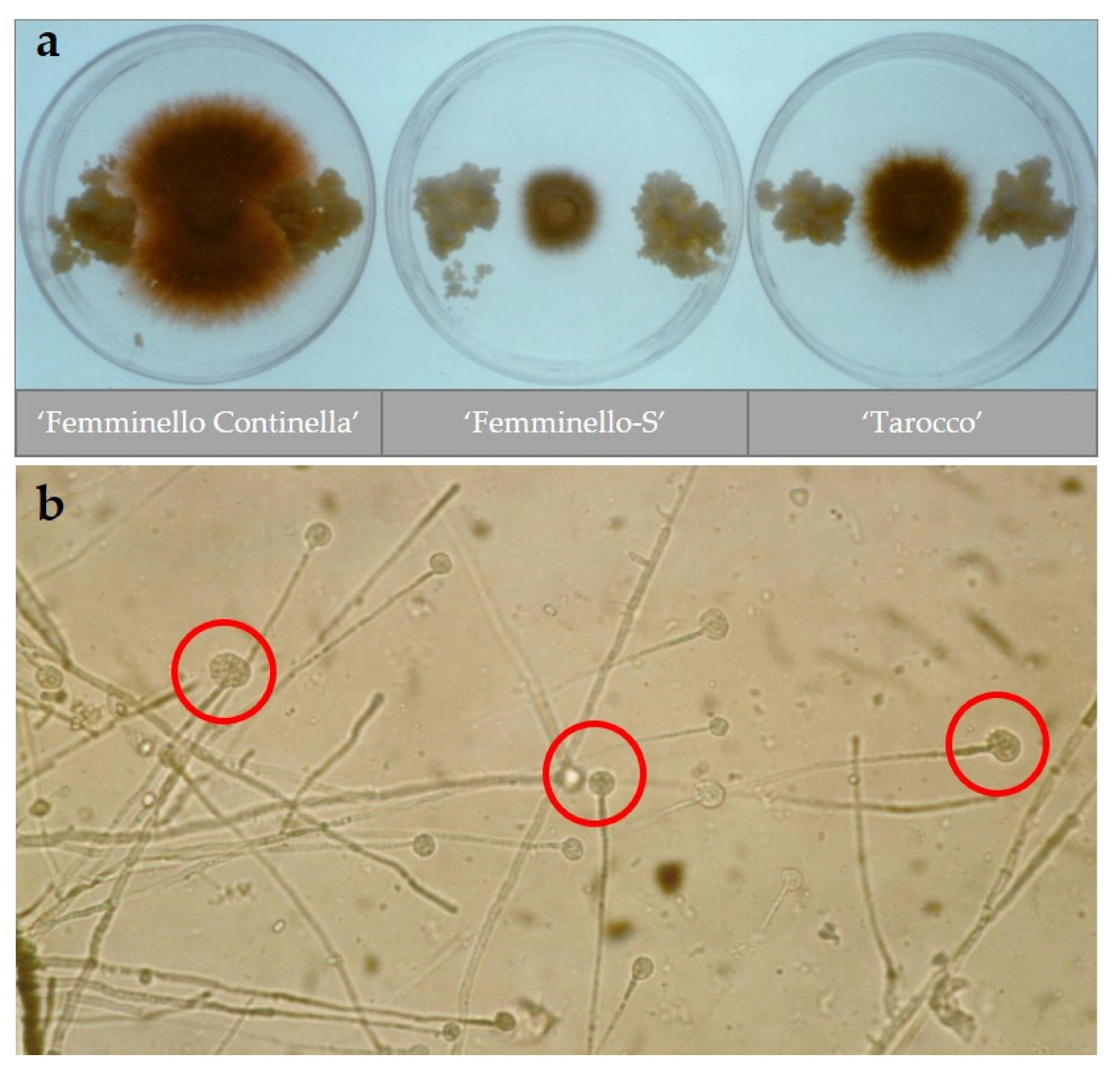
2.1. In Vitro Selection
2.2. Somatic Hybridization
2.3. Genetic Transformation
3. New Biotechnological Approaches: The Era of NGS (Next Generation Sequencing) and MAS (Marker-Assisted Selection)
4. Phenotyping as a Key Tool for Genotyping
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- EFSA PLH Panel. Scientific opinion on the pest categorisation of Plenodomus tracheiphilus (Petri) gruyter, aveskamp & verkley [syn. Phoma tracheiphila (Petri) L.A. Kantschaveli & Gikashvili]. EFSA J. 2014, 12, 34. [Google Scholar] [CrossRef]
- Hajlaoui, M.R.; Kalai, L.; Mnari-Hattab, M.; Guermech, A.; Ben Abdelaal, N. Occurrence of mal nero disease on mandarin and orange trees in Tunisia. Plant Pathol. 2008, 57, 784. [Google Scholar] [CrossRef]
- Migheli, Q.; Cacciola, S.O.; Balmas, V.; Pane, A.; Ezra, D.; Di San Lio, G.M. Mal secco disease caused by phoma tracheiphila: A potential threat to lemon production worldwide. Plant Dis. 2009, 93, 852–867. [Google Scholar] [CrossRef]
- Nigro, F.; Ippolito, A.; Salerno, M.G. Mal secco disease of citrus: A journey through a century of research. J. Plant Pathol. 2011, 93, 523–560. [Google Scholar] [CrossRef]
- Catara, A.; Catara, V. Il “Mal Secco” Degli Agrumi: Da Un Secolo in Sicilia. In Memorie e Rendiconti; USPI Associato all’Unione Stampa Periodica Italiana: Giovanni Battista RM, Italy, 2019; Volume 3, pp. 33–58. [Google Scholar]
- Abbate, L.; Mercati, F.; Fatta Del Bosco, S. An overview on citrus mal secco disease: Approaches and strategies to select tolerant genotypes in C. limon. Crop Breed. Genet. Genom. 2019. [Google Scholar] [CrossRef]
- Sun, L.; Nasrullah; Ke, F.; Nie, Z.; Wang, P.; Xu, J. Citrus genetic engineering for disease resistance: Past, present and future. Int. J. Mol. Sci. 2019, 20, 5256. [Google Scholar] [CrossRef]
- Barry, G.H.; Caruso, M.; Gmitter, F.G. Commercial scion varieties. In The Genus Citrus; Talon, M., Caruso, M., Gmitter, F.G., Jr., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 129–148. [Google Scholar]
- Russo, R.; Sicilia, A.; Caruso, M.; Arlotta, C.; Di Silvestro, S.; Gmitter, F.G.; Nicolosi, E.; Lo Piero, A.R. De novo transcriptome sequencing of rough lemon leaves (Citrus jambhiri Lush.) in response to Plenodomus tracheiphilus infection. Int. J. Mol. Sci. 2021, 22, 882. [Google Scholar] [CrossRef] [PubMed]
- Crino, P.; Cristinzio, G.; Gentile, A.; Haegi, A.; Rugini, E.; Soressi, G. Culture filtrates and toxins in the selection of disease resistant plants. Petria 1996, 6, 197–217. [Google Scholar]
- Nachmias, A.; Barash, I.; Solel, Z.; Strobel, G.A. Purification and characterization of a phytotoxin produced by Phoma tracheiphila, the causal agent of mal secco disease of citrus. Physiol. Plant Pathol. 1977, 10, 147–157. [Google Scholar] [CrossRef]
- Nachmias, A.; Barash, I.; Buchner, V.; Solel, Z.; Strobel, G.A. A phytotoxic glycopeptide from lemon leaves infected with Phoma fracheiphila. Physiol. Plant Pathol. 1979, 14, 135–140. [Google Scholar] [CrossRef]
- Fogliano, V.; Marchese, A.; Scaloni, A.; Ritieni, A.; Visconti, A.; Randazzo, G.; Graniti, A. Characterization of a 60 kDa phytotoxic glycoprotein produced by Phoma tracheiphila and its relation to malseccin. Physiol. Mol. Plant Pathol. 1998, 53, 149–161. [Google Scholar] [CrossRef]
- Barash, I.; Pupkin, G.; Koren, L.; Ben-Hayyim, G.; Strobel, G.A. A low molecular weight phytotoxin produced by Phoma tracheiphila, the cause of mal secco disease in citrus. Physiol. Plant Pathol. 1981, 19, 17–29. [Google Scholar] [CrossRef]
- Parisi, A.; Piattelli, M.; Tringali, C.; Di San Lio, G.M. Identification of the phytotoxin mellein in culture fluids of Phoma tracheiphila. Phytochemistry 1993, 32, 865–867. [Google Scholar] [CrossRef]
- Nachmias, A.; Barash, I.; Solel, Z.; Strobel, G.A. Translocation of mal secco toxin in lemons and its effect on electrolyte leakage, transpiration, and citrus callus growth. Phytoparasitica 1977. [Google Scholar] [CrossRef]
- Surico, G.; Iacobellis, N.S. Produzione di fitotossine di Phoma tracheiphila (Petri) Kanc. et Ghik. I. Influenza delle condizioni colturali e ricerca di idonei saggi biologici. Phytopath. Medit. 1980, 19, 173–174. [Google Scholar]
- Pennisi, A.M.; Graniti, A. Alterazioni della permeabilità cellulare in tessuti di agrumi infetti da Phoma tracheiphila (Petri) Kanc. et Ghik. Phytopath. Medit. 1987, 26, 143–145. [Google Scholar]
- Pennisi, A.M.; Di Pasquale, G.; Bonforte, M.; Sesto, F. Phytotoxic metabolites of ipo-virulent and virulent Phoma tracheiphila isolates. In Proceedings of the Sixth International Citrus Congress, Tel Aviv, Israel, 6–11 March 1988; pp. 817–827. [Google Scholar]
- Kashakashvili, T.S.; Goliadze, S. Reaction of tissue of lemon hybrids to treatment with toxin of the fungus causing mal secco. Subtrop. Kul’tury 1990, 2, 55–61. [Google Scholar]
- Surico, G.; De Cicco, V.; Iacobellis, N.S. Osservazioni sulla patogenicità di Phoma tracheiphila (Petri) Kanc. et Ghik. in relazione alla produzione in vitro di metaboliti fitotossici. Phytopath. Medit.1 1981, 20, 17–22. [Google Scholar]
- Rosciglione, B.; Burgio, A.; Bottalico, A.; Laviola, C. Relationship between phytotoxicity of metabolites produced in vitro by strains of Phoma tracheiphila (Petri) Kanc. et Ghik. and their virulence. J. Phytopathol. 1991, 133, 23–28. [Google Scholar] [CrossRef]
- Orshanskaya, V.N.; Ordzhhonikidze, N.P. An accelerated laboratory method of testing Citrus resistance to mal secco. Agrobiology 1956, 5, 35–44. [Google Scholar]
- Goliadze, S.K. Methods for determining the resistance of citrus plants to mal secco. Ref. Zh. Biol. 1957, 4, 204. [Google Scholar]
- Geraci, G.; Tusa, N.; Somma, V. Culture filtrates of Phoma tracheiphila (Petri) Kanc. et Ghik. to test lemon resistance to mal secco disease. In Proceedings of the Sixth International Citrus Congress, Tel Aviv, Israel, 6–11 March 1988; pp. 829–832. [Google Scholar]
- Geraci, G.; Tusa, N.; Buiatti, M.; Scala, A. Correlation between resistance in vivo to mal secco and in vitro to culture filtrate of Phoma tracheiphila on calli and ovules of citrus genotypes. In Proceedings of the 2nd International Meeting on Mediterranean Tree Crops, Chania, Crete, Greece, 2–4 November 1988; pp. 94–97. [Google Scholar]
- Sesto, F.; Grimaldi, V.; Pennisi, A.M. Sensitivity of different citrus and non citrus species protoplasts towards “Mal secco” toxin. Adv. Hort. Sci. 1990, 4, 97–102. [Google Scholar]
- Traversa, E.; Ippolito, A.; De Cicco, V. Failure to evaluate lemon resistance against mal secco disease using culture filtrates of the pathogen. In Proceedings of the 7th International Citrus Congress, Acireale, Italy, 8–13 March 1992; pp. 881–883. [Google Scholar]
- Marchese, A.; Graniti, A. Reactions of three citrus selection to Phoma tracheiphila toxins. In Proceedings of the 10th Congress of the Mediterranean Phytopathological Union, Montpellier, France, 1–5 June 1997; pp. 403–406. [Google Scholar]
- Nadel, B.; Spiegel-Roy, P. Selection of citrus limon cell culture variants resistant to the mal secco toxin. Plant Sci. 1987. [Google Scholar] [CrossRef]
- Gentile, A.; Tribulato, E.; Continella, G.; Vardi, A. Differential responses of citrus calli and protoplasts to culture filtrate and toxin of Phoma tracheiphila. Theor. Appl. Genet. 1992, 83, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Gentile, A.; Tribulato, E.; Deng, Z.N.; Vardi, A. In vitro selection of nucellar lemon callus and regeneration of plants tolerant to Phoma tracheiphila toxin. Adv. Hortic. Sci. 1992, 6, 151–154. [Google Scholar]
- Gentile, A.; Tribulato, E.; Deng, Z.N.; Galun, E.; Fluhr, R.; Vardi, A. Enhanced release of chitinase and glucanase into the culture medium by ‘Femminello’ lemon nucellar callus tolerant to the toxin of Phoma tracheiphila. In Proceedings of the 7th International Citrus Congress, Acireale, Italy, 8–13 March 1992; pp. 154–159. [Google Scholar]
- Gentile, A.; Tribulato, E.; Deng, Z.N.; Galun, E.; Fluhr, R.; Vardi, A. Nucellar callus of “Femminello” lemon, selected for tolerance to Phoma tracheiphila toxin, shows enhanced release of chitinase and glucanase into the culture medium. Theor. Appl. Genet. 1993, 86, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Gentile, A.; Tribulato, E.; Deng, Z.N.; Domina, F.; Galun, E.; Fluhr, R.; Vardi, A. Rilascio di proteine PR, in vitro e in vivo, in diversi genotipi di agrumi con noto comportamento nei confronti del mal secco. Italus Hortus 1994, 1, 59–64. [Google Scholar]
- Gentile, A.; Domina, F.; Deng, Z.N. Toxin of Phoma tracheiphila elicits PR-protein accumulation in citrus cell culture. In Proceedings of the International Society of Citriculture, Sun City, South Africa, 12–17 May 1996; pp. 1021–1023. [Google Scholar]
- Gentile, A.; Deng, Z.N.; Tribulato, E.; Albanese, G.; Grimaldi, V.; Catara, A.; Vardi, A. Evaluation of lemon somaclones for tolerance to mal secco disease by artificial inoculation. Acta Hortic. 2000, 535, 259–263. [Google Scholar] [CrossRef]
- Grimaldi, V.; Gentile, A.; Domina, F.; Catara, A. In vitro evaluation of lemon somaclones for mal secco tolerance. In Proceedings of the 5th Congress of the European Foundation for Plant Pathology, Taormina, Italy, 18–22 September 2000; pp. 398–400. [Google Scholar]
- Russo, R.; Caruso, M.; Arlotta, C.; Lo Piero, A.R.; Nicolosi, E.; Di Silvestro, S. Identification of Field Tolerance and Resistance to Mal Secco Disease in a Citrus Germplasm Collection in Sicily. Agronomy 2020, 10, 1806. [Google Scholar] [CrossRef]
- Deng, Z.N.; Gentile, A.; Domina, F.; Nicolosi, E.; Tribulato, E. Selecting lemon protoplasts for insensitivity to Phoma tracheiphila toxin and regenerating tolerant plants. J. Am. Soc. Hortic. Sci. 1995. [Google Scholar] [CrossRef]
- Bas, B.; Koç, N.K. In vitro selection of Kütdiken lemon 20b to candidate for resistance to Phoma tracheiphila. Plant Pathol. J. 2006, 5, 35–40. [Google Scholar] [CrossRef]
- Gentile, A.; Deng, Z.; La Malfa, S.; Distefano, G.; Domina, F.; Vitale, A.; Polizzi, G.; Lorito, M.; Tribulato, E. Enhanced resistance to and Botrytis cinerea in transgenic lemon plants expressing a Trichoderma harzianum chitinase gene. Plant Breed. 2007. [Google Scholar] [CrossRef]
- Remotti, P.C. Somaclonal Variation and In-Vitro Selection for Crop Improvement; Springer: Dordrecht, The Netherlands, 1998. [Google Scholar] [CrossRef]
- Faddetta, T.; Abbate, L.; Renzone, G.; Palumbo Piccionello, A.; Maggio, A.; Oddo, E.; Scaloni, A.; Puglia, A.M.; Gallo, G.; Carimi, F.; et al. An integrated proteomic and metabolomic study to evaluate the effect of nucleus-cytoplasm interaction in a diploid citrus cybrid between sweet orange and lemon. Plant Mol. Biol. 2018. [Google Scholar] [CrossRef]
- Grosser, J.W.; Gmitter, F.G.; Tusa, N.; Recupero, G.R.; Cucinotta, P. Further evidence of a cybridization requirement for plant regeneration from citrus leaf protoplasts following somatic fusion. Plant Cell Rep. 1996. [Google Scholar] [CrossRef] [PubMed]
- Grosser, J.W.; Gmitter, F.G. Protoplast fusion and citrus improvement. Plant Breed. Rev. 1990. [Google Scholar] [CrossRef]
- Tusa, N.; Grosser, J.W.; Gmitter, F.J.J. Plant regeneration of “Valencia” sweet orange, “Femminello” lemon, and the interspecific somatic hybrid following protoplasm fusion. J. Am. Soc. Hortic. Sci. 1990, 115, 1043–1046. [Google Scholar] [CrossRef]
- Tusa, N.; Grosser, J.W.; Gmitter, F.G.; Louzada, E.S. Production of tetraploid somatic hybrid breeding parents for use in lemon cultivar improvement. HortScience 1992. [Google Scholar] [CrossRef]
- Tusa, N.; Fatta Del Bosco, S.; Nardi, L.; Lucretti, S. Obtaining triploid plants by crossing citrus lemon cv. “Femminello” 2n x 4n allotetraploid somatic hybrids. In Proceedings of the International Society of Citriculture, Sun City, South Africa, 12–17 May 1996; pp. 133–136. [Google Scholar]
- Tusa, N.; Del Bosco, S.F.; Nigro, F.; Ippolito, A. Response of cybrids and a somatic hybrid of lemon to Phoma tracheiphila infections. HortScience 2000. [Google Scholar] [CrossRef]
- Deng, Z.N.; Gentile, A.; Nicolosi, E.; Domina, F.; Vardi, A.; Tribulato, E. Identification of in vivo and in vitro lemon mutants by RAPD markers. J. Hortic. Sci. 1995. [Google Scholar] [CrossRef]
- Peña, L.; Cervera, M.; Fagoaga, C.; Perez, R.; Romero, J.; Juarez, J.; Pina, J.A.; Navarro, L. Agrobacterium-Mediated Transformation of Citrus. Transgenic Crop. World 2004, 145–158. [Google Scholar] [CrossRef]
- Ghorbel, R.; Dominguez, A.; Navarro, L.; Pena, L. High efficiency genetic transformation of sour orange (Citrus aurantium) and production of transgenic trees containing the coat protein gene of citrus tristeza virus. Tree Physiol. 2000, 20. [Google Scholar] [CrossRef] [PubMed]
- Omar, A.A.; Song, W.Y.; Grosser, J.W. Introduction of Xa21, a Xanthomonas-resistance, gene from rice, into “Hamlin” sweet orange [Citrus sinensis (L.) Osbeck] using protoplast- GFP co-transformation or single plasmid transformation. J. Hortic. Sci. Biotechnol. 2007, 82. [Google Scholar] [CrossRef]
- Cardoso, S.C.; Barbosa-Mendes, J.M.; Boscariol-Camargo, R.L.; Christiano, R.S.C.; Filho, A.B.; Vieira, M.L.C.; Mendes, B.M.J.; Filho, F.; de Assis Alves Mourão Filho, F. Transgenic sweet orange (Citrus sinensis L. Osbeck) expressing the attacin a gene for resistance to Xanthomonas citri subsp. Citri. Plant Mol. Biol. Report. 2010, 28. [Google Scholar] [CrossRef]
- La Malfa, S.; Distefano, G.; Domina, F.; Nicolosi, E.; Toscano, V.; Gentile, A. Evaluation of citrus rootstock transgenic for rolABC genes. Acta Hortic. 2011, 892, 131–140. [Google Scholar] [CrossRef]
- La Malfa, S.; Domina, F.; Distefano, G.; Vitale, A.; La Rosa, G. Cloni transgenici di limone: Una nuova via per ottenere la resistenza al mal secco. Riv. Fruttic. Ortofloric. 2007, 1, 52–55. [Google Scholar]
- Bolar, J.P.; Norelli, J.L.; Wong, K.W.; Hayes, C.K.; Harman, G.E.; Aldwinckle, H.S. Expression of endochitinase from Trichoderma harzianum in transgenic apple increases resistance to apple scab and reduces vigor. Phytopathology 2000, 90, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Gentile, A.; Polizzi, G.; La Malfa, S.; Domina, F.; Vitale, A.; Distefano, G.; Lorito, M.; Tribulato, E. Espressione genica del sistema di difesa in piante di limone transgeniche per l’endochitinasi di Trichoderma harzianum. Italus Hortus 2007, 14, 12–14. [Google Scholar]
- Distefano, G.; La Malfa, S.; Vitale, A.; Lorito, M.; Deng, Z.; Gentile, A. Defence-related gene expression in transgenic lemon plants producing an antimicrobial Trichoderma harzianum endochitinase during fungal infection. Transgenic Res. 2008. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; La Malfa, S.; Abbate, C.; Gentile, A.; Cirvilleri, G.; Catara, A. Transgenic endochitinase troyer citrange shows higher chitinase and glucanase activity and reduced colonization by fungi. J. Plant Pathol. 2009, 91, 84–86. [Google Scholar]
- Oliveri, C.; Distefano, G.; La Malfa, S.; La Rosa, R.; Deng, Z.N.; Gentile, A. Lemon fruits from endochitinase transgenic plants exhibit resistance against postharvest fungal pathogens. Acta Hortic. 2015, 1065, 1639–1645. [Google Scholar] [CrossRef]
- Muccilli, V.; Vitale, A.; Sheng, L.; Gentile, A.; Cardullo, N.; Tringali, C.; Oliveri, C.; La Rosa, R.; Di Guardo, M.; La Malfa, S.; et al. Substantial equivalence of a transgenic lemon fruit showing postharvest fungal pathogens resistance. J. Agric. Food Chem. 2020, 68, 3806–3816. [Google Scholar] [CrossRef] [PubMed]
- Reforgiato Recupero, G.; Gentile, A.; Russo, M.P.; Domina, F. Genetic analysis of resistance to in three Citrus and Poncirus progenies. Plant Breed. 1997, 116, 198–200. [Google Scholar] [CrossRef]
- Deng, Z.N.; Gentile, A.; Domina, F.; Tribulato, E.; Messina, A. Identification of stress resistance genes by PCR-select cDNA subtraction in Citrus. In Proceedings of the 5th Congress of the European Foundation for Plant Pathology, Taormina, Italy, 18–22 September 2000; pp. 261–263. [Google Scholar]
- Koutsioumari, E.M.; Voloudakis, A.E. Isolation and expression analysis of differentially expressed genes in stem tissue of the Greek lemon cv. Adamopoulou. J. Hortic. Sci. Biotechnol. 2017, 92, 48–56. [Google Scholar] [CrossRef]
- Imai, A.; Nonaka, K.; Kuniga, T.; Yoshioka, T.; Hayashi, T. Genome-wide association mapping of fruit-quality traits using genotyping-by-sequencing approach in citrus landraces, modern cultivars, and breeding lines in Japan. Tree Genet. Genomes 2018, 14, 24. [Google Scholar] [CrossRef]
- Minamikawa, M.F.; Nonaka, K.; Kaminuma, E.; Kajiya-Kanegae, H.; Onogi, A.; Goto, S.; Yoshioka, T.; Imai, A.; Hamada, H.; Hayashi, T.; et al. Genome-wide association study and genomic prediction in citrus: Potential of genomics-assisted breeding for fruit quality traits. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Di Guardo, M.; Bink, M.C.A.M.; Letschka, T.; Lozano, L.; Busatto, N.; Poles, L.; Tadiello, A.; Bianco, L.; Visser, R.G.F.; van de Weg, E.; et al. Deciphering the genetic control of fruit texture in apple by multiple family-based analysis and genome-wide association. J. Exp. Bot. 2017, 68, 1451–1466. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Huang, M.; Roose, M.L.; Yu, Q.; Du, D.; Yu, Y.; Zhang, Y.; Deng, Z.; Stover, E.; Gmitter, F.G. Construction of high-density genetic maps and detection of QTLs associated with huanglongbing tolerance in citrus. Front. Plant Sci. 2018, 9, 1694. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics—Technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Dhondt, S.; Wuyts, N.; Inzé, D. Cell to whole-plant phenotyping: The best is yet to come. Trends Plant Sci. 2013, 18, 428–439. [Google Scholar] [CrossRef]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Willocquet, L.; Savary, S.; Yuen, J. Multiscale phenotyping and decision strategies in breeding for resistance. Trends Plant Sci. 2017, 22, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 2018, 23, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.R.; Reynolds, M.; Pinto, F.; Khan, M.A.; Bhat, M.A. High-throughput phenotyping for crop improvement in the genomics era. Plant Sci. 2019, 282, 60–72. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop phenomics and high-throughput phenotyping: Past decades, current challenges, and future perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef] [PubMed]
- Houle, D.; Govindaraju, D.R.; Omholt, S. Phenomics: The next challenge. Nat. Rev. Genet. 2010, 11, 855–866. [Google Scholar] [CrossRef]
- Afonnikov, D.A.; Genaev, M.A.; Doroshkov, A.V.; Komyshev, E.G.; Pshenichnikova, T.A. Methods of high-throughput plant phenotyping for large-scale breeding and genetic experiments. Russ. J. Genet. 2016, 52, 688–701. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef]
- Poles, L.; Gentile, A.; Giuffrida, A.; Valentini, L.; Endrizzi, I.; Aprea, E.; Gasperi, F.; Distefano, G.; Artioli, G.; La Malfa, S.; et al. Role of fruit flesh cell morphology and MdPG1 allelotype in influencing juiciness and texture properties in apple. Postharvest Biol. Technol. 2020, 164, 111161. [Google Scholar] [CrossRef]
- Singh, A.; Jones, S.; Ganapathysubramanian, B.; Sarkar, S.; Mueller, D.; Sandhu, K.; Nagasubramanian, K. Challenges and opportunities in machine-augmented plant stress phenotyping. Trends Plant Sci. 2021, 26, 53–69. [Google Scholar] [CrossRef]
- Mutka, A.M.; Bart, R.S. Image-based phenotyping of plant disease symptoms. Front. Plant Sci. 2015, 5. [Google Scholar] [CrossRef]
- Loprieno, N.; Bugiani, A. Conservazione della forma picnidica del Deuterophoma tracheiphila Petri in coltura e innovazione della tecnica di infezione su piantine di Arancio amaro. Phytopath. Z 1958, 32, 341–351. [Google Scholar] [CrossRef]
- Scaramuzzi, G.; Salerno, M.; Catara, A. Ricerche sul “mal secco” degli agrumi (Deuterophoma tracheiphila Petri). II. Influenza delle basse temperature sul decorso della malattia. Riv. Patol. Veg. 1964, 4, 319–327. [Google Scholar]
- Luisi, N.; De Cicco, V.; Cutuli, G.; Salerno, M. Factors in early testing for citrus mal secco resistance. In Proceedings of the International Society of Citriculture, Sidney, Australia, 15–23 August 1978; pp. 197–200. [Google Scholar]
- Lima, G.; Nigro, F.; Santomauro, A.; Ippolito, A. Ulteriori tentativi di lotta biologica contro il mal secco degli agrumi mediante isolati ipovirulenti del patogeno. Dif. Delle Piante 1994, 17, 135–144. [Google Scholar]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. CRC. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Bock, C.H.; Parker, P.E.; Cook, A.Z.; Gottwald, T.R. Visual rating and the use of image analysis for assessing different symptoms of citrus canker on grapefruit leaves. Plant Dis. 2008, 92, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Pourreza, A.; Lee, W.; Lu, J.; Roberts, P. Development of a multiband sensor for citrus black spot disease detection. In Proceedings of the 13th International Conference on Precision Agriculture, St. Louis, MO, USA, 31 July–3 August 2016. [Google Scholar]
- Salerno, M.; Catara, A. Ricerche sul mal secco degli Agrumi (Deuterophoma tracheiphila Petri). VI Indagini sulla riproduzione sperimentale della malattia. Riv. Patol. Veg. 1967, 3, 89–97. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catalano, C.; Di Guardo, M.; Distefano, G.; Caruso, M.; Nicolosi, E.; Deng, Z.; Gentile, A.; La Malfa, S.G. Biotechnological Approaches for Genetic Improvement of Lemon (Citrus limon (L.) Burm. f.) against Mal Secco Disease. Plants 2021, 10, 1002. https://doi.org/10.3390/plants10051002
Catalano C, Di Guardo M, Distefano G, Caruso M, Nicolosi E, Deng Z, Gentile A, La Malfa SG. Biotechnological Approaches for Genetic Improvement of Lemon (Citrus limon (L.) Burm. f.) against Mal Secco Disease. Plants. 2021; 10(5):1002. https://doi.org/10.3390/plants10051002
Chicago/Turabian StyleCatalano, Chiara, Mario Di Guardo, Gaetano Distefano, Marco Caruso, Elisabetta Nicolosi, Ziniu Deng, Alessandra Gentile, and Stefano Giovanni La Malfa. 2021. "Biotechnological Approaches for Genetic Improvement of Lemon (Citrus limon (L.) Burm. f.) against Mal Secco Disease" Plants 10, no. 5: 1002. https://doi.org/10.3390/plants10051002
APA StyleCatalano, C., Di Guardo, M., Distefano, G., Caruso, M., Nicolosi, E., Deng, Z., Gentile, A., & La Malfa, S. G. (2021). Biotechnological Approaches for Genetic Improvement of Lemon (Citrus limon (L.) Burm. f.) against Mal Secco Disease. Plants, 10(5), 1002. https://doi.org/10.3390/plants10051002










